Abstract
The Hia autotransporter proteins are highly immunogenic surface adhesins expressed by nontypeable Haemophilus influenzae (NTHI). The objective of our study was to assess the opsonophagocytic activity of anti-Hia antibodies against homologous and heterologous NTHI. A segment of the hia gene that encodes a surface-exposed portion of the H. influenzae strain 11 Hia protein was cloned into a pGEMEX-2 expression vector. Escherichia coli JM101 was transformed with the resulting pGEMEX-Hia BstEII del recombinant plasmid, and recombinant fusion protein was recovered. An immune serum against recombinant GEMEX-Hia (rGEMEX-Hia)-mediated killing of the homologous NTHI strain 11 at a 1:160 titer and five heterologous Hia-expressing strains at titers of ≥1:40. Immune serum did not mediate killing of two Hia-knockout strains whose hia genes were inactivated but did mediate killing of one knockout strain at a high titer after the strain was transformed with a plasmid containing the hia gene. Immune serum did not mediate killing of HMW1/HMW2-expressing NTHI strains, which do not express the Hia adhesin. However, when two representative HMW1/HMW2-expressing strains were transformed with the plasmid containing the hia gene, they expressed abundant Hia and were susceptible to killing by the immune serum. Immune serum did not mediate killing of HMW1/HMW2-expressing strains transformed with the plasmid without the hia gene. Our results demonstrate that the Hia proteins of NTHI are targets of opsonophagocytic antibodies and that shared epitopes recognized by such antibodies are present on the Hia proteins of unrelated NTHI strains. These data argue for the continued investigation of the Hia proteins as vaccine candidates for the prevention of NTHI disease.
Otitis media remains a significant health problem for children in this country and elsewhere in the world (10, 11). Most children in the United States have had at least one episode of otitis by their third birthdays, and one-third have had three or more episodes (34). In addition to the short-term morbidity and costs of this illness, the potential for delay or disruption of normal speech and language development in children with persistent middle ear effusions is a subject of considerable concern (33, 41). Experts in the field have strongly recommended that efforts be made to develop safe and effective vaccines for the prevention of otitis media in young children (20). Although the total prevention of disease will be a difficult goal to achieve, the prevention of even a portion of cases would be beneficial, given the magnitude and costs of the problem.
Bacteria, usually in pure culture, can be isolated from middle ear exudates in approximately two-thirds of the cases of acute otitis media (16, 35). Streptococcus pneumoniae is the most common bacterial pathogen recovered in all age groups, with isolation rates commonly ranging from 35% to 40% (16, 35). Nontypeable Haemophilus influenzae (NTHI) is the second-most-common bacterium recovered and accounts for 20% to 30% of the cases of acute otitis media and a larger percentage of the cases of chronic and recurrent disease (26). Interestingly, since the introduction of the pneumococcal conjugate vaccine as part of the regular childhood vaccine schedule, NTHI has become an even more common cause of acute and recurrent middle ear disease, often surpassing S. pneumoniae in the frequency of recovery from middle ear specimens (12, 26). Many different antigens have been suggested as possible NTHI vaccine candidates (1, 3, 18, 29, 30, 42). Outer membrane proteins appear to be the principal targets of bactericidal and protective antibodies (22), and as a group, they have been the major focus of vaccine development efforts. Table 1 summarizes the relevant characteristics of some of the leading vaccine candidates currently under active investigation.
TABLE 1.
Potential vaccine antigens of NTHI
| Antigen | Molecular mass (kDa) | Osonophagocytic or bactericidal antibody | Protects animals | Adherence factor | Reference(s) or source |
|---|---|---|---|---|---|
| LOS | 3-5 | Yes | Yes | Yes | 23, 46 |
| PilA | 16 | Not known | Yes | Yes | 2 |
| OMP P6 | 16 | Yes | Yes | No | 17, 30 |
| OMP 26 | 26 | Not known | Yes | No | 18 |
| OMP P5 fimbrin | 36 | Not known | Yes | Yes | 4 |
| OMP P2 | 36-42 | Yes | Yes | Yes | 29 |
| Protein D | 42 | Not known | Yes | No | 36 |
| HMW1/HMW2 | 100-150 | Yes | Yes | Yes | 5, 44 |
| Hia | 100->250 | Yes | Not known | Yes | This study |
In our early work, we demonstrated that the development of bactericidal antibodies in the sera of children recovered from acute NTHI otitis media was associated with the appearance of serum antibodies directed against highly immunogenic high-molecular-weight proteins (6). This work subsequently led to the identification and characterization of the HMW1/HMW2 family of proteins (7). The HMW1/HMW2 proteins have subsequently been shown to be major adhesins of NTHI (37), as well as targets of opsonophagocytic (43, 44) and protective antibodies (5). The HMW1/HMW2-like proteins are expressed by approximately 75% of NTHI strains (7, 38). The 25% of NTHI strains that do not express HMW1/HMW2-like proteins express immunogenic high-molecular-weight proteins that are recognized by human convalescent-phase serum antibodies (6). Almost all such HMW1/HMW2-negative strains have subsequently been shown to express a second distinct class of adhesin known as Hia (9). Nearly all NTHI strains that lack HMW1/HMW2 proteins contain an hia gene and express an Hia protein, and conversely, strains that express HMW1/HMW2 proteins lack an hia gene (9, 38).
