Abstract
AIM: To investigate the molecular pathways involved in human cholangiocarcinogenesis by gene expression profiling.
METHODS: Oligonucleotide arrays (Affymetrix U133A) were used to establish a specific gene expression profile of intrahepatic CCC in comparison to corresponding non-malignant liver tissue. To validate the expression values of the most overexpressed genes, RT-PCR experiments were performed.
RESULTS: Five hundred and fifty-two statistically differentially expressed genes/ESTs (221 probes significantly up-regulated, 331 probes down-regulated; P < 0.05; fold change > 2; ≥ 70%) were identified. Using these data and two-dimensional cluster analysis, a specific gene expression profile was obtained allowing fast and reproducible differentiation of CCC, which was confirmed by supervised neuronal network modelling. The most consistently overexpressed gene (median fold change 33.5, significantly overexpressed in 100%) encoded osteopontin. Furthermore, an association of various genes with the histopathological grading could be demonstrated.
CONCLUSION: A highly specific gene expression profile for intrahepatic CCC was identified, allowing for its fast and reproducible discrimination against non-malignant liver tissue and other liver masses. The most overexpressed gene in intrahepatic CCC was the gene encoding osteopontin. These data may lead to a better understanding of human cholangiocarcinogenesis.
Keywords: Cholangiocarcinoma, Oligonucleotide arrays, Osteopontin, Cell cycle regulation, Gene expression
INTRODUCTION
Cholangiocarcinoma (CCC) is the second most common primary hepatic malignancy next to hepatocellular carcinoma (HCC), arising from cholangiocytes of intra- or extrahepatic bile ducts. In contrast to HCC, cholangiocarcinomas are, in most cases, adenocarcinomas and relatively hypovascularised. Two principal forms of these tumors are recognized based on the clinical presentation[1]. The most common form is a highly desmoplastic cancer with a growth pattern characterized by periductal extension and infiltration, leading to an early onset of jaundice as one of the first clinical signs of this disease (perihilar and distal extrahepatic CCC). The other form of CCC grows as a mass lesion within the liver, very often leading to the misdiagnosis of hepatocellular carcinoma or metastatic adenocarcinoma (intrahepatic CCC).
Despite recent progress in various imaging modalities the neoplasm lesion is, in most cases, advanced by the time of diagnosis. Thus, curative treatment options by surgical resection are limited, leading to a poor prognosis with median survival times of only three to six months, especially in patients with intrahepatic CCCs[2,3].
Recent epidemiologic studies have shown an increasing incidence of CCC in western countries[4,5]. In most patients with CCC, no risk factors for cholangiocarcinogenesis can be identified. However, established risk factors for ductal cholangiocarcinoma include diseases leading to chronic inflammation of the bile ducts, such as infections with Clonorchis sinensis, Opisthorchis viverrini (liver flukes) and chronic viral hepatitis, chronic intrahepatic lithiasis, primary sclerosing cholangitis (PSC), congenital diseases (Caroli disease, congenital choledochal cysts) and exposure to the radiopaque medium thorium dioxide (Thorotrast)[6–8]. In particular, patients with PSC have an increased risk for cholangiocarcinogenesis and epidemiologic studies have shown a lifetime risk for CCC of approximately 1.5% per year of disease[6].
Despite this knowledge about the major role of chronic inflammation in cholangio-carcinogenesis, little is known about the molecular pathways involved in CCC that lead to uncontrolled cell growth, downregulated apoptosis and, as a result, to tissue invasion.
Therefore, it is important to identify the genes involved in these molecular pathomechanisms, as this may shed light on the specific tumor biology and help to establish prognosis patterns as well as specific markers for screening in patients with a known increased risk for CCC (for example, patients with PSC). At present, the most commonly used markers in CCC include tumor antigens or products, such as CA 19-9, cytokines (for example, interleukin-6), (epi-)genetic lesions (for example, K-ras and p53 mutations), and metabolic products as lactate and proteases (for example, trypsinogen-2). However, none of these have been proven to be a useful diagnostic tool with high specificity and sensitivity for CCC[9].
Today, high-throughput techniques using cDNA or oligonucleotide arrays for the simultaneous and fast analysis of thousands of genes are available, potentially leading to the identification of new markers and subclassification of various human carcinomas[10–12]. In the present study oligonucleotide arrays (HU 133A, Affymetrix) containing more than 20 000 genes and expressed sequence tags (ESTs) were used to analyze gene expression in intrahepatic CCC tissues. The aim was first to detect specific genes (by comparison with genes expressed in non-malignant liver tissue) that may be helpful for discrimination between non-malignant and different malignant liver masses, and to establish a specific diagnostic genetic profile of intrahepatic CCC for clinical routine, and second, to elucidate the genetic pathways that are typically altered in cholangiocarcinogenesis.
MATERIALS AND METHODS
Acquisition of samples
Surgical specimens were obtained from 10 patients who underwent surgical treatment with curative intention for intrahepatic CCC between 2003 and 2004 (Table 1). In 8 cases, the adjacent corresponding non-malignant liver tissue was also acquired for microarray analyses. No patient received preoperative or adjuvant chemo- or radiotherapy.
Table 1.
Patient demographics and histopathological data
| Patient No. | Sex | Age | Histology | Stage | Grading |
| 1 | F | 57 | Adenocarcinoma | pT3pNxpM0 | G3 |
| 2 | F | 65 | Adenocarcinoma | pT3pN0pM0 | G2 |
| 3 | F | 68 | Adenocarcinoma | pT2pNxpM0 | G2 |
| 4 | M | 74 | Adenocarcinoma | pT3pN1pM0 | G3 |
| 5 | F | 62 | Adenocarcinoma | pT3pN0pM0 | G2 |
| 6 | F | 47 | Adenocarcinoma | pT3pNxpM0 | G2 |
| 7 | M | 73 | Adenocarcinoma | pT3pN0pM0 | G2 |
| 8 | F | 71 | Adenocarcinoma | pT2pNxpM0 | G2 |
| 9 | M | 52 | Adenocarcinoma | pT3pN2pM0 | G2 |
| 10 | M | 65 | Adenocarcinoma | pT3pNxpM0 | G1 |
Samples were resected after appropriate informed consent was obtained and the genetic analysis of the tumor and liver tissue was conducted in accordance with the guidelines of the Declaration of Helsinki (1995) and approved by the local ethic committee. Samples of sufficient weight (> 400 mg) from malignant and corresponding non-malignant liver tissue were excised and snap-frozen in liquid nitrogen within 20 min after excision. Until RNA isolation, samples were stored constantly at -80°C.
Histopathological evaluation
The various architectural patterns and cytological variants of cholangiocarcinoma and especially hepatocellular may complicate the differentiation of primary liver tumors and even that between primary liver tumors and metastases. Therefore, pathological reports with tumor typing, staging (performed using UICC criteria) and grading as well as clinical data were obtained for each tissue sample. Hematoxylin-eosin staining was performed for detection of features like bile canalicular structure and Mallory hyaline bodies. Additional histochemical and immunohistochemical stainings with a panel of antibodies were used routinely (HEP-PAR-1, CK7, CA19-9, CD10, CEA, AFP; Figure 1A and B) in all resected tissue biopsies to confirm the histogenesis of cholangiocellular carcinoma and to exclude hepatocellular carcinoma or metastases. In particular, CK7 is used as an important marker for CCC, as it is significantly overexpressed in CCC compared with HCC and most cases of other adenocarcinoma[13].
