Abstract
Free-living protozoan communities in water supplies may include hosts for Legionella pneumophila and other undesired bacteria, as well as pathogens. This study aimed at identifying free-living protozoa in two unchlorinated groundwater supplies, using cultivation-independent molecular approaches. For this purpose, samples (<20°C) of treated water, distributed water, and distribution system biofilms were collected from supply A, with a low concentration of natural organic matter (NOM) (<0.5 ppm of C), and from supply B, with a high NOM concentration (7.9 ppm of C). Eukaryotic communities were studied using terminal restriction fragment length polymorphism and clone library analyses of partial 18S rRNA gene fragments and a Hartmannella vermiformis-specific quantitative PCR (qPCR). In both supplies, highly diverse eukaryotic communities were observed, including free-living protozoa, fungi, and metazoa. Sequences of protozoa clustered with Amoebozoa (10 operational taxonomic units [OTUs]), Cercozoa (39 OTUs), Choanozoa (26 OTUs), Ciliophora (29 OTUs), Euglenozoa (13 OTUs), Myzozoa (5 OTUs), and Stramenopiles (5 OTUs). A large variety of protozoa were present in both supplies, but the estimated values for protozoan richness did not differ significantly. H. vermiformis was observed in both supplies but was not a predominant protozoan. One OTU with the highest similarity to Acanthamoeba polyphaga, an opportunistic human pathogen and a host for undesired bacteria, was observed in supply A. The high level of NOM in supply B corresponded with an elevated level of active biomass and with elevated concentrations of H. vermiformis in distributed water. Hence, the application of qPCR may be promising in elucidating the relationship between drinking water quality and the presence of specific protozoa.
Free-living protozoa are ubiquitous in natural freshwater environments (7, 38, 51, 71) but also proliferate in engineered water systems, including water treatment systems (3, 47, 70), distribution systems (6, 75), and tap water installations inside buildings (54, 69). Concentrations of protozoa, determined using cultivation methods and microscopy, range from <1 to 104 cells liter−1 in treated water (3, 47, 70, 75) and from <1 to 7 × 105 cells liter−1 in distribution systems (6, 61, 64, 75). Genera of free-living protozoa commonly observed in these systems and in tap water installations include Acanthamoeba, Echinamoeba, Hartmannella, Platyamoeba, Vahlkampfia, and Vannella (47, 58, 69, 70). In warm water systems, certain free-living protozoa, e.g., Acanthamoeba spp. (57), Balamuthia mandrillaris (62), Echinamoeba exandans (16), Hartmannella spp. (39, 56), Naegleria spp. (49, 57), Tetrahymena spp. (18, 33), and Vahlkampfia jugosa (56), serve as hosts for Legionella pneumophila, the etiologic agent of Legionnaires' disease. High concentrations of L. pneumophila are generally associated with the proliferation of host protozoa in biofilms (38, 53). In addition, other amoeba-resistant, potentially pathogenic bacteria, e.g., Burkholderia spp. (28) and Mycobacterium spp. (37), have been observed in man-made aquatic environments (24). Free-living protozoa may enhance the multiplication of bacteria, serve as a transmission vector, or serve as a shelter against unfavorable environmental conditions, such as the presence of disinfectants. Furthermore, certain free-living protozoa are human pathogens, e.g., Naegleria fowleri (81), Balamuthia mandrillaris (77), and Acanthamoeba spp. (12) can cause encephalitis. Acanthamoeba spp. have also been associated with keratitis in persons wearing contact lenses (31).
Free-living protozoa feed on bacteria, algae, fungi, other protozoa, and organic detritus in biofilms or in the planktonic phase, thereby affecting the structure of microbial communities. In turn, the community of free-living protozoa depends on the diversity and abundance of bacteria in the biofilm and in the planktonic phase (26, 50, 51, 55, 63, 65). Water quality is a critical factor for biofilm formation in distribution systems and tap water installations and therefore will affect the abundance and diversity of free-living protozoa in these systems (72, 78). However, information about the presence and identity of free-living protozoa in water supplies in relation to the quality of treated water is scarce, which may be attributed to the limitations of microscopic techniques and cultivation methods for detection and identification of these organisms, e.g., low detection limits and selectivity for specific groups (19).
