Abstract
Uridine insertion/deletion RNA editing is a unique form of posttranscriptional RNA processing that occurs in mitochondria of kinetoplastid protists. We have carried out 3D structural analyses of the core editing complex or “L (ligase)-complex” from Leishmania tarentolae mitochondria isolated by the tandem affinity purification procedure (TAP). The purified material, sedimented at 20–25S, migrated in a blue native gel at 1 MDa and exhibited both precleaved and full-cycle gRNA-mediated U-insertion and U-deletion in vitro activities. The purified L-complex was analyzed by electron tomography to determine the extent of heterogeneity. Three-dimensional structural comparisons of individual particles in the tomograms revealed that a majority of the complexes have a similar shape of a slender triangle. An independent single-particle reconstruction, using a featureless Gaussian ball as the initial model, converged to a similar triangular structure. Another single-particle reconstruction, using the averaged tomography structure as the initial model, yielded a similar structure. The REL1 ligase was localized on the model to the base of the apex by decoration with REL1-specific IgG. This structure should prove useful for a detailed analysis of the editing reaction.
Keywords: editing, electron microscopy, kinetoplast, Leishmania, trypanosome
Uridine (U) insertion/deletion RNA editing is a posttranscriptional process in mitochondria of kinetoplastid protists that involves the modification of mRNA transcripts of mitochondrial cryptogenes by precise insertion and deletion of U residues to create translatable sequences (1–3). We previously proposed a model for the mechanism of this editing process involving a cleavage, addition or deletion of U's and ligation progressing in a 3′ to 5′ polarity (4), and this model has been since experimentally validated in almost every detail (5–10).
Editing involves multiple multiprotein complexes associated by RNA linkers. The core editing complex, known as the L-complex, “20S complex,” “editosome,” or “∼20S editosome,” was detected by following the sedimentation and gel filtration of 2 adenylatable RNA ligases using Leishmania tarentolae and Trypanosoma brucei mitochondrial extracts (11, 12). The activity sedimented around 20–25S and migrated as a single band in a native gel (13). A ∼20S multiprotein complex that showed both U-insertion and U-deletion in vitro activities was also purified from T. brucei mitochondria by column chromatography. An improved isolation of the L-complex from L. tarentolae was achieved by the tandem affinity purification procedure (TAP) (14); TAP-tagged plasmid-expressed REL1 became incorporated into the L-complex in vivo, allowing isolation of tagged complexes.
Fourteen proteins were initially identified by mass spectrometry of the L. tarentolae REL1-TAP pull-down (14). All of the L. tarentolae proteins had homologs in T. brucei, and several additional proteins found in the T. brucei L-complex (15) were later identified in L. tarentolae (Tables S1 and S2).
Two 3-protein subcomplexes containing the 2 RNA ligases have been described (16). The REL1-TAP pull-down showed a minor amount of the endogenous REL1 in addition to the tagged REL1 (14). Similar results were obtained for another component of the REL1 subcomplex—the LC4 (MP63) protein (17). These results suggest that there is a minor percentage of the REL1 subcomplexes that are dimeric, but the majority are single-copy.
A limited amount of L-complex protein heterogeneity has been reported from analysis of REN1, REN2, and REN3 TAP pull-downs. This data suggests the presence of at least 3 classes of L-complex particles that differ by a few proteins (18, 19).
There are in addition a growing number of multiprotein complexes that interact with the L-complex in substoichiometric amounts via RNA linkers (14, 20).
In this paper, we present chemical, functional, and structural analyses of TAP-purified L-complexes from L. tarentolae.
Results
TAP Isolation of LC8-tagged L-complex.
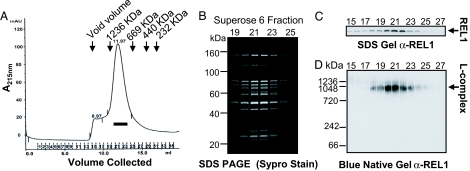
We chose the LC8 core L-complex protein for the TAP pull-down. The TAP pull-down was run on a Superose 6 column. The L-complex eluted as a small retarded peak in fractions 20–23 (Fig. 1A), as shown by characteristic polypeptide profiles (Fig. 1B) and by the distribution of the REL1 RNA ligase (Fig. 1C). Blue native gel analysis of the Superose 6 peak fractions showed a single major REL1-containing band migrating around 1 MDa with a very minor band around 1.2 MDa (Fig. 1D) (21). Little if any degradation was detected by blue native gel analysis. Degradation, however, did occur if the TAP pull-down was frozen-thawed or kept at 4 °C for more than a few hours. Therefore the preparation of grids for electron microscopy analysis was performed with freshly isolated Superose 6 fractions.