Figure 1.
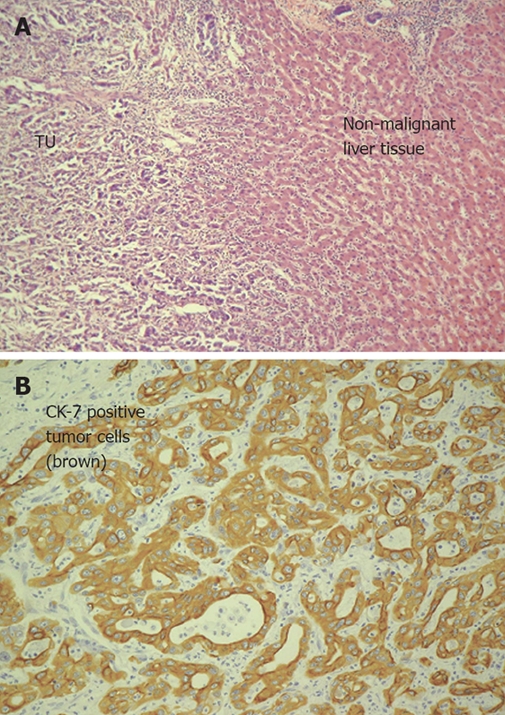
A: HE staining of infiltrative intrahepatic cholangiocarcinoma (Patient No. 6; left tumor tissue with cells of a moderate (G2) differentiated adenocarcinoma, right non-malignant liver tissue); B: Detection of cytokeratin 7 in intrahepatic cholangiocarcinoma (same patient as in A) using immunohistochemistry. Cytokeratin 7 was detected in over 80% of all analyzed CCCs strongly overexpressed (14.6-fold change).
Oligonucleotide array analysis
Sample preparation: Samples were processed with only minor modifications according to the Affymetrix GeneChip® Expression Analysis Manual (Santa Clara, CA/USA). For the extraction of total RNA frozen samples were homogenized in Trizol (Invitrogen, USA). The total RNA yield for each sample was 200-400 μg. After washing with DEPC-treated water (Ambion, USA), total RNA was cleaned using RNEasy mini kits (Quiagen). The amount of total RNA was measured photometrically using a BIO-Photometer (Eppendorf; OD1 E260 = 40 μg/mL).
Preparation of labeled cRNA and hybridization to oligonucleotide arrays: For transcription of total RNA into cDNA, 8 to 10 μg of total RNA was used for cRNA preparation using the SuperScript Choice system (Invitrogen, USA). First-strand cDNA synthesis was primed using a T7-(dT24) oligonucleotide primer with an RNA polymerase promoter site added to the 3’ end. After second-strand synthesis, in vitro transcription was performed in the presence of biotin-11-CPT and biotin-16-UTP (Enzo Diagnostics) to produce biotin-labeled cRNA. After fragmentation of the cRNA products (20 μg at 94°C for 35 min) to lengths of 35-200 bp, the samples were added to a hybridization solution to a final cRNA concentration of 0.05 mg/mL. Hybridization was performed by incubation (18-20 h) of 200 μL of the sample to an Affymetrix human GeneChip (Hu 133A) containing 22 283 probe sets for known genes/ESTs and stained with streptavidin-phycoerythrin. A Gene Array scanner G2500A (Hewlett Packard, ID) was used to scan the arrays according to procedures described in the Affymetrix manual.
To validate the relative change in gene expression of the most consistently overexpressed gene in all tumor samples (osteopontin, OPN), further analysis with real-time PCR using the LightCycler© system (Roche Diagnostics, Mannheim, Germany) was performed in 8 samples (tumor tissue vs corresponding non-malignant liver tissue).
Gene-specific primers corresponding to the coding region were designed using OLIGO software and were obtained from Biomers.net (Ulm, Germany; forward primer OSTEO: 696 U; GGACAGCCGTGGGAAGG, reverse primer OSTEO 810 L; TCAATCACATCGGAATGCTCA). Preliminary experiments were performed to test for the specificity of product formation, determine annealing temperatures, and check for Tm(Product-Primer Dimer) > 3°C. Validation experiments were performed within a fluorescence signal window excluding primer-dimer formation. The correct PCR efficiency for each target was determined by constructing relative standard curves using five-point half-logarithmic RNA dilutions from one sample. Template concentrations were given arbitrary values of 1, 0.316, 0.1, 0.0316 and 0.01. RT-PCR reactions were performed using the LC RNA Amplification Kit containing SYBR Green I, (Roche Diagnostics; Mannheim, Germany). Amplification was followed by melting curve analysis. Relative values for the initial target concentration in each sample were determined using Light Cycler software 3.5. Crossing points (Cp) were computed using the “2nd derivative maximum method”, part of the software above. The relative change in gene expression was computed by pairwise comparison of tumor samples to samples of adjacent normal tissue for each patient.
Statistical analysis
The statistical analyses and presentation of the obtained data were performed in accordance with the MIAME criteria and will be published on the web page (www.matrigene.de/geneprofiles/CCC).
Preliminary data analysis: Raw data analysis was conducted using the software of the Affymetrix microarray suite (MAS vs 5.0.1). MAS produced an expression value plus an index parameter indicating positive or negative detection (present call index) for each of the 22 283 probe sets (known genes/ESTs) on the array. Statistical analysis and post-processing were performed using GeneSpring (vs 6.1; Silicon Genetics, Redwood City, CA) and GeneExplore (vs 1.1; AppliedMaths, Saint-Martens-Latem, Belgium) software. Mismatch probes acted as specific controls on each array and allowed the direct subtraction of both background and cross-hybridization signals. Only Chip results with scaling factors of 0.5 to 1.8 were accepted for further analyses. Expression values were log2 transformed on the basis of the signal log ratio, given by the comparison of two array results between tumor and non-tumorous tissues.
A P-value cut off of < 0.05 (t-test) and a fold change difference of ≥ 2 in 70% or more of all analyzed samples were considered to be significant.
Second analysis step - Hierarchical clustering and supervised analysis: To identify differentially expressed genes in CCC, normalized expression data on statistically different genes were used for a paired data analysis by hierarchical clustering with gene trees/experiment trees using either Pearson’s correlation or standard correlation algorithms (Genespring vs 6.1; GeneExplore vs 1.1).
The class predictor program (supplied by GeneSpring 6.1) was used for supervised learning and training (neuronal training) to identify unknown probes as tumor or non-tumorous tissue. A list of the most predictive genes was made for each class and an equal number of genes with the lowest P-value using Fischer’s exact test were taken from each list. To make a prediction, the class predictor uses the k-nearest-neighbour method.
To detect statistically differentially expressed genes in well and poor differentiated intrahepatic CCC (genetic subclassification), a 1-way ANOVA test was performed. In a second step significantly dysregulated genes (P < 0.05) in tumor samples were used for a weighted two-dimensional clustering.