The Hia proteins are members of a large family of bacterial proteins known as autotransporters that are found in many gram-negative bacteria (24, 48). Autotransporters are typically expressed as precursor proteins with three functional domains, an N-terminal signal peptide, an internal “passenger domain,” and a C-terminal translocator or beta domain (24, 27). The signal peptide directs the protein across the inner bacterial membrane, and the translocator or beta domain forms a β-barrel structure in the outer membrane through which the passenger domain is extruded to the bacterial surface (24, 27). On the bacterial surface, the passenger domain is usually cleaved, but in the case of the Hia protein, the protein remains uncleaved and cell associated, and it functions on the cell surface as an important adhesin for Hia-expressing NTHI strains (25, 48). At present, members of the autotransporter family expressed by other gram-negative bacteria are under active investigation as possible vaccine candidates (13, 28, 40).
No information is currently available concerning the functional activity of antibodies directed against the Hia proteins of NTHI. The objective of the present study was to assess the ability of antibodies directed against the Hia proteins to mediate opsonophagocytic activity. In the work described here, we demonstrated that the Hia proteins are indeed targets of opsonophagocytic antibodies, and furthermore, we demonstrated that epitopes recognized by such antibodies are also present on the Hia proteins of heterologous NTHI strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 2 . Escherichia coli strains DH5α and JM101 have been described previously (45, 47). The NTHI strains examined have been described in previous work from our laboratory (6, 9, 31, 43, 44). NTHI strains 3A, 5, 11, 15, 1512A, 3179B, 3248A, and 3640A were each isolated in pure culture from middle-ear fluid specimens from children with acute otitis media. Each strain was identified as H. influenzae by standard methods and was classified as nontypeable by its failure to agglutinate with a panel of typing antisera for H. influenzae types a to f (Burroughs Wellcome Co., Research Triangle Park, NC) and to show lines of precipitation with these antisera in counterimmunoelectrophoresis assays.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Reference(s) or source |
|---|---|---|
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 phoA | Invitrogen Life Technologies |
| JM101 | supE thi Δ(lac-proAB) F′ (traD36 proAB lacIqZΔM15) | New England Biolabs |
| H. influenzae | ||
| 3A | Clinical isolate; nontypeable; Hia-expressing strain | 6, 31 |
| 5 | Clinical isolate; nontypeable; HMW1/HMW2-expressing strain | 6, 37 |
| 11 | Clinical isolate; nontypeable; Hia-expressing strain | 6, 9 |
| 11 hia::kan | Strain 11 derivative hia::kan | 9 |
| 15 | Clinical isolate; nontypeable; HMW1/HMW2-expressing strain | 6, 37 |
| 1512A | Clinical isolate; nontypeable; Hia-expressing strain | 6, 9 |
| 1512A hia::kan | Strain 1512A derivative hia::kan | 9 |
| 3179B | Clinical isolate; nontypeable; Hia-expressing strain | 31 |
| 3248A | Clinical isolate; nontypeable; Hia-expressing strain | 31 |
| 3640A | Clinical isolate; nontypeable; Hia-expressing strain | 31 |
| Plasmids | ||
| pHMW8-4 | pT7-7 derivative containing the full-length hia gene from NTHI strain 11 | 9 |
| pHMW8-6 | pT7-7 derivative containing the hia gene from NTHI strain 11 interrupted by a kanamycin resistance cassette | 9 |
| pGEMEX-2 | T7 expression plasmid for construction of in-frame T7 gene 10 fusions; Ampr | Promega |
| pGEMEX-Hia | pGEMEX-2 derivative with an in-frame fusion of T7 gene 10 and the portion of the hia gene encoding amino acids 200 to 1098 of the strain 11 Hia protein | This study |
| pGEMEX-Hia BstEII del | pGEMEX-2 derivative with an in-frame fusion of T7 gene 10 and the portion of the hia gene encoding amino acids 200 to 788 of the strain 11 Hia protein | This study |
| pSPEC1 | E. coli/H. influenzae shuttle vector; Sper | 2 |
| pHia | pSPEC1 derivative containing the full-length hia gene from NTHI strain 11 | This study |
Spe, spectinomycin.
E. coli strains were grown on Luria-Bertani (LB) agar or in LB broth and were stored at −70°C in LB broth with 10% glycerol. Strain DH5α was used for preparation of the plasmid constructs described below, and strain JM101 was used for generation of the recombinant protein. All H. influenzae organisms were stored at −70°C in skim milk within two or three subpassages of the initial clinical isolation. H. influenzae cells were grown on chocolate agar or in brain heart infusion broth supplemented with hemin and NAD, as previously described (6). Antibiotics used to select for plasmids included ampicillin (100 μg/ml) and spectinomycin (200 μg/ml).
Construction of plasmids.
