Abstract
The tumor suppressor p53 preserves genome integrity by inducing transcription of genes controlling growth arrest or apoptosis. Transcriptional activation involves nucleosomal perturbation by chromatin remodeling enzymes. Mammalian SWI/SNF remodeling complexes incorporate either BRG1 or Brm as the ATPase subunit. The observation that tumor cell lines harboring wild-type p53 specifically maintain expression of BRG1 and that BRG1 complexes with p53 prompted us to examine the role of BRG1 in regulation of p53. Remarkably, RNAi depletion of BRG1 but not Brm led to activation of endogenous wild-type p53 and cell senescence. We found a proline rich region (PRR) unique to BRG1 was required for binding to the histone acetyl transferase (HAT) protein CBP as well as to p53. Ectopic expression of a PRR deletion mutant BRG1 that is defective for CBP binding inhibited p53 destabilization. Importantly, RNAi knockdown of BRG1 and CBP reduced p53 poly-ubiquitination in vivo. In support of p53 inactivation by the combined activities of BRG1 and CBP, we show that DNA damage signals promoted disassociation of BRG1 from CBP thereby allowing p53 accumulation. Our data demonstrate a novel function of the evolutionarily conserved chromatin remodeling subunit BRG1, which cooperates with CBP to constrain p53 activity and permit cancer cell proliferation.
Keywords: Chromatin remodeling, p53, proteasomal degradation
Introduction
Diverse stress signaling pathways converge on the tumor suppressor p53, which orchestrates a transcription program to maintain genome integrity by inducing growth arrest, replicative senescence or apoptosis (Lane, 1992; Vousden and Lane, 2007). Cancer cells circumvent this proliferative restriction by selecting for loss or mutation of the p53 gene (Soussi and Wiman, 2007). Nonetheless, nearly 50% of human malignancies express wild-type p53 but often exhibit other changes such as epigenetic silencing of p14ARF or amplification of the mdm2 locus (Oliner et al., 1992; Sherr, 2001). Activity of p53 is strictly coordinated as response to genomic insult or aberrant oncogenic signaling; otherwise p53 is rapidly degraded by the proteasome. The major regulator of p53 catabolism is the E3 ubiquitin ligase mdm2, but other factors including Pirh2, ARF-BP1, Cop1, E4F1 and HAUSP were reported to destabilize p53 (Brooks and Gu, 2006; Le Cam et al., 2006). HAT proteins p300 and pCAF were also reported to posses HAT independent ubiquitin ligase activity (Grossman et al., 2003). HAT proteins are best known as histone and transcription factor acetylates that alter chromatin structure and activate transcription (Lee and Workman, 2007). Degradation of p53 is counteracted by acetylation of multiple lysine residues (Brooks and Gu, 2003), which interferes with ubiquitination and underscores the complexity of p53 inactivation mechanisms.
Eukaryotic genomes are wrapped around histones that restrict stochastic access to the genetic information embedded in DNA. Chromatin remodeling enzymes (CREs) facilitate entry of transcription complexes to specific segments of DNA by disrupting the histone - DNA contacts. Though the components of CRE complexes are conserved from yeast to humans, the interplay between the CREs and p53 in integrating stress signals and cell survival has remained enigmatic. Mammalian SWI/SNF incorporates either one of the highly related ATPase subunits Brahma (Brm) or Brahma related gene 1 (BRG1), which share 75% identity at the amino acid level. Brm knockout mice are normal with a slight increase in body weight; paradoxically, BRG1 knockout caused pre-implantation lethality (Bultman et al., 2000; Reyes et al., 1998). These contrasting BRG1 and Brm knockout phenotypes led us to investigate the distinguishing functions of these proteins and their impact on p53 based on our observation that BRG1 is uniformly expressed in cancer derived cell lines that harbor wild-type p53, while BRG1 deficient tumor cells contain p53 mutations or do not express p53 (Table S1). From these inferences we hypothesized that BRG1 may constrain p53 activity in proliferating cells.