Identification of significantly altered metabolic pathways involved in cholangiocarcinogenesis: To identify specific signaling or metabolic pathways involved in cholangiocarcinogenesis data on significantly differentially expressed genes were transferred to a public domain software program (GenMAPP 2.0 beta©Gladstone Institutes, 2000-2004).
RESULTS
Gene expression profiling of intrahepatic CCC
Primary chip data were screened for RNA quality by 5’ to 3’ degradation. Of the 22 283 probes (genes/ESTs) present on the chip, on average, 43.3% (SD ± 4.53) and 39.0% (SD ± 3.48) of genes were expressed in CCC and corresponding non-malignant tissue samples.
Based on the primary data an algorithm was developed to identify and rank the most consistently up- and downregulated genes in CCC compared with non-malignant tissue. All statistically different genes (P < 0.05) with a fold change of at least a 2-fold increase or decrease in 70% or higher percentage of the CCC tumor samples were used to generate a databank of 552 genes/ESTs. Of these, 221 were upregulated and 331 were downregulated. This set of data was used for two-dimensional clustering. Ninety percent of all tumor samples showed a consistent pattern of up- and downregulated genes; only 1 case (No. 7) showed characteristics of both tumor and non-tumor tissue (Figure 2).
Figure 2.
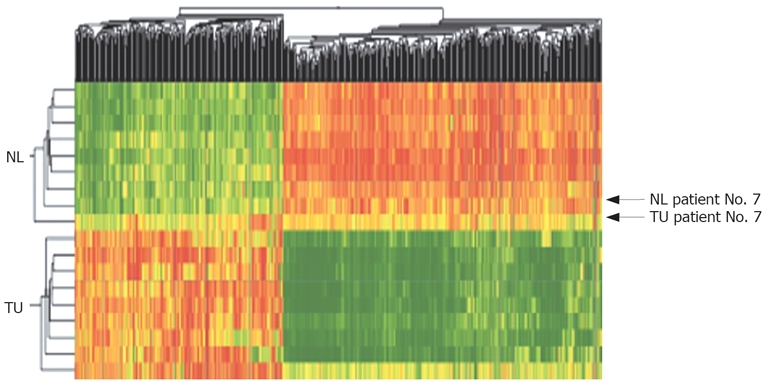
Two-dimensional cluster analysis of CCC (TU) and corresponding non-malignant liver tissues (NL) using 552 dysregulated genes/ESTs (red: overexpressed genes, green: downregulated genes; Pearson’s correlation). Dysregulation was defined as different genetic expression in > 70% of all probes with a fold change > 2.0. Horizontal lines show all dysregulated genes in one singular tissue probe, vertical lines show the expression of one gene in all analyzed tumor and non-malignant tissue probes. Using cluster analysis a fast differentiation between tumor tissue and non-malignant tissue was possible in 90% of cases (tumor tissue of Patient No. 7 showed many genetic similarities to non-malignant liver tissue).
To further test the robustness of this cluster, a supervised learning method (SLM) based on neuronal networking was applied. An optimal prediction with subsequent cross-validation was obtained using all significantly dysregulated genes/ESTs and 8 neighbors; using this, all 10 tumor samples could be correctly identified. This resulted in a positive prediction value of 90% (P < 0.001) and a negative predictive value of 100% (P < 0.00023) of the Gene cluster with the expression pattern of the 552 probes (Table 2).
Table 2.
Statistical results obtained using a supervised neuronal training method for discrimination between CCC and non-malignant liver tissue
| Condition | True value | Prediction | P value |
| Pat. No. 1, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 2, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 3, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 4, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 5, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 6, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 7, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 8, non-malignant | Normal liver tissue | Non-malignant | 0.00023 |
| Pat. No. 1, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 2, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 3, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 4, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 5, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 6, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 7, CCC | CCC | Malignant | 0.16100 |
| Pat. No. 8, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 9, CCC | CCC | Malignant | 0.00103 |
| Pat. No. 10, CCC | CCC | Malignant | 0.00103 |
Statistical results of the ability to differentiate between normal and malignant liver tissue (CCC) using a supervised neuronal training method. In all cases, a fast and correct differentiation was possible with a high positive (P < 0.001 in 90%) and negative (100%, P < 0.00023) predictive value.
Using these data, even a specific and fast discrimination between intrahepatic CCC and other malignant liver tumors (HCC, metastases; data not shown), with a high predictive value (P < 0.001), was possible by two-dimensional cluster analysis and SLM.
Dysregulated genes and specifically involved metabolic pathways in intrahepatic CCC
CCC gene expression profile data were imported into GeneMapp software to identify specific changes in molecular pathways. Of the 552 dysregulated genes, a total of 364 genes could be assigned to specific metabolic or signaling pathways. Most of the consistently and strongly overexpressed genes were related to cell cycle regulation and DNA replication (15 genes, including ribonucleoside-diphosphate reductase M2, calgizzarin, calcyclin, BUB1B) or intracellular signaling (15 genes, including CD24 and MARCKS), genes encoding transcription factors (6 genes, such as SOX9), or genes involved in nuclear organization and nucleic metabolism (13 genes, such as thymidylate synthetase). Most of the other up-regulated genes could be attributed to gene families, such as genes coding for extracellular matrix and cell adhesion molecules (37 genes, for example, OPN, ADAM9, thymosin beta-10, integrin alpha-6), cytoskeleton structure (16 genes, such as tropomyosin2, cytokeratin 7 and 19) or protein biosynthesis (4 genes).
According to an upregulation of genes involved in cell cycle regulation, most of the genes encoding proteins involved in cellular apoptosis (7 genes, such as growth-arrest specific protein 2, CIDE-B) were found to be downregulated in intrahepatic CCC. Furthermore, a significant suppression of genes encoding proteins important for metabolic pathways like amino acid metabolism (39 genes), carbohydrate and fat metabolism (13 and 11 genes) or electron transport (27 genes) was noted. For clarity, only the 41 most consistently dysregulated (increased or decreased) genes (P < 0.001; 4-fold change in 100%) are listed in Table 3.
Table 3.