Fig. 1.
Isolation of L-complex. (A) Superose 6 chromatography of LC8-TAP pull-down. Fractions were monitored at 215 nm. (B) Peak fractions from A were subjected to SDS gel analysis. Gel was stained with Sypro Ruby stain. (C) Western blot analysis of SDS gel in B, using anti-REL1 monoclonal antibody as probe. (D) Blue native gel analysis of indicated peak fractions from A. The gel was blotted and probed with anti-REL1 antibody.
Polypeptide Composition of the TAP-isolated L-complex.
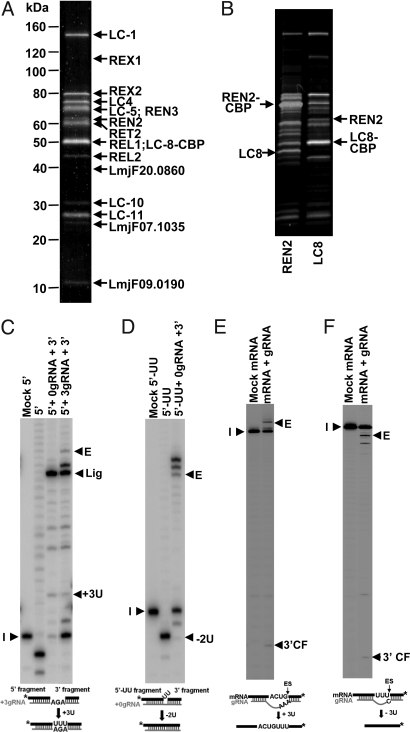
A comparison of the LC8-TAP polypeptide pattern with the previously obtained REL1-TAP profile (14) showed that in addition to the known bands seen in the REL1-TAP material, there were 4 previously undetected bands of varying intensity (Fig. 2A). Two of the four bands were homologous to proteins previously reported from T. brucei—REN3 and MP19—and two are reported here (Table S2).
Fig. 2.
Analysis of polypeptides in isolated L-complex and in vitro activities. (A) SDS gel was stained with colloidal Coomassie blue, and the bands were excised for MS-MS mass spectrometry analysis. The 2 proteins identified are indicated. (B) Comparison of LC8 and REN2 TAP pull-downs. Note the absence of endogenous LC8 in the LC8 pull-down and endogenous REN2 in the REN2 pull-down. (C) Precleaved U-insertion assay. (D) Precleaved U-deletion assay. (E) Full-cycle U-insertion assay. (F) Full-cycle U-deletion assay. The 3′ end-labeled input RNAs (I) and the edited products (E) are indicated. The presence of a minor unguided −6U product is puzzling, but does not affect the conclusions.
The LC8-TAP pull-down contains only the tagged LC-8 and not the endogenous LC-8 (Fig. 2B), indicating that it is single-copy in the complex. REN2 was shown to be single-copy by the same method (Fig. 2B).
Functional Activities of the Isolated L-complex.
To assay the functionality of the LC8-TAP-isolated L-complex, several in vitro assays were performed. The purified L-complex showed gRNA-mediated precleaved U-insertion and U-deletion activities (Fig. 2 C and D). It also showed robust gRNA-mediated full-cycle U-insertion and U-deletion editing activities (Fig. 2 E and F).
Structural Classification of the TAP-isolated L-complexes by Electron Tomography.