Results
BRG1 restricts endogenous p53 and senescence
To address whether tumor cell lines that express wild-type p53 require BRG1, we used RNA silencing to specifically deplete BRG1 in HeLa, G401, and RKO cell lines originating from cervical, rhabdoid, and colon cancers respectively. BRG1 depletion resulted in stabilization of p53 protein in each of these cell lines and was accompanied by induction of the p53 transcriptional target p21Cip1 (Fig. 1A, B & C). In contrast, RNAi knockdown of the BRG1 paralogue Brm did not increase p53 protein levels (Fig. S1). In HeLa cells, the complex of the human papillomavirus E6 protein and the ubiquitin ligase E6AP stimulates p53 protein degradation, while mdm2 mediated p53 ubiquitination was reported to be inactive (Hengstermann et al., 2001). BRG1 depletion did not alter levels of E6 protein (Fig. S7). We also observed that RNA silencing of BRG1 in HPV negative p53 wild-type cell lines G401 and RKO led to a moderate increase in p53 protein and robust induction of p21Cip1 (Fig. 1B&C). BRG1 silencing also increased wild-type p53 protein levels in U2OS cells (Fig. S2). To confirm the effects of BRG1 depletion on p53 dependent transcription, we performed quantitative real time PCR (qRT-PCR) of p21 transcripts. There was two-fold increase in p21 transcripts in BRG1 depleted cells compared to control cells (Fig. 1E). A luciferase reporter under the control of p21 promoter was also used to assess the transcriptional activity of p53 in BRG1 depleted RKO cells. BRG1 knockdown led to a 2.7 fold increase in luciferase activity (Fig. 1F).
Figure 1. BRG1 depletion activates p53 and senescence in tumor derived cell lines.
Immunoblots of HeLa (A), G401 (B) and RKO (C) cells transfected with control shRNA or shRNA to BRG1. (D) HeLa cells transfected with control shRNA or shRNA to BRG1 and stained for SA-ß-Gal. 1000 cells from each of the three independent transfection were counted. (E) Quantitative real time PCR showing relative levels of p21 RNA in control and BRG1 depleted HeLa cells. (F) Luciferase reporter activity under control of p21 promoter in control or BRG1 depleted RKO cells.
Importantly, BRG1 depletion in HeLa cells led to a striking increase in senescence-associated-ß-galactosidase (SA-ß-Gal), an indicator of the senescent phenotype (Fig. 1D) and consistent with the induction of p21. While we predict p21 induction was due to activation of p53, it might be a consequence of reduced levels of this SWI/SNF subunit affecting another pathway. To address the p53 dependency of p21 induction in BRG1 depleted cells, we generated a HeLa cell line constitutively expressing dominant negative p53 mutant (dnp53). These cells were resistant to p21 induction and cell senescence following BRG1 depletion, confirming the p53 dependence of these effects (Fig. S3). Taken together, these data suggest that tumor cells must retain BRG1 expression to restrict transcriptional activation and growth arrest by p53.
BRG1 partners with CBP in regulation of p53
Two lines of evidence hinted that CBP complexes with BRG1. First, CBP recruits BRG1 to the interferon ß promoter (Agalioti et al., 2000). Second, BRG1 and CBP proteins were co-purified in a complex with an activated form of the TGF-ß pathway effector Smad2 (He et al., 2006). These findings prompted us to test whether HATs cooperate with BRG1 to inactivate p53 in proliferating cells. G401 cells were transfected with shRNA targeting CBP or p300. Knockdown of CBP reduced levels of CBP protein and conversely increased p53 and p21Cip1 proteins (Fig. 2A). Activation of p53 was also observed in U2OS and HeLa cells following depletion of CBP (data not shown). CBP knockdown did not alter the levels of BRG1 but there was a slight increase in p300 levels. In contrast, CBP and BRG1 levels remained unchanged and p53 was not activated following silencing of p300. To test p53 transcriptional activity under these conditions, we transfected G401 cells with a luciferase reporter under the control of the p21 promoter along with control, CBP or p300 siRNAs (Fig. 2B). There was a four-fold induction of luciferase in CBP knockdown cells compared to control and p300 siRNA transfections. To test the biological impact on the p53 pathway, we performed colony formation assays in G401 cells after RNAi knockdown of CBP and p300. CBP depletion inhibited colony formation in accord with activation of p53, whereas p300 shRNA did not (Fig. 2C). Furthermore knockdown of p53 in colony assays revealed that cell growth inhibition due to CBP knockdown was p53 dependent. These results imply that both BRG1 and CBP are required for proliferation of cells that express wild-type p53.
Figure 2. Depletion of CBP activates p53.
(A) Immunoblots of G401 cells transfected with control, CBP or p300 shRNA plasmids. (B) Luciferase reporter under control of p21 promoter co-transfected with control, CBP or p300 siRNAs in G401 cells and assayed for luciferase activity. (C) Colony formation in G401 cells co-transfected with hygromycin resistance plasmid along with control, CBP, p300 or CBP plus p53 shRNA plasmids, selected with hygromycin for 2 weeks and stained with crystal violet.