Most significantly dysregulated genes (> 4-fold change in 100% of all cases) in intrahepatic cholangiocarcinoma - relation to known cellular functions and metabolic pathways (using the GenMAPP® software)
| Swiss prot | GenMAPP ID | Gene ID | Dysregulated in % | Name | fc inc (median) | fc dec (median) |
| Cell adhesion (cell-cell/cell-matrix)-extracellular matrix | ||||||
| OSTP_HUMAN | P10451 | SPP1 | 100 | Osteopontin precursor (Bone sialoprotein 1/SPP-1 1) | 33.5 | |
| Q13443 | Q13443 | ADAM9 | 100 | A disintegrin and metalloproteinase domain 9 (meltrin γ) | 10.4 | |
| TYBO_HUMAN | P13472 | TMSB10 | 100 | Thymosin beta-10 | 5.3 | |
| ITA6_HUMAN | P23229 | ITGA6 | 100 | Integrin alpha-6 precursor | 4.5 | |
| Signal transduction - G protein signaling | ||||||
| CD24_HUMAN | P25063 | CD24 | 100 | Signal transducer CD24 precursor | 11.8 | |
| IQG1_HUMAN | P46940 | IQGAP1 | 100 | Ras GTPase-activating-like protein IQGAP1 | 9.1 | |
| ECT2_HUMAN | Q9H8V3 | ECT2 | 100 | ECT2 protein (Epithelial cell transforming 2 oncogene) | 6.3 | |
| MACS_HUMAN | P29966 | MACS | 100 | Myristolated alanine-rich C-kinase substrate (MARCKS) | 4.6 | |
| Q9BUV5 | Q9BUV5 | INSIG1 | 100 | Similar to insulin induced gene 1 | 5 | |
| Cell cycle - DNA replication | ||||||
| RIR2_HUMAN | P31350 | RRM2 | 100 | Ribonucleoside-diphosphate reductase M2 chain | 16.2 | |
| S111_HUMAN | P31949 | S100A11 | 100 | Calgizzarin (S100C protein) | 15.5 | |
| O00496 | O00496 | IPL | 100 | Tumor suppressing subtransferable candidate 3 | 14.1 | |
| S106_HUMAN | P06703 | S100A6 | 100 | Calcyclin (Growth factor inducible protein 2A9) | 6.9 | |
| Transcription | ||||||
| SOX9_HUMAN | P48436 | SOX9 | 100 | Transcription factor SOX9 | 4.1 | |
| NRI3_HUMAN | Q14994 | NR1/3 | 100 | Orphan nuclear receptor NR1/3 | 6.8 | |
| Cell motility – cytoskeleton | ||||||
| SPT2_HUMAN | O53291 | SPINT2 | 100 | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) | 5.8 | |
| Amino acid metabolism | ||||||
| BHMT_HUMAN | Q93088 | BHMT | 100 | Betaine-homocysteine S-methyltransferase | 48.5 | |
| METL_HUMAN | Q00266 | MAT1A | 100 | S-adenosylmethionine synthetase alpha and beta forms | 38.5 | |
| ATTY_HUMAN | P17735 | TAT | 100 | Tyrosine aminotransferase | 27.0 | |
| SPYA_HUMAN | P21549 | AGXT | 100 | Serine-pyruvate aminotransferase | 22.0 | |
| HUTH_HUMAN | P42357 | HAL | 100 | Histidine ammonia-lyase | 16.7 | |
| CGL_HUMAN | P32929 | CTH | 100 | Cystathione gamma-lyase (Gamma-Cystathionase) | 9.9 | |
| GLSL_HUMAN | Q9UI32 | GA | 100 | Glutaminase, liver isoform, mitochondrial precursor | 9.5 | |
| GAMT_HUMAN | Q14353 | GAMT | 100 | Guanidinoacaetate N-methyltransferase | 8.4 | |
| SDHL_HUMAN | P20132 | SDS | 100 | L-serine dehydratase | 7.1 | |
| Carbohydrate/monosaccharide metabolism/catabolism | ||||||
| KPY2_HUMAN | P14786 | PKM2 | 100 | Pyruvate kinase, M2 isozyme | 26.7 | |
| ALFA_HUMAN | P04075 | ALDOA | 100 | Fructose biphosphate aldolase A (Lung cancer antigen) | 16.3 | |
| PPCC_HUMAN | Q16822 | PCK1 | 100 | Phosphoenolpyruvate carboxykinase, cytosolic | 16.7 | |
| Q9H277 | Q9H277 | Q9H227 | 100 | Cytosolic beta-glucosidase | 7.6 | |
| Electron transport | ||||||
| ACDB_HUMAN | P45954 | ACADSB | 100 | Acyl-CoA dehydrogenase, branched chain specific | 14.1 | |
| CPA6_HUMAN | X13929 | CYP2A6 | 100 | Cytochrome P450 2A6 | 11.9 | |
| CP12_HUMAN | P05177 | CYP1A2 | 100 | Cytochrome P450 1A2 | 10.0 | |
| Protein metabolism/proteolysis | ||||||
| BAE2_HUMAN | Q9Y5Z0 | BACE2 | 100 | Beta secretase 2 precursor | 10.5 | |
| Q9UJ28 | Q9UJ28 | GALNT7 | 100 | UDP-GalNAc: N-acetylgalactosaminyltransferase 7 | 5.4 | |
| CATC_HUMAN | P53634 | CTSC | 100 | Dipeptidyl-peptidase I precursor | 4.7 | |
| GLMT_HUMAN | Q14749 | GNMT | 100 | Glycine N-methyltransferase | 49.5 | |
| Transport | ||||||
| CHLR_HUMAN | P17516 | AKR1C4 | 100 | Chlordecone reductase | 22.0 | |
| APF_HUMAN | Q13790 | APOF | 100 | Apolipoprotein F precursor | 15.5 | |
| Lipid binding/metabolism | ||||||
| Q9Y2P5 | Q9Y2P5 | Q9Y2P5 | 100 | Very-long-chain AcylCoA synthetase homolog 2 | 23.7 | |
| VLCS_HUMAN | O14975 | FACVL1 | 100 | Very-long-chain AcylCoA synthetase | 16.6 | |
| MMSA_HUMAN | Q02252 | MMSDH | 100 | Methylmalonate-semialdehyde dehydrogenase | 8.1 | |
Separation of well and poorly differentiated intrahepatic CCC by 2-dimensional cluster analysis
For subclassification of intrahepatic CCC in relation to histopathological differentiation (grading) a weighted two-dimensional clustering of the tumor samples was performed. All expressed genes/ESTs in the tumor samples (43.3%, approx. 9650 genes/ESTs) were grouped according to the histological grading ranging from 1 to 3 and then ranked according to their significantly differential expression values. Differentiation of the gene expression profiles of tumor samples, according to the histopathological grading of 1 and 3, identified a total of 136 dysregulated genes (P < 0.05 and two-fold change upregulation or downregulation in ≥ 70), which were used for a weighted two-dimensional cluster analysis (Figure 3). Interestingly, upregulation of specific cell surface antigens such as cytokeratins (for example, cytokeratin 6, 7, 13 and 15) and specific membrane proteins (such as EMP1, EVA1 and proteoglycan 2) was detected in well differentiated tumor samples (G1, G2) when compared with more dedifferentiated tumor samples. In contrast, G3-tumor samples showed a relative overexpression of genes involved in G-protein signaling and genes involved in transcription (Table 4).
Figure 3.
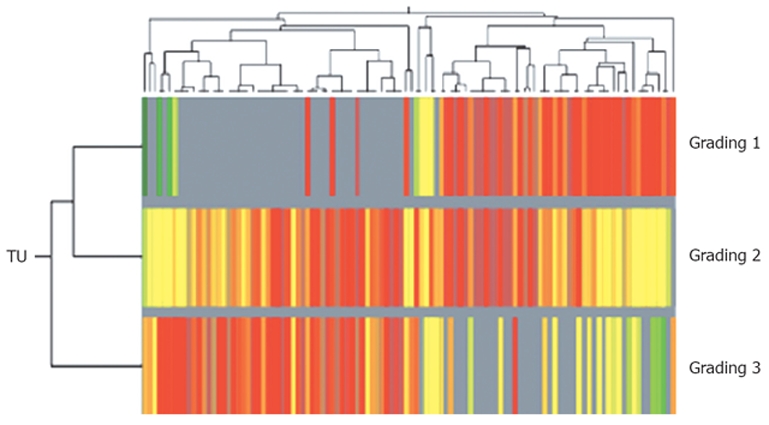
Subclassification of intrahepatic CCC using two-dimensional cluster analysis of 136 different regulated genes/ESTs (dysregulation in more than 70% of all probes, fold change of genetic expression > 2.0; Pearson's correlation) in relation to histopathological findings (well to poor differentiated tumor tissue). Horizontal lines show all dysregulated genes in one singular tumor tissue probe, vertical lines show the expression of one gene in all analyzed G1-, G2- and G3-tumors. Overexpressed genes are coloured red, downregulated genes are shown in green).