Plasmid pGEMEX-Hia contains a 5′-truncated hia gene fused in-frame to the T7 gene 10 present in pGEMEX-2 (Promega). pGEMEX-Hia was constructed by excising a 4.0-kbp EcoRI fragment from pHMW8-6 (9) and ligating it into EcoRI-digested pGEMEX-2. This fragment encodes amino acids 200 to 1098 of the strain 11 Hia protein and includes the 3′ terminus of the hia gene as well as additional downstream DNA. Plasmid pGEMEX-Hia BstEII del contains the truncated hia gene just described but with an additional 3′ deletion. This latter plasmid was constructed by digesting pGEMEX-Hia with BstEII, recovering the 7.0-kbp larger fragment, and religating the free ends after the ends were blunted. Plasmid GEMEX-Hia BstEII del encodes amino acids 200 to 788 of the strain 11 Hia protein. The fidelity of the plasmid constructs was determined by selecting constructs with the expected restriction endonuclease gel profile. Plasmid pSPEC1 is an E. coli/H. influenzae shuttle vector that was a kind gift from Lauren Bakaletz and Kevin Mason of the Research Institute at Nationwide Children's Hospital (2). Plasmid pSPEC1/Hia contains a full-length hia gene from strain 11 and was constructed by excising an 8.0-kbp BamHI-EcoRI fragment from pHMW8-4 (9) and ligating it into BamHI-EcoRI-digested pSPEC1.
Generation and purification of the GEMEX-Hia recombinant fusion protein.
E. coli strain JM101 was transformed with pGEMEX-Hia BstEII del using standard methods (7). To prepare recombinant GEMEX-Hia protein, JM101/pGEMEX-Hia BstEII del was recovered from a frozen stock culture and grown overnight at 37°C on LB agar with ampicillin. Several isolated colonies from the overnight growth were used to inoculate a flask of L broth containing 50 μg of ampicillin per ml, and the cells were allowed to grow to an A600 of 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a concentration of 1 mM, and mGP1-2, an M13 phage containing the T7 RNA polymerase gene, was added at a multiplicity of infection of 10.Two to three hours later, cells were harvested. The recombinant protein exists as inclusion bodies when generated using this method. The cell pellet recovered from the induced cell culture was lysed in a Tris-HCl-, NaCl-, and EDTA-containing solution containing lysozyme and 1% Triton X-100, and DNase and RNase were added to digest DNA and RNA, respectively. The remaining pellet was washed repeatedly with a Triton X-100 solution prior to being solubilized in 3 M guanidine. Refolding and resolubilization of recombinant protein were achieved by slowly dialyzing the solution at 4°C in decreasing concentrations of guanidine in phosphate-buffered saline. Insoluble debris was removed from the solution by centrifugation, and protein purity was assessed by examination of fractions on polyacrylamide gels.
Chinchilla immunization protocol for generation of anti-Hia antisera.
Four chinchillas were immunized with the purified recombinant GEMEX-Hia protein prepared as described above. Each animal received five subcutaneous injections of 100 μg of the recombinant protein administered every 4 to 6 weeks. The first dose of the protein was mixed with Freund's complete adjuvant, and subsequent doses were mixed with incomplete Freund's. Serum antibody responses were monitored by enzyme-linked immunosorbent assay and Western immunoblotting (5), and immune serum from the animal with the highest level of serum antibody was used for the opsonophagocytic experiments described below. All animal experiments complied with federal and institutional guidelines and were approved by the local Institutional Animal Care and Use Committee.
Generation of hybridoma cell line producing MAb 1F4.
The hybridoma line producing monoclonal antibody (MAb) 1F4, a MAb that recognizes the Hia protein of prototype NTHI strain 11, was generated using techniques identical to those described previously for the generation of anti-HMW1/HMW2 MAbs (8). In the case of MAb 1F4, recombinant soluble Hia protein, prepared as described above, was used for both mouse immunization and screening of hybridoma supernatants. Determination of antibody isotypes was performed as described previously (8), and MAb IF4 was determined to be of the mouse immunoglobulin G1 (IgG1) subclass. Purified MAb 1F4 used in the Western immunoblot assay was prepared by passing 100 ml of hybridoma supernatant over a protein G (Sigma Chemical Co., St. Louis, MO) affinity column, washing the column with buffer, and then eluting the bound MAb with 0.1 M glycine (pH 2.5). The pH of the eluted antibody solution was quickly adjusted to a neutral pH with 1 M Tris [pH 8.0], and the sample was then dialyzed against phosphate-buffered saline prior to use in the Western immunoblot assay.
Western immunoblot assay for detection of Hia protein expression.
Bacteria were recovered from skim milk stocks by transfer of a loopful of thawed organisms to a chocolate agar plate and incubation for 16 h at 37°C in a 5% CO2 atmosphere. The following morning, a bacterial suspension was prepared from the overnight plate growth and adjusted to an optical density of approximately 1.0. Fifteen microliters of each bacterial suspension was solubilized in electrophoresis sample buffer, subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 7.5% acrylamide gels, and transferred to nitrocellulose with a Genie electrophoretic blotter (Idea Scientific Company, Corvallis, OR) for 45 min at 24 V. After the samples were transferred, the nitrocellulose sheet was blocked with a 3% gelatin solution in Tris-buffered saline and then probed with anti-Hia MAb 1F4, followed by alkaline phosphatase-conjugated goat anti-mouse IgG secondary antibody. Bound antibodies were detected by incubation with a nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate solution.