From RNA silencing experiments in cancer cell lines and inhibition of p53 activity by BRG1-CBP partnership, we hypothesized that BRG1 physically interacts with CBP and this complex attenuates p53. We therefore undertook a biochemical approach to test the physical interaction between BRG1 and CBP. Endogenous CBP and p300 immunoprecipitations from HeLa cell extracts revealed that each contained BRG1 (Fig. 3A). Interestingly, the CBP immunoprecipitate did not include p300 and the p300 immunoprecipitate did not pull down CBP, implying BRG1-CBP and BRG1-p300 are distinct complexes. Consistent with these results, BRG1 co-immunoprecipitated both CBP and p300 (Fig. 3A’). To ascertain whether BRG1 binds to CBP in vitro, fragments of CBP fused to glutathione S-transferase (GST) were purified from E. coli (Turnell et al., 2005). Baculoviruses were constructed to express in insect cells full-length BRG1 and an in-frame deletion (called NBRG) of amino acids 70-340 in the N-terminal proline-rich region (PRR) as this domain is not present in Brm. Binding assays showed that full-length BRG1 protein bound to the C/H3 region of CBP spanning residues 1460-1891 (Fig. 3B) while NBRG did not bind to GST-CBP fragments. It can be argued that NBRG is misfolded and on this basis is CBP binding defective. Therefore we introduced NBRG into H1299 cells and analyzed the immunoprecipitate for the presence of SWI/SNF subunit BAF47/Ini1 (Fig. S4A). NBRG immunoprecipitates contained BAF47 similar to BRG1. Moreover we introduced NBRG into SW13 cells and assayed for induction of the BRG1 transcriptional target CD44 and found that NBRG induced CD44 similar to BRG1 (Fig. S4B). BRG1 has been reported to co-immunoprecipitate with p53 though direct binding has not been proven (Lee et al., 2002; Napolitano et al., 2007). To confirm the BRG1-p53 interaction, we transfected BRG1 and p53 null H1299 cells with Flag-BRG1 and p53. Reciprocal immunoprecipitations of BRG1 and p53 indicate these proteins interact in vivo (Fig. 3C & C’). Additional in vitro binding experiments with purified and benzonase treated proteins showed that p53 bound to BRG1 and weakly to NBRG (Fig. 3D). These results imply that direct binding of BRG1 to CBP and strong binding of BRG1 with p53 requires the N-terminal PRR of BRG1.
Figure 3. BRG1 interacts with CBP/p300 and p53.
(A) Immunoprecipitations showing endogenous BRG1-CBP and BRG1-p300 interactions along with 15% total extract as input. (A’) Similar to (A) showing reverse immunoprecipitation and 10% total extract as input. (B) Binding reactions of bacterially purified GST-CBP fragments with baculovirus/insect cell produced Flag-BRG1 or Flag-NBRG (Δ70-340 amino acid), resolved by SDS-PAGE along with 15% of input protein, and blotted with Flag antibodies. (B’) Baculovirus expressed, Flag column purified proteins stained with Coomassie blue. (C-C’) Immunoprecipitation from H1299 cells transfected with p53 and Flag-BRG1 with control IgG, anti-p53 or Flag-M2 antibodies. Input is 5% of total extract. (D-D’) GST-p53 pull-down of baculovirus produced and purified Flag-BRG1 or Flag-NBRG. Input is 10% of total protein.
The N-terminal PRR of BRG1 is crucial for p53 destabilization and inactivation
Despite a high degree of sequence identity between BRG1 and Brm (Fig. 5A), RNAi depletion of BRG1 but not Brm activated p53. BRG1 but not Brm or NBRG co-precipitated p53 (Fig. 4B). We therefore inferred a role for the PRR unique to BRG1 in p53 destabilization. Moreover, as NBRG did not bind CBP, we questioned whether it would exert dominant-negative effects on steady state levels of endogenous p53 and its activation. Over-expression of NBRG or Brm, but not luciferase, increased levels of p53 protein in HeLa cells (Fig. 4C, D & E). Cycloheximide chase experiments revealed that heterologous expression of luciferase did not change the half-life of p53 protein but NBRG and Brm increased the half-life of p53 protein from <30 minutes to >60 minutes (Fig. 4E & F).
Figure 5. BRG1 and CBP promote destabilization of endogenous p53.
(A) HeLa cells transfected with control or shRNA to BRG1 or CBP. Cell lysates were immunoprecipitated with either rabbit IgG or p53 polyclonal antibodies and bound proteins blotted with ubiquitin antibodies. Lower panels show steady state input levels of indicated proteins. (B) Immunoblot of HeLa cells transfected with control shRNA or shRNA to BRG1 or CBP after treatment with cycloheximide (CHX) and harvested every 30 minutes. (C) Graph showing p53 decay normalized to actin from immunoblots of (B).