Table 4.
Expression levels of dysregulated genes in intrahepatic cholangiocarcinoma in relation to tumor differentiation according histopathological criteria (grading 1 to 3)
| Probe set | Grading 1 | Grading 2 | Grading 3 | Gene function |
| 204857_at | 3.09 | 4.14 (2.9-5.8) | No data | Hs. MAD1 (mitotic arrest deficient, yeast, homolog)-like 1 |
| 211571_s_at | 3.49 | 0.98 (0.7-1.3) | 1.411 | mRNA for proteoglycan PG-M (V3), proteoglycan 2 |
| 209016_s_at | 3.93 | 1.25 (0.3-6.7) | 0.96 (0.92-1.0) | Hs. similar to keratin 7 (KRT7) |
| 201325_s_at | 4.27 | 4.5 (3.0-6.7) | 1.08 | Hs. epithelial membrane protein 1 (EMP1), mRNA |
| 203779_s_at | 5 | No data | No data | Hs. epithelial V-like antigen 1 (EVA1), mRNA |
| 211796 | 7.82 | 1.53 (1.1-1.9) | 1.16 | Hs. T cell receptor beta chain (TCRBV13S1-TCRBJ2S1) mRNA |
| 204734_at | 7.97 | 8.5 (6.3-11.4) | No data | Hs. keratin 15 (KRT15), mRNA |
| 209126_x_at | 10.26 | 8.254 | No data | Hs. keratin 6 isoform K6f (KRT6F) mRNA |
| 207935_s_at | 19.94 | 10.4 | No data | Hs. keratin 13 (KRT13), mRNA |
| 217109_at | 22.75 | No data | No data | Hs. partial mRNA for sv7-MUC4 apomucin |
| 205157_s_at | 34.79 | 10.36 | No data | Hs. keratin 17 (KRT17), mRNA |
| 209070_s_at | No data | 3.188 | 7.54 (6.9-8.2) | Hs. regulator of G-protein signaling 5, mRNA |
| 204338_s_at | No data | 18.98 | 128.5 | Hs. regulator of G-protein signaling 4 (RGS4), mRNA |
| 203638_s_at | No data | 1.14 (0.7-2.3) | 4.16 (2.7-6.4) | Hs. FGF receptor 2, keratinocyte growth factor receptor |
| 208343_s_at | No data | 1.36 (1.1-1.8) | 2.45 (1.2-4.9) | Hs. CYP7A promoter binding factor mRNA, nuclear receptor 5 |
| 207846_at | No data | No data | 1.414 | Hs. POU domain, class 1, transcription factor 1 (Pit1) |
No data: No significant (< 2-fold change) or consistent (< 70% of all cases) change in the expression level of the gene in this subgroup of CCC (G1-, G2- or G3-tumors).
Results of RT-PCR for detection of dysregulated OPN expression
The most consistently overexpressed gene (median fold change 33.5) in all analyzed intrahepatic CCC samples was that encoding the glycoprotein OPN.
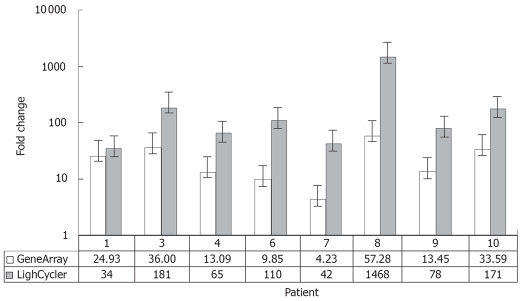
To validate this observation the expression levels of OPN were confirmed by quantitative RT-PCR in 16 tissue samples (malignant and corresponding non-malignant tissue from 8 patients) using the LightCycler© system. In general, the changes in OPN expression levels detected by microarray reflected the results obtained by RT-PCR. However, the dynamic range of real-time PCR results was greater than that of the microarray data (Figure 4).
Figure 4.
Comparison between expression values of osteopontin in 8 intrahepatic CCCs estimated by RT-PCR (LightCycler®System; grey) and gene expression data (HU 133A, Affymetrix; white). The detected changes in osteopontin expression levels measured by RT-PCR reflected very well the changes in gene expression between tumor and non-malignant liver tissue obtained by microarray analysis. The results of RT-PCR revealed larger changes than the microarray data in all cases.
DISCUSSION
The poor response of cholangiocarcinoma to therapy, along with the concerning worldwide increase in morbidity and mortality[4,5], highlights the need for increased efforts to understand the etiology and pathogenesis of this primary liver cancer.
Previous studies have examined the potential role of known genes involved in human carcinogenesis to elucidate the molecular pathways involved in cholangiocellular carcinoma, for example, members of the type I family of growth factor receptors (EGF-R, c-erbB-2)[14–16], (proto-) oncogenes (c-met, c-Ki-ras, Cyclin D)[17–19], dysregulated tumor-suppressor genes (p53, pRB, p16INK4a, p21WAF1)[20,21], and apoptosis-related genes (bcl2, FAS-L, BAX)[22,23]. Other studies have shown microsatellite instability and loss of heterogeneity (LOH) as contributing factors in the development of CCC[24,25].
Excepting the actual data there are only two other reports using gene expression analysis to detect dysregulated genes and prognostic markers in cholangiocarcinogenesis[26,27]. The present study identified a high number of differentially expressed genes and ESTs in CCC (43.6 vs 39.01%, P > 0.05). Based on a relatively conservative arbitrarily chosen algorithm, a gene expression profile of 552 dysregulated genes (221 genes upregulated, 331 genes downregulated) for CCC was generated. This profile was validated based on a neuronal training method by its ability to differentiate the CCC profile from the expression profiles of corresponding and non-corresponding non-malignant liver tissues. This method achieved a fast and reproducible differentiation between CCC and non-malignant liver tissue in all samples, as well as between CCC and other malignant tissues (HCC, liver metastases; data not shown). One tumor sample (No. 7) also showed characteristics of non-malignant liver tissue in that the tissue expressed a number of genes that were more frequently expressed in non-diseased human liver than in CCC samples. The sample was attributed to the non-malignant tissue group in two-dimensional hierarchical clustering. A potential reason for this could be a putative contamination of tumor sample with normal liver tissue, especially owing to the fact that no microdissection of tumor tissue was used. However, using a supervised neuronal training method to reanalyze all of the obtained array data, correct assignment to malignant and non-malignant tissue groups was possible for all samples (including No. 7).