Opsonophagocytic assay with NTHI strains.
The growth conditions of the bacteria, the growth and differentiation of HL-60 cells, and the opsonophagocytic assay itself were performed as described previously (43, 44), with minor modifications. The opsonophagocytic assay used in this study was performed with 5-ml capped polystyrene tubes (Sarstedt, Newton, NC) rather than with the 96-well plates described in our earlier studies. The tubes provided more-reliable and -consistent mixing of the contents in the reaction mixture than did the plates. The complement source used in this study was human serum collected from a single healthy adult rather than the guinea pig complement used in the earlier work. Complement controls were included in all assays as described previously (43, 44). Prior to use in the assay, the human serum was adsorbed to remove serum IgG by passing aliquots repeatedly over a protein-G affinity column at 4°C (32). This treatment removed >95% of the serum IgG, as determined by laser nephelometry, yet maintained functional complement activity as assessed in a standard total hemolytic complement assay. The percentage of cells killed at each serum dilution was calculated by determining the ratio of the bacterial colony count at each dilution to that of the complement control. The means and standard deviations were calculated using data from samples run in duplicate in each assay. Statistical analyses were performed using a NCSS 2000 software package (NCSS, Kaysville, UT). Opsonophagocytic titers were defined as the reciprocal of the serum or antibody dilution that resulted in the killing of ≥50% of the cells of the bacterial inoculum compared to the amount of growth in the complement control wells.
RESULTS
Construction of the Hia recombinant plasmid and preparation of recombinant GEMEX-Hia (rGEMEX-Hia).
To generate the Hia recombinant protein used in this work, we constructed the pGEMEX-Hia BstEII del recombinant plasmid described in the Materials and Methods section. Previous work has defined several important functional domains within the strain 11 Hia protein, including the signal peptide, the passenger and the beta or translocator domains, and the two binding domains (Fig. 1) (14, 25, 48). pGEMEX-Hia BstEII del encodes a large fragment of the Hia passenger domain of the mature Hia protein that is fused in-frame with the T7 gene 10 derived from the pGEMEX-2 expression plasmid. The hia gene fragment cloned into pGEMEX-Hia BstEII del encodes amino acids 200 to 788 of the Hia protein and includes the domains of the mature protein that comprise the primary binding domain (residues 549 to 705) and a portion of the secondary binding domain (residues 50 to 374) (Fig. 1) (48).
FIG. 1.
Schematic diagram of functional domains within the strain 11 Hia protein and location of the portion of the Hia gene encoded by the hia gene fragment inserted into pGEMEX-2. The signal peptide (SP) contains residues 0 to 49, the passenger domain, residues 50 to 1022, the beta domain, residues 1023 to 1098, the GEMEX insert, residues 200 to 788, the HiaBD1 binding domain, residues 549 to 705, and the HiaBD2 binding domain, residues 50 to 374.
This particular fragment was selected for cloning and recombinant protein expression because it encoded important regions of the mature Hia protein, as just described, and because it possessed useful restriction sites at the 5′ and 3′ termini. Furthermore, we were able to generate abundant amounts of soluble recombinant protein from E. coli transformed with pGEMEX-Hia BstEII del. Efforts to generate and purify recombinant protein using the pGEMEX-Hia construct that contained the larger insert (residues 200 to1098) were not successful because recombinant protein was produced in much smaller amounts and was for the most part insoluble.
Shown in Fig. 2 is an SDS-polyacrylamide electrophoretic gel demonstrating the purified soluble rGEMEX-Hia protein expressed by the pGEMEX-Hia BstEII del construct. The major protein species has a molecular mass of approximately 118 kDa, a size slightly larger than the predicted molecular mass for the GEMEX-Hia fusion protein based upon the amino acid sequence. The reason for the larger-than-predicted size is not completely clear, but it should be noted that native full-length Hia protein also demonstrates anomalous migration in SDS-polyacrylamide electrophoretic gels. Also, as can be seen in Fig. 2, minor lower-molecular-weight degradation products are present in the preparation, but they comprise a very small percentage of the overall protein present in the preparation.
FIG. 2.
An SDS-polyacrylamide electrophoretic gel with 7.5% acrylamide demonstrating the purified rGEMEX-Hia protein used for immunization. Lane 1, molecular weight standards; lane 2, purified GEMEX-hia recombinant fusion protein.
Once the rGEMEX-Hia protein was expressed and purified, we raised high-titer immune sera in chinchillas, using the multidose immunization regimen described in the Materials and Methods section. We chose the chinchilla for antiserum generation because of our previous success in employing serum from this species for the measurement of opsonophagocytic antibodies directed against other NTHI surface antigens and because our ultimate goal will be to test Hia proteins as vaccine candidates, using the chinchilla model of NTHI infection (5, 43).
Opsonophagocytic killing of NTHI strain 11 and its derivatives by rGEMEX-Hia antiserum.