In this study, we applied a variety of cultivation-independent techniques, viz., quantitative PCR, terminal restriction fragment length polymorphism (T-RFLP) analysis, and cloning and sequencing of eukaryotic 18S rRNA gene fragments, for the detection and identification of free-living protozoa predominating in two unchlorinated groundwater supplies. The concentrations of dissolved natural organic matter (NOM) in treated water at the plant were <0.5 mg C liter−1 and 7.9 mg C liter−1, covering the entire range of NOM concentrations in drinking water in The Netherlands. The objectives of the study were (i) to elucidate the identities of and diversity in the free-living protozoa predominating in these two different water supplies and (ii) to trace the presence of host protozoa for L. pneumophila and pathogenic free-living protozoa. The study revealed that treated water and biofilms in the distribution systems of both water supplies contained a large variety of free-living protozoa, including protozoan hosts for Legionella bacteria.
MATERIALS AND METHODS
Selected water supplies.
Two groundwater supplies in The Netherlands, distributing drinking water with different NOM concentrations, were selected (Table 1). In supply A, with an annual production of 5.6 × 106 m3 and a supply area of ca. 40 km2 without service reservoirs, aerobic groundwater abstracted from a sand aquifer is aerated to remove CO2, followed by limestone filtration to increase the pH and hardness of the water (see the supplemental material for details). The treated water of supply A (TW-A) contains a low concentration of NOM (<0.5 mg C liter−1), measured as nonpurgable organic carbon (NPOC). In supply B, with an average annual production of 2.5 × 107 m3 and a supply area of ca. 1,000 km2 with several service reservoirs, anaerobic groundwater abstracted from below a peat layer is treated by intensive aeration, rapid sand filtration, caustic dosage followed by pellet softening, aeration, and a second stage of rapid sand filtration (see the supplemental material for details). The two stages of rapid sand filtration remove ammonia, iron, and manganese. The NOM concentration in the treated water of supply B (TW-B) is 7.9 mg C liter−1. Both water types are treated and distributed without chemical disinfection (73).
TABLE 1.
Quality characteristics of treated water at the treatment plants of supply A and supply Ba
| Parameter | TW-A value
|
TW-B value
|
||||
|---|---|---|---|---|---|---|
| Mean | Minimum | Maximum | Mean | Minimum | Maximum | |
| Temp (°C) | 10.0 | 9.5 | 11.5 | 11.5 | 10.0 | 13.5 |
| pH | 7.8 | 7.2 | 8.2 | 7.6 | 7.4 | 8.1 |
| O2 concn (mg liter−1) | 6.4 | 5.6 | 7.8 | 5.9 | 3.9 | 8.3 |
| HCO3 concn (mg liter−1) | 98 | 92 | 124 | 282 | 273 | 308 |
| Cl concn (mg liter−1) | 13 | 11 | 14 | 28 | 27 | 31 |
| Ca concn (mg liter−1) | 35.4 | 32.9 | 39.6 | 32.7 | 25.7 | 52.8 |
| Mg concn (mg liter−1) | 2.37 | 2.07 | 2.73 | 9.72 | 8.36 | 10.9 |
| Total hardness (mmol liter−1) | 0.98 | 0.9 | 1.09 | 1.22 | 1.04 | 1.68 |
| Fe concn (μg liter−1) | <20 | <20 | <20 | 25 | <20 | 73 |
| Mn concn (μg liter−1) | <10 | <10 | <10 | <10 | <10 | <10 |
| SO4 concn (mg liter−1) | 16 | 13 | 19 | <10 | <10 | <10 |
| NH4 concn (mg liter−1) | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 |
| NPOC concn (mg C liter−1) | 0.33 | <0.3 | 0.49 | 7.9 | 7.6 | 8.3 |
Mean, minimum, and maximum values, based on routine monitoring over a period of 1 year.
Sample collection.
During all seasons of the year 2005, samples of treated water were collected at both plants. In September and October 2005, samples of the biofilms in pipe segments of both distribution systems were taken. Distances between sample locations and the treatment plant ranged from 0.4 km to 6.0 km for supply A and from 17.1 km to 35 km for supply B. In July and November 2007, treated water and water from both distribution systems were collected. The samples of the distributed water were taken at the same locations where biofilms were collected. The numbers and letters in sample names indicate the locations of samples in the distribution systems, e.g., BF-A1 and DW-A1 indicate that the biofilm sample and the distributed water sample, respectively, were collected at location 1 in supply A (see the supplemental material for details). The water samples were stored at 4°C in sterile glass containers and processed within 24 h. At seven locations in the distribution system of supply A and at eight locations in the distribution system of supply B, segments (30 cm) of unplasticized polyvinyl chloride pipes (diameter, 110 mm) were removed, after thorough cleaning of the outer surface, and subsequently placed in plastic cylinders containing water from the distribution system. The samples were stored at 4°C and processed within 24 h. The attached biomass at the inside surface was collected by swabbing ±20 cm2 with three sterile cotton swabs (Copan Innovation, Italy). These swabs were placed in 10 ml phosphate-buffered saline, and the biomass was removed from the swabs by four 2-min sonication steps in a water bath at a frequency of 40 kHz and an average power input of 0.015 W/ml (44). Total ATP concentrations in biofilms and the planktonic phase, representing the active biomass, were determined by ATP analysis as described by Magic-Knezev and van der Kooij (44).