Previous studies have shown a correlation between differentially expressed genes and tumor progression or tumor dedifferentiation in various human tumors[28,29]. In the present study we used two-dimensional cluster analysis with all expressed genes in the tumor samples (> 9000 genes/ESTs) for the detection of statistically significantly differentially expressed genes in relation to tumor differentiation (grading). This method detected 136 differentially expressed genes when tumor samples with low (G1) and high (G3) grading were compared. Because of the small number of analyzed tumor samples (one highly differentiated tumor (G1) among 10 samples) it was not possible to create an unambiguous clear correlation between all dysregulated genes and the histopathologic grading. However, an obvious trend towards a higher expression of specific cell surface proteins (EMP1, EVA1, proteoglycan2) and intermediate filaments (cytokeratin 6, 7, 13, 15, 17) in well-differentiated tumors was observed, whereas samples of high-grade intrahepatic CCC had an elevated expression of genes involved in G-protein signaling and nuclear transcription. Pathological alterations to the chemical composition, molecular structure, or spatial arrangement of the liver matrix will ultimately lead to specific changes in the intermediate filament pattern in human cholangiocytes. It can be assumed that the dedifferentiation of tumor cells in CCC results in pathological alterations of cell surface antigens, cell-to-cell and cell-to-matrix attachment, followed by a switch from physiological to pathological cell-activation. More investigations are needed to elucidate the role of dysregulated genes in tumor progression with histomorphological tumor dedifferentiation in cholangiocarcinogenesis.
To further validate the microarray data, real-time PCR was performed. The gene encoding OPN was identified as the highest and most consistently overexpressed gene (33.5-fold change) in all analyzed tumor samples. The expression profile determined by real-time PCR correlated well with the resulting ratios carried out with the gene expression data of the analyzed CCC. However, as has been shown previously for other similar comparisons[30,31], the dynamic range of real-time PCR was about 1.5-fold to 10-fold higher than that of array analysis.
Using oligonucleotide arrays we were able to identify genes already known to be dysregulated in other human cancers[32,33]. However, a significant number of the genes identified to show a significant alteration in their expression values, have not been known to be involved in cholangiocarcinogenesis until now. By incorporating the expression data into GenMAPP software, a specific pattern of signaling and metabolic pathways as well as changes in cell cycle regulators specific for CCC could be generated. For example, changes in apoptosis-regulating genes consistent with the decreased cell death rate typical of tumor cells were identified. Generally, the changes in cell cycle metabolism are well in line with a pattern of increased cell growth. However, the major focus of this study was to establish a specific genetic profile of CCC for differentiation against non-malignant liver tissue and other malignant liver tumors (HCC, metastases) for clinical routine. This approach intentionally did not take into consideration the different origins of the analyzed tissue probes (tumorous epithelium cells vs non-malignant transformed hepatocytes). Thus, it remains a possibility that our observations of the downregulation of genes involved in specific metabolic pathways, such as amino acid metabolism, may be a consequence of the different origin of malignant and non-malignant tissue rather than a specific feature of cholangiocarcinogenesis. Nevertheless, our own unpublished data revealed similarities between the genetic profiles of other malignant liver tumors (HCC, metastases of adenocarcinomas) and CCC with a downregulation of hepatocyte-specific metabolic functions and an upregulation of genes well known to be involved in human carcinogenesis.
The gene encoding OPN, a secreted adhesive glycoprotein was identified as the most consistently overexpressed gene in all CCC samples (see above). OPN has been shown to be overexpressed in excessive amounts in a variety of human carcinomas and its expression level has been linked to the recurrence of metastasis or to clinical progression[34–36].
OPN is not a typical oncogene; it is mutationally activated in cancer cells, but it is regulated by a variety of stimuli, such as the Wnt/Tcf signaling pathway, steroid receptors, growth factors, TNF-alpha and transcription factors, such as AP-1 and Ras proto-oncogene[37]. OPN is a ligand of CD44, binds to alphaV-containing integrins and has an important role in malignant cell attachment and tumor invasion[38,39]. Potential mechanisms of OPN in human carcinogenesis include the regulation of different signaling cascades through activation of various kinases[40]. Thus, OPN seems to be a potent anti-apoptotic factor via inhibition of caspase 3 activation[41]; it also induces increased activation of promatrix metalloproteinase-2 (pro-MMP2, mediated by nuclear factor kappaB) and secretion of urokinase-type plasminogen activator[42].
Using gene expression profiling, OPN has been shown to be one of the highest overexpressed genes in hepatocellular carcinoma (HCC) and it has been shown the expression of OPN correlates with earlier recurrence, poorer prognosis and metastasis in HCC[43,44]. To the best of our knowledge, there is only one report on OPN in human cholangiocarcinogenesis[45]. Interestingly, Terashi et al observed a correlation between low OPN levels and tumor aggressiveness, whereas another group found high levels of the glycoprotein in CCC in a rat model system[46]. Therefore, it remains to be shown whether OPN is a potential prognostic marker and target for anticancer treatment in CCC.
In conclusion, the present study represents one of the first attempts to establish a gene expression pattern of intrahepatic CCC by genetic comprehensive analysis. Based on the analysis of a subgroup of 364 genes, a specific pattern of changes in metabolic and signaling pathways, as well as receptors, cell cycle changes and alterations in apoptosis could be identified. The observed changes are consistent with some of the known characteristics of CCCs in particular and tumors in general. The gene expression profile appears to be useful as a diagnostic tool, especially in terms of differentiation from other liver masses as well as for the subclassification of intrahepatic CCC in comparison to histopathological findings.
Conclusively, besides a number of known overexpressed tumor-related genes in intrahepatic CCC, we describe here for the first time other strongly and consistently dysregulated genes in cholangiocarcinogenesis, which are well known to be involved in other human cancers. Although our data at present do not support a correlation between the expression of these genes and tumor stage for CCC, this gene expression pattern is in line with the usually observed bad clinical prognosis of this cancer. Another evolving application based on the profile described in this report and the observation of OPN as the most consistently overexpressed gene might be the identification of novel therapeutic targets and related genes predicting survival and outcomes of therapeutic modalities.
COMMENTS
Background
Cholangiocarcinoma (CCC) is the second most common primary hepatic malignancy with increasing incidence in western countries. Despite recent progress in various imaging modalities, intrahepatic CCC is in most cases advanced by the time of diagnosis. Aggravating circumstances leading to a poor prognosis with median survival times of only three to six months are the limited curative treatment options by surgical resection and the low effect of palliative chemotherapy in CCC. Therefore a better understanding of human cholangiocarcinogenesis is strongly needed to establish new diagnostic markers and therapeutic approaches.
Research frontiers
Despite the advances in genomic profiling of many human malignancies in recent years, there have only been a few reports of gene expression analysis in CCC. In this study, oligonucleotide arrays were used to detect differentially expressed genes and to establish a specific genetic profile of intrahepatic CCC. To subclassify between well and poorly differentiated tumors, additional analyses were performed.
Innovations and breakthroughs
A total of 41 genes that were strongly deregulated (upregulated or downregulated in 100%, fc > 4) in CCC were detected by gene expression analysis. For the first time, a subclassification of intrahepatic CCC in relation to malignant differentiation using oligonucleotide arrays was performed. The gene encoding osteopontin (OPN) was the most overexpressed gene with a high fold-change of 33.5, which was confirmed by RT-PCR.