We first examined a set of isogenic strains derived from NTHI parent strain 11 for their susceptibility to opsonophagocytic killing by the rGEMEX-Hia antiserum. Strain 11 hia::kan, whose construction was described in earlier work, is a derivative of NTHI strain 11 in which the hia gene has been inactivated with a kanamycin resistance cassette (9). Strain 11 hia::kan/pHia is a derivative of strain 11 hia::kan in which the Hia protein is expressed in trans on the pSPEC1 shuttle plasmid (2) into which the hia gene has been cloned.
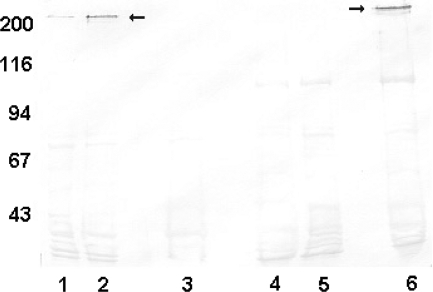
Shown in Fig. 3 is a Western immunoblot assay in which the levels of expression of the Hia protein by these strains are demonstrated. As can be seen, strain 11 demonstrates detectable but relatively low levels of Hia protein expression (Fig. 3, lane 1). In contrast, strain 11 hia::kan/pHia, the derivative in which the hia gene is expressed in trans on pSPEC1 (Fig. 3, lane 2), expresses Hia protein at a level significantly greater than that of the parent strain. We could demonstrate a roughly threefold increase in the expression level of Hia in the pHia transformant compared to that in parent strain 11 when the relative protein levels of the strains were compared by densitometry. Hia protein expression was not demonstrable at all in NTHI strain 11 hia::kan, the Hia knockout strain (Fig. 3, lane 3).
FIG. 3.
Western immunoblot assay with cell sonicates of NTHI strains probed with anti-Hia MAb 1F4. Lane 1, strain 11; lane 2, strain 11 hia::kan/pHia; lane 3, strain 11 hia::kan; lane 4, strain 5; lane 5, strain 5/pSPEC1; lane 6, strain 5/pHia. Arrows indicate Hia protein bands. The numbers on the left represent molecular weight standards.
Shown in Fig. 4 is a graph demonstrating the results of a representative opsonophagocytic assay in which the three strains just described were assessed for their susceptibility to killing mediated by the rGEMEX-Hia antiserum. Preimmune serum was unable to mediate killing of parent NTHI strain 11 and both of its derivatives (data not shown). In contrast, the rGEMEX-Hia immune serum mediated killing of the parent NTHI strain at a titer of 1:160. Strain 11 hia::kan, the knockout strain that did not express any detectable Hia protein, was not susceptible to opsonophagocytic killing mediated by the rGEMEX-Hia antiserum. In contrast, strain 11 hia::kan/pHia, the transformant that expressed considerably higher levels of Hia protein than the parent strain, was susceptible to killing at a titer of ≥1:320.
FIG. 4.
Opsonophagocytic killing assay. rGEMEX-Hia antiserum was assayed against NTHI strain 11, strain 11 hia::kan, and strain 11 hia::kan/pHia. Error bars indicate the standard deviations calculated for each serum dilution.
Opsonophagocytic killing of heterologous Hia-expressing NTHI strains and their derivatives.
Having demonstrated the ability of the rGEMEX-Hia antiserum to mediate opsonophagocytic killing of the homologous strain, we next examined the activity of the antiserum against heterologous Hia-expressing NTHI strains. Five representative Hia-expressing NTHI strains were selected for examination. These strains were all isolated in pure culture from middle ear fluid specimens of children with acute otitis media and were collected from diverse regions of the country. As was observed with NTHI strain 11, the preimmune serum was unable to mediate killing of any of the five heterologous strains (data not shown). In contrast, the rGEMEX-Hia immune serum mediated killing of each of the heterologous strains at titers ranging from 1:40 to 1:80 (Table 3).
TABLE 3.
Opsonophagocytic activity of anti-Hia antiserum against a panel of Hia-expressing NTHI strains
| Strain | Opsonophagocytic titer |
|---|---|
| 11 | 1:160 |
| 3A | 1:80 |
| 1512A | 1:80 |
| 3179B | 1:80 |
| 3248A | 1:80 |
| 3640A | 1:40 |
The hia genes from four of these five heterologous strains have been sequenced (9, 25, 39) The predicted amino acid sequences of the respective Hia proteins differ considerably from that of the prototype strain 11 Hia protein. These differences range from 97% identity/98% similarity when strain 3A and 11 Hia proteins are compared to 53% identity/62% similarity when strain 3248A and 11 proteins are compared. If one limits the comparison to the central regions corresponding to the pGEMEX-Hia BstEII del gene insert (Fig. 1), the differences range from 100% identity/100% similarity when the 3A and 11 “GEMEX insert” regions are compared to 44% identity/55% similarity when the 3248A and 11 regions are compared.