Figure 4. N-terminal PRR of BRG1 is required for p53 destabilization.
(A) Graphic representation showing the domain organization of BRG1, NBRG and Brm proteins. (B) Cell lysates prepared from H1299 cells transfected with p53 plus Flag-tagged Brm, NBRG or BRG1 plasmids were incubated with Flag-agarose beads, separated on a gel and blotted with p53 or Flag antibodies. (C, D & E) HeLa cells were transfected with increasing amounts of luciferase, NBRG or Brm plasmids and cell extracts were blotted with the indicated antibodies. (F) Representative picture of HeLa cells transfected with vector control, luciferase, NBRG or Brm and extracts prepared after cycloheximide treatment of indicated time points were analyzed for p53 levels. (G) Protein levels of p53 from three independent experiments were normalized to actin.
In another series of experiments, transfection of wild-type p53 inhibited colony formation of C33a cells that lack functional p53 and are deficient for BRG1. Growth suppression of C33a cells by p53 was neutralized by co-transfection with BRG1 but not with NBRG (Fig. S5). Interestingly p53-mediated inhibition of colony formation was reversed by the BRG1-KR point mutant that is defective for chromatin remodeling (Khavari et al., 1993). Since the PRR of BRG1 is required for binding to CBP and p53, these results imply that the PRR is crucial for p53 destabilization and inactivation.
BRG1 and CBP are required for poly-ubiquitination of p53 in vivo
While several factors are known to ubiquitinate p53, we queried whether BRG1 and CBP have critical roles in promoting p53 poly-ubiquitination. Endogenous p53 was immunoprecipitated from HeLa cell lysates and blotted with antibodies to ubiquitin. Heterogeneous forms of poly-ubiquitinated p53 were abundant in vector control transfected extracts (Fig. 5A, lane 2). RNAi depletion of BRG1 (lane 3) or CBP (lane 4) led to a dramatic reduction of ubiquitinated p53 species and accordingly increased steady-state p53 protein levels. Although CBP knockdown was not as complete, we observed a near total abrogation of p53 poly-ubiquitination in HeLa cells. An explanation how this partial CBP knockdown led to abrogation of p53 ubiquitination might be that multiple p53 ubiquitination effectors converge on CBP. Similar results were noted following shRNA silencing of BRG1 in U2OS cells (Fig. S2). Ubiquitination of an unrelated protein was slightly increased in shBRG1 treated cells, indicating that RNAi to BRG1 did not inactivate the ubiquitin pathway (data not shown). Cycloheximide chase experiments indicated prolongation of p53 protein half-life to > 2 hours following RNAi knockdown of BRG1 (Fig. 5B & C). CBP knockdown also increased the half-life of p53 protein to 120 min. These results reinforce our model that BRG1 and CBP collaborate to destabilize endogenous p53 protein in proliferating cancer cells.
DNA damage effects on BRG1 and CBP complexes
Since RNA silencing of BRG1 and CBP activated p53 and BRG1 interacts with CBP, we hypothesized that BRG1 might dissociate from CBP to allow p53 accumulation. In response to DNA damage, p53 levels began to increase after 6 hours (Fig. 6A). To test whether HAT proteins CBP and p300 dissociate from BRG1, we analyzed CBP or p300 immunoprecipitations for the presence of BRG1 seven hours post doxorubicin treatment. CBP and p300 clearly associated with BRG1 in unstressed cells (Fig. 6B lanes 3,4). Upon DNA damage, reduced amounts of BRG1 were in complex with CBP and p300 (lanes 7,8). These results support our hypothesis that association of BRG1 with CBP promotes degradation of p53, and conversely, dissociation of BRG1 from CBP following DNA damage allows p53 stabilization and activation.
Figure 6. DNA damage inhibits BRG1-CBP/p300 interaction.
(A) Immunoblots of HeLa cell extracts after treatment with doxorubicin (Dox) at indicated time points. (B) HeLa cells treated with doxorubicin for 7 hours (+ dox) or untreated (- dox), immunoprecipitated with antibodies to CBP or p300 and blotted for BRG1. Input indicates 15% of each reaction.