Applications
Besides a number of known overexpressed tumor-related genes in intrahepatic CCC, other strong and consistently dysregulated genes were described for the first time in human cholangiocarcinogenesis. Another evolving application based on the profile described in this report and the observation of OPN as the most consistently overexpressed gene might be the identification of novel therapeutic targets and related genes predicting survival and outcomes of therapeutic modalities.
Peer review
The manuscript by Hass et al describes a microarray analysis of genetic changes in human cholangiocarcinoma samples compared to non-tumor tissue. The analysis revealed very intriguing data with respect to the overexpression of a number of genes, the most consistent of which is OPN, and further analysis with respect to tumor grade was performed.
Supported by The fortüne-program of the University of Tuebingen, No. F1281305
Peer reviewer: Sharon DeMorrow, Dr, Division of Research and Education, Scott and White Hospital and the Texas A & M University System, Health Science Center, College of Medicine, Temple, Texas 76504, United States
S- Editor Zhong XY L- Editor McGowan D E- Editor Lu W
References
- 1.Suh KS, Roh HR, Koh YT, Lee KU, Park YH, Kim SW. Clinicopathologic features of the intraductal growth type of peripheral cholangiocarcinoma. Hepatology. 2000;31:12–17. doi: 10.1002/hep.510310104. [DOI] [PubMed] [Google Scholar]
- 2.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita H, Tanimura H, Uchiyama K, Tani M, Onishi H, Yamaue H. Prognostic factors of intrahepatic cholangiocarcinoma after surgical treatment. Oncol Rep. 2002;9:97–101. [PubMed] [Google Scholar]
- 4.Taylor-Robinson SD, Toledano MB, Arora S, Keegan TJ, Hargreaves S, Beck A, Khan SA, Elliott P, Thomas HC. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968-1998. Gut. 2001;48:816–820. doi: 10.1136/gut.48.6.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davila JA, El-Serag HB. Cholangiocarcinoma: the "other" liver cancer on the rise. Am J Gastroenterol. 2002;97:3199–3200. doi: 10.1111/j.1572-0241.2002.07132.x. [DOI] [PubMed] [Google Scholar]
- 6.Bergquist A, Ekbom A, Olsson R, Kornfeldt D, Loof L, Danielsson A, Hultcrantz R, Lindgren S, Prytz H, Sandberg-Gertzen H, et al. Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. J Hepatol. 2002;36:321–327. doi: 10.1016/s0168-8278(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 7.Kato I, Kido C. Increased risk of death in thorotrast-exposed patients during the late follow-up period. Jpn J Cancer Res. 1987;78:1187–1192. [PubMed] [Google Scholar]
- 8.Gores GJ. Cholangiocarcinoma: current concepts and insights. Hepatology. 2003;37:961–969. doi: 10.1053/jhep.2003.50200. [DOI] [PubMed] [Google Scholar]
- 9.Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin Liver Dis. 2004;24:139–154. doi: 10.1055/s-2004-828891. [DOI] [PubMed] [Google Scholar]
- 10.Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Hashimoto K, Tangoku A, Hamada K, et al. Differential gene expression in distinct virologic types of hepatocellular carcinoma: association with liver cirrhosis. Oncogene. 2003;22:3007–3014. doi: 10.1038/sj.onc.1206401. [DOI] [PubMed] [Google Scholar]
- 11.Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC, et al. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin Cancer Res. 2003;9:4695–4704. [PubMed] [Google Scholar]
- 12.Takahashi M, Yang XJ, Sugimura J, Backdahl J, Tretiakova M, Qian CN, Gray SG, Knapp R, Anema J, Kahnoski R, et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene. 2003;22:6810–6818. doi: 10.1038/sj.onc.1206869. [DOI] [PubMed] [Google Scholar]
- 13.Xiao SY, Wang HL, Hart J, Fleming D, Beard MR. cDNA arrays and immunohistochemistry identification of CD10/CALLA expression in hepatocellular carcinoma. Am J Pathol. 2001;159:1415–1421. doi: 10.1016/S0002-9440(10)62528-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito Y, Takeda T, Sasaki Y, Sakon M, Yamada T, Ishiguro S, Imaoka S, Tsujimoto M, Higashiyama S, Monden M, et al. Expression and clinical significance of the erbB family in intrahepatic cholangiocellular carcinoma. Pathol Res Pract. 2001;197:95–100. doi: 10.1078/0344-0338-00016. [DOI] [PubMed] [Google Scholar]
- 15.Endo K, Yoon BI, Pairojkul C, Demetris AJ, Sirica AE. ERBB-2 overexpression and cyclooxygenase-2 up-regulation in human cholangiocarcinoma and risk conditions. Hepatology. 2002;36:439–450. doi: 10.1053/jhep.2002.34435. [DOI] [PubMed] [Google Scholar]
- 16.Sirica AE, Lai GH, Endo K, Zhang Z, Yoon BI. Cyclooxygenase-2 and ERBB-2 in cholangiocarcinoma: potential therapeutic targets. Semin Liver Dis. 2002;22:303–313. doi: 10.1055/s-2002-34507. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi K, Nakajima Y, Kanehiro H, Tsutsumi M, Taki J, Aomatsu Y, Yoshimura A, Ko S, Kin T, Yagura K. Ki-ras mutations and p53 protein expressions in intrahepatic cholangiocarcinomas: relation to gross tumor morphology. Gastroenterology. 1995;109:1612–1617. doi: 10.1016/0016-5085(95)90650-9. [DOI] [PubMed] [Google Scholar]
- 18.Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito Y, Takeda T, Sasaki Y, Sakon M, Yamada T, Ishiguro S, Imaoka S, Tsujimoto M, Matsuura N. Expression and clinical significance of the G1-S modulators in intrahepatic cholangiocellular carcinoma. Oncology. 2001;60:242–251. doi: 10.1159/000055325. [DOI] [PubMed] [Google Scholar]
- 20.Taniai M, Higuchi H, Burgart LJ, Gores GJ. p16INK4a promoter mutations are frequent in primary sclerosing cholangitis (PSC) and PSC-associated cholangiocarcinoma. Gastroenterology. 2002;123:1090–1098. doi: 10.1053/gast.2002.36021. [DOI] [PubMed] [Google Scholar]
- 21.Tannapfel A, Weinans L, Geissler F, Schutz A, Katalinic A, Kackerling F, Hauss J, Wittekind C. Mutations of p53 tumor suppressor gene, apoptosis, and proliferation in intrahepatic cholangiocellular carcinoma of the liver. Dig Dis Sci. 2000;45:317–324. doi: 10.1023/a:1005412626515. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Takeda T, Sasaki Y, Sakon M, Yamada T, Ishiguro S, Imaoka S, Tsujimoto M, Matsuura N. Expression of Fas and Fas ligand reflects the biological characteristics but not the status of apoptosis of intrahepatic cholangiocellular carcinoma. Int J Mol Med. 2000;6:581–586. doi: 10.3892/ijmm.6.5.581. [DOI] [PubMed] [Google Scholar]