To further investigate the importance of the Hia protein as a target of opsonophagocytic antibodies, we also compared the susceptibility of strains 1512A and 1512A hia::kan to killing by the rGEMEX-Hia antiserum (9). In results that mirror those described for strain 11 and its isogenic Hia knockout, the rGEMEX-Hia antiserum mediated killing of strain 1512A at a titer of 1:80 but was unable to support opsonophagocytic killing of the corresponding 1512A Hia-knockout strain (Table 4).
TABLE 4.
Opsonophagocytic activity of anti-Hia antiserum against a panel of NTHI strains
| Strain | Opsonophagocytic titer |
|---|---|
| Hia-expressing | |
| 11 | 1:160 |
| 11 hia::kan | <1:10 |
| 11 hia::kan/pHia | 1:320 |
| 1512A | 1:80 |
| 1512A hia::kan | <1:10 |
| HMW-expressing | |
| 5 | <1:10 |
| 5/pHia | 1:160 |
| 5/pSPEC1 | <1:10 |
| 15 | <1:10 |
| 15/pHia | 1:160 |
| 15/pSPEC1 | <1:10 |
Opsonophagocytic killing of representative HMW1/HMW2-expressing NTHI strains and their respective Hia transformants.
To further explore the specificity of opsonophagocytic killing mediated by the rGEMEX-Hia antiserum, we assessed the ability of the antiserum to support killing of representative NTHI strains that express HMW1/HMW2-like proteins. As noted earlier, NTHI strains almost uniformly express an HMW1/HMW2-like adhesin(s) or an Hia-like adhesin, but not both (9, 38). The Hia immune serum was unable to mediate opsonophagocytic killing of two representative HMW1/HMW2-expressing NTHI strains, strain 5 and strain 15, that did not express an Hia adhesin (Table 4).
We next transformed these two HMW1/HMW2-expressing strains with either pHia, the shuttle plasmid containing the hia gene, or pSPEC1, the shuttle plasmid without an insert. The two pHia transformants expressed abundant Hia protein, and the pSPEC1 transformants expressed none. Lanes 4 to 6 of the Western immunoblot shown in Fig. 3 demonstrate Hia protein expression seen in NTHI strain 5 and its two transformants. As can be seen, the pHia transformant expressed abundant Hia protein, whereas none was seen in the pSPEC1 transformant.
We next examined the transformants for their susceptibility to killing in the opsonophagocytic assay. The rGEMEX-Hia immune serum mediated killing of both strain 5/pHia and strain 15/pHia at a titer of 1:160. In contrast, the immune serum did not support killing of either of these strains when they were transformed with pSPEC1 plasmid without an insert (Table 4).
DISCUSSION
The Hia proteins are members of a family of critical adhesins expressed by approximately 25% of NTHI (9, 38). Elegant molecular studies performed subsequent to their initial identification have demonstrated these proteins to be so-called trimeric autotransporters, gram-negative bacterial proteins defined by their short trimeric C-terminal translocator domain (14, 15). The C-terminal domains of the trimeric proteins are hypothesized to translocate the associated passenger domains across the bacterial outer membrane, where the latter are extensively intertwined on the surface of the bacterium in a position in which they interact with the external environment (48). In the work presented here, we demonstrated that the Hia proteins are present on the bacterial surface in a configuration that makes them readily accessible to antibodies capable of mediating opsonophagocytic activity. This observation has important implications in terms of better understanding host immunity to NTHI and potentially in terms of development of vaccines for prevention of NTHI disease.
In previously published work, we and our collaborators sequenced the hia genes of nine different NTHI strains (9, 25, 39). The sequences of these genes and the predicted amino acid sequences of their respective Hia proteins demonstrate considerable strain-to-strain variability, as might be expected of a bacterial surface protein that interacts with the host immune system (21). This variability could be seen as a hindrance to the development of the Hia proteins as viable vaccine candidates. However, it would be premature to come to such a conclusion. The Hia proteins are known to contain highly conserved regions within their overall structures, including within the previously identified binding domains (25, 48) and within the β-domain responsible for translocation of the passenger domain to the bacterial surface (39). In addition, Yeo and coworkers have reported that other regions of the Hia protein also demonstrate significant sequence homology with sequences of the primary binding domains (48). While the β domains of the Hia proteins are likely not accessible to host antibodies, given their probable location within the bacterial outer membrane layer, the binding domains are predicted to be more accessible to antibody as are other conserved regions within the passenger domain. The results we have presented in this work confirm the existence of shared immunogenic epitopes on the Hia proteins, given the ability of the rGEMEX-Hia antiserum to kill both homologous and representative heterologous NTHI strains. It will be important in future studies to more precisely localize the specific domains of the protein that are capable of eliciting cross-reactive functional antibodies, as these domains could serve as the basis for future recombinant or peptide-based vaccines.