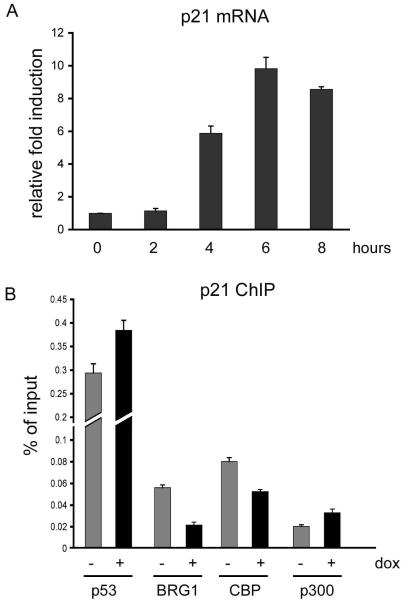
Since BRG1 and CBP modify chromatin fluidity, we reasoned that their inactivation of p53 might occur at a p53 transcriptional target promoter region. To confirm p21 transcript induction by activated p53, we treated U2OS cells with doxorubicin and assayed p21 transcripts by quantitative real time PCR. Compared to untreated cells, levels of p21 RNA increased beginning at four hours post-treatment (Fig. 7A). Subsequently we performed quantitative chromatin immunoprecipitation (qChIP) to determine occupancy of the p21 promoter by p53, BRG1, CBP and p300 in doxorubicin treated versus untreated U2OS cells. Commonly, p53 ChIP assays are conducted using the p53 monoclonal antibody DO-1 that recognizes an epitope on the transactivation region of p53 where mdm2 and other transcription complexes bind. Coincidently this region is subjected to phosphorylation in response to DNA damage signal. Therefore we deliberately chose to use rabbit antibodies that were raised against the full-length p53 protein to capture p53 at the p21 promoter. The qChIP assay revealed that total p53 and p300 present at the p21 promoter were only increased slightly subsequent to DNA damage (Fig. 7B). In contrast, there was reduced CBP and BRG1 after DNA damage compared to untreated cells. These results support our hypothesis that p53 is inactivated at the site of recruitment by a BRG1-CBP complex in proliferating cells. Conversely, p53 activation corresponded with reduced levels of BRG1-CBP at the p21 promoter.
Figure 7. DNA damage reduces the levels of BRG1 and CBP on p21 promoter.
(A) Quantitative real time PCR depicting relative levels of p21 RNA at time points following doxorubicin exposure of U2OS cells. (B) U2OS cells treated with doxorubicin for 5 hours (black bar) or untreated (gray bar) were subjected to chromatin immunoprecipitation at the distal p53 binding site in the p21Cip1 promoter and assayed using quantitative real time PCR by normalizing to control IgG from three independent experiments.
Discussion
ATP dependent multi-protein SWI/SNF complexes disrupt the nucleosomal barrier and allow transcription to initiate and proceed (Li et al., 2007). Chromatin remodeling enzymes are conserved from yeast to human and have a recognized role in stress responses in yeast (Downs et al., 2007; Papamichos-Chronakis et al., 2006; Tsukuda et al., 2005). We noticed a striking correlation that several cancer cell lines harboring wild-type p53 uniformly maintain expression of the SWI/SNF subunit BRG1. Because p53 induces expression of proteins that restrict cell cycle progression, it must remain transcriptionally inactive for continuous proliferation of tumor cells. BRG1 has been reported to bind p53 but the biological significance of their interaction remains unresolved. These observations led us to investigate whether BRG1 is merely a chromatin remodeling engine necessary for transcriptional activation but in addition may counter p53-induced growth suppression. The biochemical and RNA silencing experiments reported here support a novel role for BRG1 in cooperation with the HAT protein CBP in p53 destabilization that is necessary for inactivation of wild-type p53 in malignant cells.
BRG1 and CBP inactivate p53
Multiple cellular proteins including Mdm2, Pirh2, ARF-BP1 Cop1, E4F1 and HAUSP influence stability of the p53 protein (Brooks and Gu, 2006; Le Cam et al., 2006). We show that RNA silencing of BRG1 decreased p53 ubiquitination and turnover, whereas knockdown of its SWI/SNF paralogue Brm did not affect p53 stability. The resulting elevated levels of the p53 transcriptional target p21 efficiently induced the senescence program, indicating that depletion of BRG1 did not interfere with these downstream events. Furthermore, the fact that knockdown of BRG1 activated p53 despite the presence of the other reported p53 ubiquitin ligases implies BRG1 is a critical regulator of p53. These effects may explain why cancer cells that retain wild-type p53 must express BRG1; conversely pre-neoplastic cells deficient for BRG1 might select for p53 mutations. Accordingly, a recent report found elevated levels of BRG1 correlated with aggressive prostate cancer (Sun et al., 2007). In contrast, Brm expression is often silenced in tumor derived cell lines (Glaros et al., 2007; Yamamichi et al., 2007), consistent with our data that Brm over-expression stabilized p53.