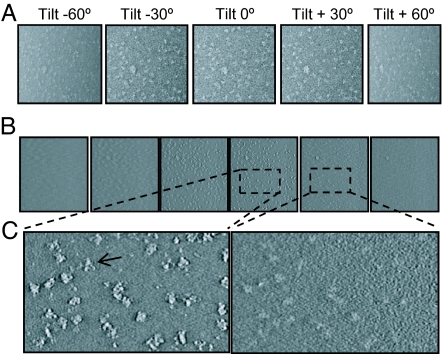
To analyze the purified L-complex particles, a total of 74 tilt series were collected, each spanning an angular range of −70 ° to 70 °, as shown in a representative tilt series in Fig. 3A. Among those, we selected the 21 best series that have uniform stain penetration for in-depth 3D structural analysis. Representative density slices through a single tomogram are shown in Fig. 3 B and C. Segmentation of the 3D tomogram produced individual L-complex particles for further 3D structural comparison at a resolution of ≈30–40 Å (Fig. S1).
Fig. 3.
Image data showing tilt series and representative 2D slices from the reconstructed tomogram. (A) Representative tilt series of a −70° to 70° tilt series. (B and C) 2D density slices extracted from the 3D reconstruction. The arrow in the 5× magnified view in C indicates the characteristic triangular shape of a single L-complex.
During electron tomography, images in a tilt series are limited due to the sample support film to a useful tilt range typically ranging from +70 ° to −70 °, thus giving rise to anisotropic resolutions and different structural distortion of particles at different orientations, a limitation commonly known as the “missing-wedge” problem. Despite this limitation, individual particles segmented from the tomograms all have a similar triangular shape and dimensions (Fig. 4A).
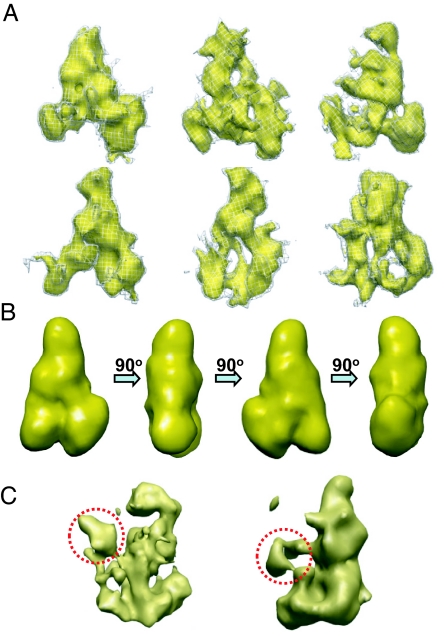
Fig. 4.
Segmented L-complexes and averaged structure. (A) Representative particles segmented from 3D tomogram. (B) Averaged major class particle at a resolution of ≈40 Å shown by rotating through 90°. (C) Average minor class particles showing extra density protrusions from central region (circled). The particle obtained from 2 structures (Left) and the particle obtained from 3 structures (Right) are shown.
To minimize the structural distortion arising from the missing-wedge problem and to improve the signal-to-noise ratio, 3D structural alignment of these particles was performed, and particles from the same class were averaged to eliminate noise and decrease the structure distortion. This structural analysis revealed one major class and a few minor classes. The model obtained by averaging 71 particles has a shape of a slender triangle (Fig. 4B). The minor classes were not studied extensively, but 2 examples of averaged particles are shown in Fig. 4C. Interestingly, the averages resemble the slender triangular shape of the major class, but have an additional density protrusion extending from the central region (Fig. 4C, dotted circles).
Electron Microscopy and Single-Particle Reconstruction.
To improve the resolution of the 3D reconstruction, we carried out single-particle analysis of 5,149 nontilted images (Fig. 5A). To prevent possible model bias, the initial single-particle reconstruction was obtained by refining against a featureless Gaussian shape as the initial model. The refinement converged to a single and stable structure after 8 cycles of iterative model-based refinement, and the effective resolution of this final structure is 24 Å based on the 0.5 criterion of the Fourier shell correlation coefficient (Fig. 5B). The class averages and 2D projections computed from the final 3D reconstruction match with one another (Fig. S2), which validates the reconstruction and suggests again that there is a single representative structure of the L-complex. The orientation distribution of the single-particle reconstruction is shown in Fig. S3.
Fig. 5.