In the studies we have presented, we examined a relatively small set of heterologous NTHI strains, but they represent a diverse collection of isolates that were collected from children across the country. Comparisons of the predicted amino acid sequences of both the full-length Hia proteins and the regions of the individual proteins corresponding to the GEMEX insert demonstrated significant differences between our prototype protein and the proteins from the heterologous strains. Nevertheless, given the great diversity of NTHI (19, 31), in future studies it will be important to examine a much larger collection of Hia-expressing strains for their susceptibility to killing by anti-Hia immune sera. It will also be of interest to examine encapsulated H. influenzae for susceptibility to killing by anti-Hia antibodies. While the encapsulated H. influenzae species do not express an Hia protein per se, they do express the closely related protein known as Hsf (for Haemophilus surface fibrils) (15). This protein family demonstrates a binding specificity very similar to that of the Hia protein family and contains highly conserved sequences within the binding domains (15), suggesting that immunogenic surface epitopes might also be shared between members of the Hia and Hsf protein families.
The Hia proteins are expressed by only a subset of NTHI strains (9, 38). If used alone, the Hia proteins would not be particularly attractive as a vaccine component. However, as was noted earlier, almost all NTHI strains that do not express an Hia adhesin protein do express an HMW1/HMW2-like adhesion protein(s). In our earlier work, we demonstrated that antibodies directed against the HMW1/HMW2 family of proteins are also opsonophagocytic for NTHI strains that express such proteins and that these antibodies are active against both homologous and heterologous HMW1/HMW2-expressing strains (43, 44). A vaccine based upon a combination of Hia and HMW1/HMW2 proteins could potentially provide protection against infections caused by most or all NTHI strains. These data argue for the continued investigation of both the Hia and HMW1/HMW2 protein families as vaccine candidates for the prevention of NTHI disease.
Acknowledgments
This work was supported by Public Health Service grant AI 48066 from the National Institute of Allergy and Infectious Diseases and grant-in-aid no. 0755643Z from the American Heart Association.
Footnotes
Published ahead of print on 27 May 2009.
REFERENCES
- 1.Akkoyunlu, M., H. Janson, M. Ruan, and A. Forsgren. 1996. Biological activity of serum antibodies to a nonacylated form of lipoprotein D of Haemophilus influenzae. Infect. Immun. 644586-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 731635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O., B. J. Kennedy, L. A. Novotny, G. Duquesne, J. Cohen, and Y. Lobet. 1999. Protection against development of otitis media induced by nontypeable Haemophilus influenzae by both active and passive immunization in a chinchilla model of virus-bacterium superinfection. Infect. Immun. 672746-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., E. R. Leake, J. M. Billy, and P. T. Kaumaya. 1997. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine 15955-961. [DOI] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J. 1996. Immunization with high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae modifies experimental otitis media in chinchillas. Infect. Immun. 641246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barenkamp, S. J., and F. F. Bodor. 1990. Development of serum bactericidal activity following nontypable Haemophilus influenzae acute otitis media. Pediatr. Infect. Dis. J. 9333-339. [DOI] [PubMed] [Google Scholar]
- 7.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 601302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of surface-exposed B-cell epitopes on high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae. Infect. Immun. 643032-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 191215-1223. [DOI] [PubMed] [Google Scholar]
- 10.Berman, S. 1995. Otitis media in children. N. Engl. J. Med. 3321560-1565. [DOI] [PubMed] [Google Scholar]
- 11.Berman, S. 1995. Otitis media in developing countries. Pediatrics 96126-131. [PubMed] [Google Scholar]
- 12.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23824-828. [DOI] [PubMed] [Google Scholar]
- 13.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 1951445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, S. E., N. K. Surana, S. Grass, and J. W. St. Geme III. 2006. Trimeric autotransporters require trimerization of the passenger domain for stability and adhesive activity. J. Bacteriol. 1885400-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotter, S. E., H.-J. Yeo, T. Juehne, and J. W. St. Geme III. 2005. Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 1874656-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagan, R., A. Hoberman, C. Johnson, E. L. Leibovitz, A. Arguedas, F. V. Rose, B. R. Wynne, and M. R. Jacobs. 2001. Bacteriologic and clinical efficacy of high dose amoxicillin/clavulanate in children with acute otitis media. Pediatr. Infect. Dis. J. 20829-837. [DOI] [PubMed] [Google Scholar]
- 17.DeMaria, T. F., D. M. Murwin, and E. R. Leake. 1996. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 645187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Adhami, W., J. M. Kyd, D. A. Bastin, and A. W. Cripps. 1999. Characterization of the gene encoding a 26-kilodalton protein (OMP26) from nontypeable Haemophilus influenzae and immune responses to the recombinant protein. Infect. Immun. 671935-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erwin, A. L., K. L. Nelson, T. Mhlanga-Mutangadura, P. J. Bonthuis, J. L. Geelhood, G. Morlin, W. C. T. Unrath, J. Campos, D. W. Crook, M. M. Farley, F. W. Henderson, R. F. Jacobs, K. Muhlemann, S. W. Satola, L. van Alphen, M. Golomb, and A. L. Smith. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 735853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giebink, G. S., L. O. Bakaletz, S. J. Barenkamp, B. Green, X. X. Gu, T. Heikkinen, M. Hotomi, P. Karma, Y. Kurono, J. M. Kyd, T. F. Murphy, P. L. Ogra, J. A. Patel, and S. I. Pelton. 2005. Report of the vaccination panel: eighth international otitis media research conference. Ann. Otol. Rhinol. Laryngol. 114S86-S103. [Google Scholar]