Our RNA silencing experiments reveal that CBP but not p300 was required for proliferation and colony formation of cells expressing wild-type p53. This phenomenon is exemplified in the RKO colon cancer cell line (Figure 1C), which harbors wild-type p53 and maintains normal levels of CBP, yet is p300 deficient (Ionov et al., 2004). We hypothesize that mitogenic signals promote CBP-BRG1 mediated destabilization of p53 in proliferating cells. In contrast, in response to genotoxic damage, CBP and p300 switch to become activators by inducing p53 acetylation and activation of its transcription functions (Barlev et al., 2001). These dual roles for CBP are therefore context dependent. This interpretation may relate to the finding that CBP is required for self-renewal of hematopoietic stem cells (Rebel et al., 2002). The ability of BRG1 and CBP to inactivate p53 is also consistent with their requirement for maintenance of pleuripotent embryonic stem cells and proliferation of pancreatic β-cells (Hussain et al., 2006; Matsumoto et al., 2006; Miyabayashi et al., 2007; Rebel et al., 2002).
The proline-rich region (PRR) unique to the N-terminus of BRG1 and not present in Brm (Kadam and Emerson, 2003) is necessary for binding to the PHD type zinc finger domain. The PHD domain of MAPK kinase MEKK1 acts as an E3 ligase and promotes ubiquitination of Erk1 (Lu et al., 2002). Over-expression of the CBP binding defective deletion mutant of BRG1, NBRG, increased levels of the endogenous p53 protein. Similarly heterologous expression of Brm, which shares all domains with BRG1 except the PRR, inhibited p53 degradation. We suspect that NBRG and Brm stabilize p53 as these proteins bind to subunits of the multi-protein BAF (BRG1 associated factor) complex and thereby compete with endogenous BRG1. Similar to BRG1, the SWI/SNF family member p400 was reported to inhibit p21 induction by p53 (Chan et al., 2005). Ini1/BAF47/Snf5 is also a component of BRG1 and Brm containing SWI/SNF complexes. Deletion of Ini1 resulted in activation of the p53-p21 pathway in mouse embryonic fibroblast cells (Isakoff et al., 2005; Klochendler-Yeivin et al., 2006). However, assembly of the BRG1 SWI/SNF complex and BRG1 dependent transcription were unaltered in Ini1 deficient cells (Doan et al., 2004). RNAi knockdown of BRG1 in Ini1 deficient G401 cells led to activation of p53 and induction of p21 in our experiments, suggesting Ini1 is dispensable for p53 inactivation by BRG1.
Context dependent, divergent roles of BRG1 in control of cell growth
BRG1 and Brm genes are mutated or not expressed in a subset of tumor derived cell lines, suggesting tumor suppressor function (Muchardt and Yaniv, 2001). The SWI/SNF subunit BRG1 binds to the tumor suppressor retinoblastoma protein Rb through a LXCXE motif and the resultant transcriptional repression complex prevents cell cycle progression (Dahiya et al., 2000; Dunaief et al., 1994; Zhang et al., 2000). BRG1 haploinsufficient mice developed mammary tumors with a penetrance of 9% but did not phenocopy Rb mutations (Bultman et al., 2008). Furthermore, the Rb binding site (LXCXE) is not conserved in Drosophila Brm and a BRG1 mutant lacking the Rb binding motif retained ability to induce growth arrest (Kang et al., 2004). A specific role for BRG1 in oncogenic transformation is supported by the observation that Ras transformed NIH3T3 cells down-regulated Brm expression but maintained high levels of BRG1 (Muchardt et al., 1998). Our conclusion that BRG1 restricts p53 activity may explain the apparent duality of BRG1 functions in development and cell transformation.