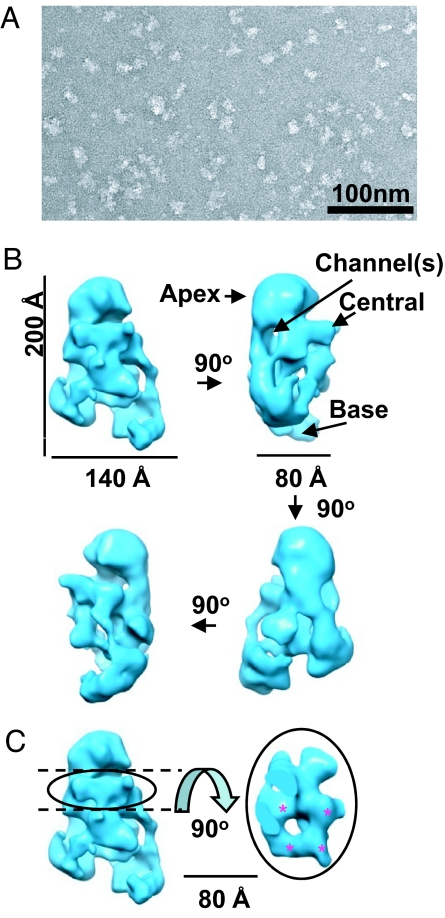
Electron microscopy and single-particle reconstruction. (A) Electron microscopy images of purified L-complexes stained by uranyl acetate. (B) Unbiased single-particle reconstruction from negative stained images. Four shaded surface views are shown rotated consecutively at 90° intervals along the vertical axis. The Apex, Central, and Base regions of the structure are indicated. (C) A region with the appearance of quasi-4-fold symmetry is located in the central region (red stars).
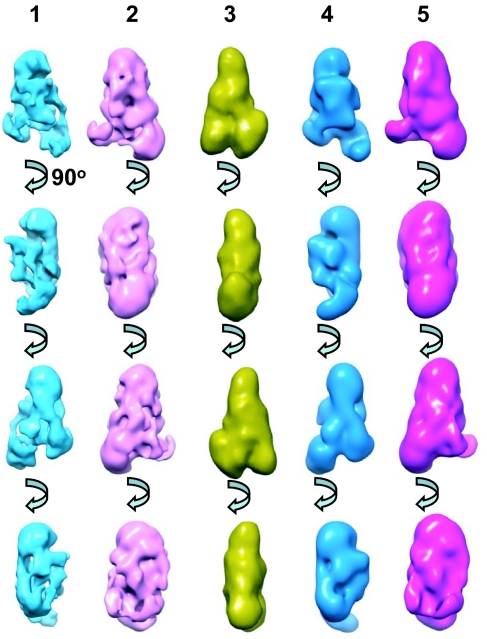
To further establish the validity of the single-particle reconstruction, a second single-particle reconstruction (Fig. 6, lane 2) was obtained by using the low-resolution average from tomography reconstruction as an initial model. This reconstruction exhibits a striking similarity to that of the unbiased reconstruction (Fig. 6, compare lanes 1 and 2). Another comparison between the averaged tomographic model (Fig. 6, lane 3) and the 2 single-particle reconstructions filtered to comparable resolution (Fig. 6, lanes 4 and 5) shows that the overall shape and architecture of the L-complex is similar to that revealed in the low-resolution tomographic model. This provides additional validation for the single-particle reconstruction as representing the major L-complex structure. The structure exhibits an overall triangular shape with dimensions of ≈200 × 140 × 80 Å and can be divided into 3 regions, “Apex,” “Central,” and “Base” (Fig. 5B). The Apex and Base regions of the complex have a globular appearance, and the Central region has an open network of densities and channels. There is an interesting central density displaying a quasi-4-fold symmetry (Fig. 5C, stars).
Fig. 6.
Comparison of single-particle and tomographic reconstructions. Lane 1, unbiased single-particle reconstruction. Lane 2, single-particle reconstruction using averaged tomography structure as initial model. Lane 3, averaged tomographic reconstruction. Lane 4, reconstruction from lane 1 filtered to 40 Å. Lane 5, reconstruction from lane 2 filtered to 40 Å.
Localization of REL1 Ligase.