- 23.Ito Y, Takeda T, Sasaki Y, Sakon M, Monden M, Yamada T, Ishiguro S, Imaoka S, Tsujimoto M, Matsuura N. Bcl-2 expression in cholangiocellular carcinoma is inversely correlated with biologically aggressive phenotypes. Oncology. 2000;59:63–67. doi: 10.1159/000012139. [DOI] [PubMed] [Google Scholar]
- 24.Pineau P, Marchio A, Nagamori S, Seki S, Tiollais P, Dejean A. Homozygous deletion scanning in hepatobiliary tumor cell lines reveals alternative pathways for liver carcinogenesis. Hepatology. 2003;37:852–861. doi: 10.1053/jhep.2003.50138. [DOI] [PubMed] [Google Scholar]
- 25.Liengswangwong U, Nitta T, Kashiwagi H, Kikukawa H, Kawamoto T, Todoroki T, Uchida K, Khuhaprema T, Karalak A, Srivatanakul P, et al. Infrequent microsatellite instability in liver fluke infection-associated intrahepatic cholangiocarcinomas from Thailand. Int J Cancer. 2003;107:375–380. doi: 10.1002/ijc.11380. [DOI] [PubMed] [Google Scholar]
- 26.Obama K, Ura K, Li M, Katagiri T, Tsunoda T, Nomura A, Satoh S, Nakamura Y, Furukawa Y. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology. 2005;41:1339–1348. doi: 10.1002/hep.20718. [DOI] [PubMed] [Google Scholar]
- 27.Hansel DE, Rahman A, Hidalgo M, Thuluvath PJ, Lillemoe KD, Shulick R, Ku JL, Park JG, Miyazaki K, Ashfaq R, et al. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am J Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren B, Yu YP, Jing L, Liu L, Michalopoulos GK, Luo JH, Rao UN. Gene expression analysis of human soft tissue leiomyosarcomas. Hum Pathol. 2003;34:549–558. doi: 10.1016/s0046-8177(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 29.Watson MA, Perry A, Budhraja V, Hicks C, Shannon WD, Rich KM. Gene expression profiling with oligonucleotide microarrays distinguishes World Health Organization grade of oligodendrogliomas. Cancer Res. 2001;61:1825–1829. [PubMed] [Google Scholar]
- 30.Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajeevan MS, Ranamukhaarachchi DG, Vernon SD, Unger ER. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods. 2001;25:443–451. doi: 10.1006/meth.2001.1266. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Lin YM, Hasegawa S, Shimokawa T, Murata K, Kameyama M, Ishikawa O, Katagiri T, Tsunoda T, Nakamura Y, et al. Genes associated with liver metastasis of colon cancer, identified by genome-wide cDNA microarray. Int J Oncol. 2004;24:305–312. [PubMed] [Google Scholar]
- 33.Thomas R, True LD, Bassuk JA, Lange PH, Vessella RL. Differential expression of osteonectin/SPARC during human prostate cancer progression. Clin Cancer Res. 2000;6:1140–1149. [PubMed] [Google Scholar]
- 34.Rittling SR, Chambers AF. Role of osteopontin in tumour progression. Br J Cancer. 2004;90:1877–1881. doi: 10.1038/sj.bjc.6601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Lin D, Yuan J, Xiao T, Zhang H, Sun W, Han N, Ma Y, Di X, Gao M, et al. Overexpression of osteopontin is associated with more aggressive phenotypes in human non-small cell lung cancer. Clin Cancer Res. 2005;11:4646–4652. doi: 10.1158/1078-0432.CCR-04-2013. [DOI] [PubMed] [Google Scholar]
- 36.Rudland PS, Platt-Higgins A, El-Tanani M, De Silva Rudland S, Barraclough R, Winstanley JH, Howitt R, West CR. Prognostic significance of the metastasis-associated protein osteopontin in human breast cancer. Cancer Res. 2002;62:3417–3427. [PubMed] [Google Scholar]
- 37.Casson AG, Wilson SM, McCart JA, O'Malley FP, Ozcelik H, Tsao MS, Chambers AF. ras mutation and expression of the ras-regulated genes osteopontin and cathepsin L in human esophageal cancer. Int J Cancer. 1997;72:739–745. doi: 10.1002/(sici)1097-0215(19970904)72:5<739::aid-ijc6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 38.Harada N, Mizoi T, Kinouchi M, Hoshi K, Ishii S, Shiiba K, Sasaki I, Matsuno S. Introduction of antisense CD44S CDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91:67–75. doi: 10.1002/1097-0215(20010101)91:1<67::aid-ijc1011>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 39.El-Tanani MK, Campbell FC, Kurisetty V, Jin D, McCann M, Rudland PS. The regulation and role of osteopontin in malignant transformation and cancer. Cytokine Growth Factor Rev. 2006;17:463–474. doi: 10.1016/j.cytogfr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty G, Jain S, Behera R, Ahmed M, Sharma P, Kumar V, Kundu GC. The multifaceted roles of osteopontin in cell signaling, tumor progression and angiogenesis. Curr Mol Med. 2006;6:819–830. doi: 10.2174/156652406779010803. [DOI] [PubMed] [Google Scholar]
- 41.Graessmann M, Berg B, Fuchs B, Klein A, Graessmann A. Chemotherapy resistance of mouse WAP-SVT/t breast cancer cells is mediated by osteopontin, inhibiting apoptosis downstream of caspase-3. Oncogene. 2007;26:2840–2850. doi: 10.1038/sj.onc.1210096. [DOI] [PubMed] [Google Scholar]
- 42.Das R, Philip S, Mahabeleshwar GH, Bulbule A, Kundu GC. Osteopontin: it's role in regulation of cell motility and nuclear factor kappa B-mediated urokinase type plasminogen activator expression. IUBMB Life. 2005;57:441–447. doi: 10.1080/15216540500159424. [DOI] [PubMed] [Google Scholar]
- 43.Pan HW, Ou YH, Peng SY, Liu SH, Lai PL, Lee PH, Sheu JC, Chen CL, Hsu HC. Overexpression of osteopontin is associated with intrahepatic metastasis, early recurrence, and poorer prognosis of surgically resected hepatocellular carcinoma. Cancer. 2003;98:119–127. doi: 10.1002/cncr.11487. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Ye QH, Ren N, Zhao L, Wang YF, Wu X, Sun HC, Wang L, Zhang BH, Liu YK, et al. The prognostic significance of preoperative plasma levels of osteopontin in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132:709–717. doi: 10.1007/s00432-006-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terashi T, Aishima S, Taguchi K, Asayama Y, Sugimachi K, Matsuura S, Shimada M, Maehara S, Maehara Y, Tsuneyoshi M. Decreased expression of osteopontin is related to tumor aggressiveness and clinical outcome of intrahepatic cholangiocarcinoma. Liver Int. 2004;24:38–45. doi: 10.1111/j.1478-3231.2004.00886.x. [DOI] [PubMed] [Google Scholar]
- 46.Takemura F, Inaba N, Miyoshi E, Furuya T, Terasaki H, Ando S, Kinoshita N, Ogawa Y, Taniguchi N, Ito S. Optimization of liver biopsy RNA sampling and use of reference RNA for cDNA microarray analysis. Anal Biochem. 2005;337:224–234. doi: 10.1016/j.ab.2004.10.041. [DOI] [PubMed] [Google Scholar]