Although BRG1 deficiency correlates with the absence of wild-type p53 in malignant cells, some BRG1 mutations found in cancer cell lines may retain the ability to counter wild-type p53. It is particularly noteworthy that the BRG1 ATPase mutant retains ability to inactivate p53, implying that p53 inactivation by BRG1 is chromatin remodeling independent. Moreover our model that BRG1 is required for restricting p53 in dividing cells provides an explanation for embryonic lethality of BRG1 knockout, as absence of BRG1 would result in p53 activation. A critical role for BRG1 in neuronal stem cell maintenance has been proposed as conditional deletion of BRG1 resulted in precocious induction of differentiation (Matsumoto et al., 2006). Remarkably, a recent report asserted a key role for several chromatin regulators including Tip60-p400 and BRG1 in preservation of embryonic stem cell characteristics (Fazzio et al., 2008). The HAT protein Tip60 was shown to acetylate and activate the transcription function of Myc (Awasthi et al., 2005; Frank et al., 2003; Patel et al., 2004). Although Tip60 was initially claimed to serve as a Myc co-activator that promotes cell proliferation, subsequent reports revealed its tumor suppressor function is required for oncogene induced DNA damage response (Gorrini et al., 2007; Squatrito et al., 2006). A recent study showed opposing roles in cell cycle for SWI/SNF complexes that differed by a single variant subunit (Nagl et al., 2007). Taken together, these effects illustrate that specific components of multiprotein SWI/SNF and HAT complexes participate in cell proliferation and growth suppression.
In response to DNA damage, less CBP was in complex with BRG1, which correlated with p53 stabilization. Furthermore, there was reduced BRG1 and CBP at the distal p53-binding site of p21 promoter, while in contrast, levels of p53 and p300 were slightly increased. It is relevant to note that in stem cells, proteasome and transcription complexes including BRG1 were localized to regulatory regions of cryptic promoters to maintain the genes potentiated for future activation (Szutorisz et al., 2006). Based on our results and compelling evidence from other laboratories (Chan et al., 2005; Espinosa et al., 2003; Kaeser and Iggo, 2002; Thomas and Chiang, 2005), we propose a model in which p53 is continuously recruited to its target promoters where it is inactivated by the BRG1 complex containing CBP, and that proliferating cancer cells expressing wild-type p53 retain BRG1-CBP function to circumvent growth arrest. While chromatin remodeling defective BRG1 restrains p53 activation, p53 may also be regulated in a chromatin independent fashion by the SWI/SNF protein complex.
Materials and Methods
Antibodies
p53 DO-1, rabbit anti-p53, anti-mdm2, anti-p21Cip1, anti-actin, rabbit anti-E6AP, anti-ubiquitin, anti-CBP and anti-p300 antibodies were purchased from Santa Cruz; Flag M2, BRG1, and anti-BRG1 and anti-Brm antibodies (de la Serna et al., 2001). Monoclonal antibody C1P5 was used to detect HPV 18 E6 (Banks et al., 1987).
Constructs
A Kozak consensus sequence was introduced at the translation initiation site of BRG1 and Brm. The N-terminal PRR deletion NBRG construct was constructed by deletion of the internal NcoI fragment. BRG1 and NBRG coding regions were transferred into pFASTBAC (Invitrogen) and recombined in vivo to obtain bacmids. CBP-myc and p300-HA constructs are described elsewhere (Eckner et al., 1994; Kazantsev et al., 1999).
shRNA constructs
The sequences for shRNA targeting were 5′-GATTTGCGAACCAAAGCGA for BRG1, 5′-CCAAGTCCTGGACCTCCAA for Brm, for CBP 5′-TAGTAACTCTGGCCATAGC, 5′-TCATTTCACACTGGAAGAA for p300, shRNA to GFP described in (Berns et al., 2004). These inverted DNA sequence fragments along with U6 or H1 promoter were cloned to EBV-based episomal pREP4 (Invitrogen) plasmids.
Transfection
Lipofectamine 2000 (Invitrogen) was mixed at 1:1 with plasmid DNA for 20 min and added to cells. 24 hours after transfection, fresh medium without antibiotics was added.
Immunoprecipitations
Immunoprecipitation (IP) buffer [20 mM Tris pH8, 150 mM KCl, 0.5% Triton X-100, 20% glycerol, 10 mM NaF, 2 mM Na-orthovanadate, 5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol and protease inhibitor cocktail (Roche)] was added to the cells after a PBS rinse and frozen at -80°C. Cells were briefly sonicated, supernatants incubated with antibodies and Protein A or G Sepharose, bound protein complexes washed with IP buffer and separated on acrylamide-SDS gels. For IP of polyubiquitinated p53, cells were lysed in 50 mM Tris pH 8.0, 150 mM NaCl, 0.5% SDS and protease cocktail and frozen at -80°C. An equal volume of lysis buffer without SDS was added to the extract prior to brief sonication. Lysates were diluted to final concentration of 0.1 %SDS and incubated with rabbit anti-p53. Immunoprecipitated p53 proteins were separated on a SDS gel and immunoblotted with ubiquitin antibodies.