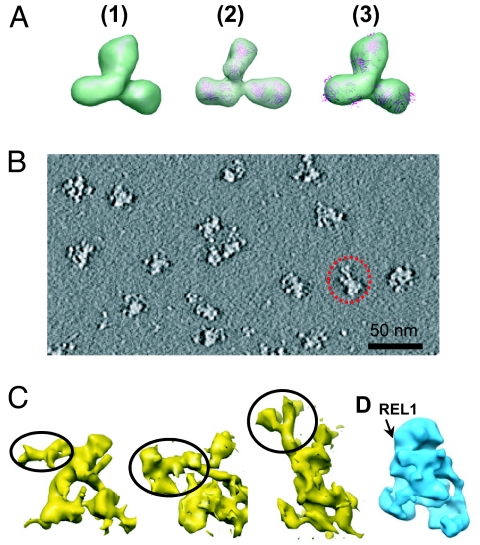
To validate the use of tomography to visualize complexes decorated with IgG, a tomographic reconstruction of the anti-REL1 IgG molecule was successfully performed (Fig. 7A and Fig. S4 A and B). To localize REL1 on the L-complex, anti-REL1 IgG was reacted with purified L-complex particles before the Superose 6 chromatography. Tomography was performed on negative stained particles. The unaveraged particles segmented from the tomogram are noisy (Fig. 7B), but it is clear that the IgG is attached near the base of the Apex in each case (Fig. 7C).
Fig. 7.
Decoration of L-complex reconstructions segmented from 3D tomograms with anti-REL1 IgG. (A) Comparison of purified anti-REL1 IgG structures obtained from electron tomography (1 and 3) and the crystal structure of IgG (2). (1) Averaged anti-REL1 IgG structure (from 26 particles segmented out from 3D tomogram). (2) IgG crystal structure (PDB number = 1IGT). The structure is shown in ribbon format superimposed with a semitransparent surface view of the same structure blurred to 40-Å resolution. (3) Anti-REL1 IgG structure from 1 superimposed with the docked ribbon representation of the IgG crystal structure from 2. (B) A central slice from the 3D tomogram of L-complex sample incubated with IgG. One L-complex particle with bound IgG is indicated by the dotted circle. (C) Shaded surface representation of 3 individual anti-REL1 IgG-decorated L-complex particles segmented out from the 3D tomogram. The third particle is the one shown circled in B. The putative IgG densities are indicated by circling. (D) Approximate location of REL1 on unbiased single-particle reconstruction.
Discussion
The L-complex is the core of the uridine insertion/deletion RNA editing machinery. There currently are 3 different systems of nomenclatures for L-complex proteins (Table S1). A TAP isolation procedure followed by gel filtration was used to obtain intact L-complex material from LC8-TAP-expressing L. tarentolae cells for electron microscopic analysis. The LC8 (MP44/KREPB5) protein was selected for the TAP procedure, because this protein has been shown to be required for viability and for stability of the L-complex (22), was a common component of all TAP pull-downs (18), and was not present in the REL1 and REL2 subcomplexes. We show that the LC8 and the REN2 proteins are single-copy in the L-complex, and it is possible that all L-complex proteins are single-copy except for a minor percentage of particles that contain 2 copies of the REL1 subcomplex. Consistent with this hypothesis, the calculated molecular weight sum of the proteins of each of the 3 classes of L-complexes (REN1–REN3), assuming single-copy, is consistent with the molecular weight determined by blue native gel electrophoresis and by gel filtration (Table S1).
We found that a final FPLC Superose 6 gel-filtration step of the LC8-TAP pull-down was necessary to obtain homogeneous complexes. The intactness of the material was established by blue native gel analysis, which showed the presence of a major single REL1 containing band migrating at 1 MDa. We show elsewhere that the very minor band migrating at 1.2 MDa is artifactual (21). The nature of this modification is not understood, but at most it represents a very minor fraction of the isolated material and does not interfere with the structural analysis.
The polypeptide profile of the isolated L-complex is similar to that previously reported, except for the presence of 2 proteins that are homologous to Leishmania major mitochondrial proteins of unknown function (Table S2). The TAP-isolated L-complex material showed several functional activities in vitro: gRNA-mediated precleaved U-insertion and U-deletion activities, and robust full-cycle U-insertion and U-deletion activities. All these data indicate that the isolated L-complex is intact and functional.