- 21.Gilsdorf, J. R., C. F. Marrs, and B. Foxman. 2004. Haemophilus influenzae: genetic variability and natural selection to identify virulence factors. Infect. Immun. 722457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnehm, H. E., S. I. Pelton, S. Gulati, and P. A. Rice. 1985. Characterization of antigens from nontypable Haemophilus influenzae recognized by human bactericidal antibodies. Role of Haemophilus outer membrane proteins. J. Clin. Investig. 751645-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu, X. X., S. F. Rudy, C. Chu, L. McCullagh, H. N. Kim, J. Chen, J. Li, J. B. Robbins, C. Van Waes, and J. F. Battey. 2003. Phase I study of a lipooligosaccharide-based conjugate vaccine against nontypeable Haemophilus influenzae. Vaccine 212107-2114. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St. Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46731-743. [DOI] [PubMed] [Google Scholar]
- 26.Leibovitz, E., M. R. Jacobs, and R. Dagan. 2004. Haemophilus influenzae: a significant pathogen in acute otitis media. Pediatr. Infect. Dis. J. 231142-1152. [PubMed] [Google Scholar]
- 27.Linke, D., T. Riess, I. B. Autenrieth, A. Lupas, and V. A. Kempf. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14264-270. [DOI] [PubMed] [Google Scholar]
- 28.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 201754-1760. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, T. F., and L. C. Bartos. 1988. Human bactericidal antibody response to outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 562673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, T. F., L. C. Bartos, P. A. Rice, M. B. Nelson, K. C. Dudas, and M. A. Apicella. 1986. Identification of a 16,600-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J. Clin. Investig. 781020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neary, J. M., and T. F. Murphy. 2006. Antibodies directed at a conserved motif in loop 6 of outer membrane protein P2 of nontypeable Haemophilus influenzae recognize multiple strains in immunoassays. FEMS Immunol. Med. Microbiol. 46251-261. [DOI] [PubMed] [Google Scholar]
- 33.Paradise, J. L., T. F. Campbell, C. A. Dollaghan, H. M. Feldman, B. S. Bernard, D. K. Colborn, H. E. Rockette, J. E. Janosky, D. L. Pitcairn, M. Kurs-Lasky, D. L. Sabo, and C. G. Smith. 2005. Developmental outcomes after early or delayed insertion of tympanostomy tubes. N. Engl. J. Med. 353576-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paradise, J. L., H. E. Rockette, D. K. Colborn, B. S. Bernard, C. G. Smith, M. Kurs-Lasky, and J. E. Janosky. 1997. Otitis media in 2253 Pittsburgh-area infants: prevalence and risk factors during the first two years of life. Pediatrics 99318-333. [DOI] [PubMed] [Google Scholar]
- 35.Piglansky, L., E. Leibovitz, S. Raiz, D. Greenberg, J. Press, A. Leiberman, and R. Dagan. 2003. Bacteriologic and clinical efficacy of high dose amoxicillin for therapy of acute otitis media in children. Pediatr. Infect. Dis. J. 22405-413. [DOI] [PubMed] [Google Scholar]
- 36.Prymula, R., P. Peeters, V. Chrobok, P. Kriz, E. Novakova, E. Kaliskova, I. Kohl, P. Lommel, J. Poolman, J. P. Prieels, and L. Schuerman. 2006. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study. Lancet 367740-748. [DOI] [PubMed] [Google Scholar]
- 37.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 902875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St. Geme III. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 27914679-14685. [DOI] [PubMed] [Google Scholar]
- 40.Tan, T. T., J. J. Christensen, M. H. Dziegiel, A. Forsgren, and K. Riesbeck. 2006. Comparison of the serological responses to Moraxella catarrhalis immunoglobulin D-binding outer membrane protein and the ubiquitous surface proteins A1 and A2. Infect. Immun. 746377-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teele, D. W., J. O. Klein, C. Chase, P. Menyuk, and B. A. Rosner, et al. 1990. Otitis media in infancy and intellectual ability, school achievement, speech, and language at age 7 years. J. Infect. Dis. 162685-694. [DOI] [PubMed] [Google Scholar]
- 42.Webb, D. C., and A. W. Cripps. 1999. Immunization with recombinant transferrin binding protein B enhances clearance of nontypeable Haemophilus influenzae from the rat lung. Infect. Immun. 672138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter, L. E., and S. J. Barenkamp. 2006. Antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are opsonophagocytic for both homologous and heterologous strains. Clin. Vaccine Immunol. 131333-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter, L. E., and S. J. Barenkamp. 2003. Human antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae mediate opsonophagocytic activity. Infect. Immun. 716884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, T.-H., and X.-X. Gu. 1999. Outer membrane proteins as a carrier for detoxified lipooligosaccharide conjugate vaccines for nontypeable Haemophilus influenzae. Infect. Immun. 675508-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 48.Yeo, H. J., S. E. Cotter, S. Laarmann, T. Juehne, J. W. St. Geme III, and G. Waksman. 2004. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 231245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]