GST pull-down assay
GST fusion proteins were purified from E. coli. Baculovirus expressed Flag-BRG1 and Flag-NBRG proteins were purified on M2 beads and eluted with Flag peptide. Proteins were treated with benzonase to remove nucleic acid contaminants then incubated in 20 mM Tris pH8.0, 100 mM NaCl, 0.5% NP40 and 0.5 mM DTT at 4°C, washed in binding buffer containing 150 mM NaCl, and analyzed by PAGE.
Senescence associated β-gal assay
Senescence assay was conducted as described elsewhere (Dimri et al., 1995). Briefly, cells were fixed for 5 min with 2% formaldehyde and 0.2% glutaraldehyde. Cells were rinsed with phosphate buffer and developed with 0.1% x-gal solution in sodium citrate buffer (pH 6.0) incubated at 37° C 24 hours to 36 hours.
Chromatin immunoprecipitation
Cells were treated with 1.5% formaldehyde at for 8 min 37°C. Crosslinking was stopped by addition of glycine to 0.125 mM for 10 min. Cells were rinsed with PBS and 1 ml of Tris-EDTA buffer and sonicated. Chromatin extracts were cleared by centrifugation followed by incubation with antibodies in chromatin ChIP buffer (0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate and 140 mM NaCl). Bound chromatin fragments were washed with ChIP buffer with 300 mM NaCl. Crosslinking was reversed at 65°C in 300 mM NaCl and 1% SDS. Released DNA fragments were purified on a spin column (Qiagen). Quantitative real time PCR was performed targeting the distal p53 binding site of the p21Cip1 promoter with primers 5′-GGCTGAGCCTCCCTGCATCC and 5′-CCTGGGGTCTTTAGAGGTCTCCTGTC. DNA amplification was measured using Cyber-green.
Supplementary Material
Acknowledgements
We thank Marius Pop and Andrew King for CBP and p300 shRNA, Andrew Turnell for GST-CBP fusion constructs, Yuval Bibi Nitzan for comments and assistance with the figures, and Neal Silverman for assistance with baculovirus protein production. R01 CA107532 to S.R.G., R01 GM56244 to A.N.I., and R01 CA107394 to E.J.A supported this work.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–78. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Awasthi S, Sharma A, Wong K, Zhang J, Matlock EF, Rogers L, et al. A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol Cell Biol. 2005;25:6178–98. doi: 10.1128/MCB.25.14.6178-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks L, Spence P, Androphy E, Hubbert N, Matlashewski G, Murray A, et al. Identification of human papillomavirus type 18 E6 polypeptide in cells derived from human cervical carcinomas. J Gen Virol. 1987;68(Pt 5):1351–9. doi: 10.1099/0022-1317-68-5-1351. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–54. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–7. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–71. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–8. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 --> p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya A, Gavin MR, Luo RX, Dean DC. Role of the LXCXE binding site in Rb function. Mol Cell Biol. 2000;20:6799–805. doi: 10.1128/mcb.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–90. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DN, Veal TM, Yan Z, Wang W, Jones SN, Imbalzano AN. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene. 2004;23:3462–73. doi: 10.1038/sj.onc.1207472. [DOI] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–8. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–30. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–84. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–27. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–74. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–80. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–66. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–4. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–41. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc Natl Acad Sci USA. 2001;98:1218–23. doi: 10.1073/pnas.031470698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MA, Porras DL, Rowe MH, West JR, Song WJ, Schreiber WE, et al. Increased pancreatic beta-cell proliferation mediated by CREB binding protein gene activation. Mol Cell Biol. 2006;26:7747–59. doi: 10.1128/MCB.02353-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y, Matsui S, Cowell JK. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc Natl Acad Sci U S A. 2004;101:1273–8. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–50. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci U S A. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–99. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–9. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–4. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–74. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–88. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277:22330–7. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945–56. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–83. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2007;104:5668–73. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. Embo J. 1998;17:223–31. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. When the SWI/SNF complex remodels...the cell cycle. Oncogene. 2001;20:3067–75. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- Nagl NG, Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. Embo J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano MA, Cipollaro M, Cascino A, Melone MA, Giordano A, Galderisi U. Brg1 chromatin remodeling factor is involved in cell growth arrest, apoptosis and senescence of rat mesenchymal stem cells. J Cell Sci. 2007;120:2904–11. doi: 10.1242/jcs.004002. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–3. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–49. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–34. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci U S A. 2002;99:14789–94. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17:6979–91. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–7. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–12. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–42. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67:203–13. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–88. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17:251–64. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–83. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, et al. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–5. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, et al. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–35. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.