The first structural question we addressed is whether, in view of the reported minor compositional heterogeneity associated with the REN1, REN2, and REN3 proteins (18, 19), the functionally active L-complexes have a uniform structural organization. It is possible that such a preparation might contain many structural species that migrate similarly in the native gel and gel filtration column. The LC8-TAP pull-down should contain all three of the REN-specific complexes, but the relative amounts of each are uncertain. In fact, the REN1-TAP pull-down procedure for L. tarentolae is much less efficient than the LC8-TAP pull-down procedure, suggesting either that the REN1 tag is not exposed on the surface of the complex or that the REN1 complexes are relatively less abundant. Another possibility that would affect a structural analysis is that the complexes might be dynamic, adopting different conformations in solution. To address these questions, structure data were first obtained by electron tomography, a method in which individual L-complex particles are visualized independently without averaging and use of models. The majority of these particles showed a uniform triangular shape. Interestingly, a minority of particles contained an additional density extending from the central region, which may represent L-complexes with an additional component.
Our structural analysis was further substantiated by single-particle reconstruction. A featureless Gaussian ball was used for the initial model, and therefore this reconstruction analysis was not biased by the tomographic model. This method also yielded a triangular structure at a resolution of 24 Å. Another single-particle reconstruction was performed using the tomographic structure as the initial model, and this also converged to a similar structure. These results point to a single representative architecture for the majority of the L-complex particles. The structure exhibits an overall triangular shape with dimensions of ≈200 × 140 × 80 Å and can be divided into 3 regions (Fig. 5B). The Apex and Base regions of the complex have a globular appearance. In contrast, the Central region has a network of channels, which may provide a possible way of organizing substrate RNA traffic. The nature of the region of quasi-4-fold symmetry at the Apex base (Fig. 5C) is not known.
A preliminary localization of the REL1 RNA ligase to the base of the apex region was performed by decoration with anti-REL1 IgG. This suggests that REL1 and the REL1 subcomplex may be localized in the vicinity of the base of the Apex region of the complex. There may also be additional locations for REL1 since it is known that there can be 2 copies of the REL1 subcomplex, but this remains to be investigated.
Materials and Methods
Details of materials and methods used are given in SI Materials and Methods. Methods include cell culture and transfection, TAP isolation of L-complex, western blotting, mass spectrometry analysis, electron tomography, transmission electron microscopy, single-particle reconstruction, and 3D segmentation and visualization.
Supplementary Material
Acknowledgments.
We thank all members of the Simpson and Zhou labs for discussion, Greg Richmond for participation in the initial effort of this project, and Jun Liu and Hanspeter Winkler for advice in using the Protomo software package. This research was supported in part by National Institutes of Health Grants AI09102 (to L.S.) and GM071940 (to Z.H.Z.).
Note Added in Proof.
During the revision of our paper, a paper by Golas et al. was published describing 3D structures of the editing complexes of T. brucei (23).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901754106/DCSupplemental.
References
- 1.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 5.Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA [see comments] Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 6.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 7.Byrne EM, Connell GJ, Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 8.Panigrahi AK, et al. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom. 2003;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 10.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell GJ, Byrne EM, Simpson L. Guide RNA-independent and guide RNA-dependent uridine insertion into cytochrome b mRNA in a mitochondrial extract from Leishmania tarentolae. J Biol Chem. 1997;272:4212–4218. doi: 10.1074/jbc.272.7.4212. [DOI] [PubMed] [Google Scholar]
- 12.Frech GC, Bakalara N, Simpson L, Simpson AM. In vitro RNA editing-like activity in a mitochondrial extract from Leishmania tarentolae. EMBO J. 1995;14:178–187. doi: 10.1002/j.1460-2075.1995.tb06988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peris M, et al. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol Biochem Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- 14.Aphasizhev R, et al. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart K, Panigrahi AK, Schnaufer A. Identification and characterization of trypanosome RNA-editing complex components. Methods Mol Biol. 2004;265:273–291. doi: 10.1385/1-59259-775-0:273. [DOI] [PubMed] [Google Scholar]
- 16.Schnaufer A, et al. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 17.Kang X, et al. Disruption of the zinc finger motifs in the leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex. J Biol Chem. 2003;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- 18.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panigrahi AK, et al. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng J, et al. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osato D, et al. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009 May 15;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, et al. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golas MM, et al. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.