Water filtration and DNA extraction.
Samples of 1.25 to 3 liters of treated water and 0.5 liter of distributed water were filtered through a 1.2-μm-pore-size and 55-mm-diameter RTTP Isopore membrane (Millipore, Molsheim, France). Samples of biomass suspended in phosphate-buffered saline were filtered through a 1.2-μm-pore-size and 25-mm-diameter RTTP Isopore membrane (Millipore). DNA was isolated and purified using a Fast DNA spin kit for soil (Bio 101, Carlsbad, CA) following the instructions of the manufacturer, with the exception that 2-ml tubes containing lysing matrix E, sodium phosphate, MT buffer, and filter with sample were processed in a FastPrep instrument (Bio 101) two times for 30 s each at speed setting 5.5. The isolated DNA was eluted in 200 μl DNase- and pyrogen-free water. Distilled water (DNase and RNase free) was used as a negative control in each experiment to check for possible DNA contamination during filtration, DNA extraction, and PCR amplification. In addition, all samples were spiked with Hartmannella vermiformis ATCC 50237 to check for the presence of inhibitors in the samples. DNA was subsequently used for the characterization of eukaryotic community composition and for quantification of H. vermiformis populations.
Detection of H. vermiformis by quantitative PCR.
Quantitative PCR assays were performed in 96-well plates, using an iCycler iQ multicolor real-time PCR detection system (Bio-Rad, Veenendaal, The Netherlands) as described by Kuiper et al. (38). Experiments were performed in duplicate, using undiluted and 10-fold diluted DNA extracts as templates. Quantification was based on a calibration curve for a suspension with a known number of H. vermiformis cells that was analyzed in different DNA dilutions with each series of samples. The detection limit was one H. vermiformis cell per reaction.
PCR for T-RFLP analysis and cloning.
PCR was performed with a GeneAmp PCR 9700 system (Applied Biosystems, Nieuwerkerk aan de IJsel, The Netherlands), using a reaction mixture (50 μl) with 10 μl template DNA. PCR was performed with 5% and 6.7% of the total DNA extracted from the treated water and biofilms, respectively. Fragments of the 18S rRNA gene were amplified with the eukaryotic primers (labeled at the 3′ end with 6-carboxyfluorescein) Euk1a-f (68) and Euk516-r (2). Amplification conditions were as follows: preheating at 94°C for 130 s, 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 45 s, and extension at 72°C for 130 s, and a terminal extension at 72°C for 7 min.
T-RFLP analysis.
Fluorescently labeled PCR products (45 μl) were purified by using a DNA Clean & Concentrator-5 kit (BaseClear, Leiden, The Netherlands) and redissolved in 20 μl of distilled water. The digestion reaction mixture (20 μl) contained 5 U of HhaI (Promega), 2 μl of buffer C (Promega), 12.5 μl of distilled water, and 5 μl of the PCR product and was incubated at 37°C for 6 h. The mixture was cleaned as described above and redissolved in 15 μl distilled water. The restriction digestion product (5 μl) was mixed with 15 μl loading buffer (Hi-Di formamide [Applied Biosystems] and GS-500 ROX [Applied Biosystems], 15:1 [vol/vol], as an internal standard). The injection time was 5 s for analysis of terminal restriction fragments, and the run time was 35 min. The fluorescently labeled terminal restriction fragments were analyzed by electrophoresis on an ABI Prism 310 genetic analyzer (Applied Biosystems) in Genescan mode. Electropherograms were imported into a genomic fingerprint analysis program, Bionumerics v. 4.6 (Applied Maths, Sint-Martens-Latem, Belgium), and fragment sizes were calculated. Banding patterns were compared using a densitometric curve-based method that evaluates the positions and intensities of bands to generate pairwise similarity scores (Pearson coefficients), and these were subsequently used for cluster analysis.
Cloning and sequencing of PCR products.
The identities of the predominant eukaryotes in the treated water at the plant and in the biofilm of the distribution system were determined by cloning and sequence analysis of approximately 550-bp 18S rRNA gene fragments amplified with the primers Euk1a-f and Euk516-r. The PCR products were cloned using the pGEM-T Easy II vector system. The DNA inserts of randomly selected positive clones were sequenced using the Euk1a-f and Euk516-r primers (BaseClear). One hundred thirty-four and 136 clones of the treated water samples and 43 to 50 clones of the biofilm samples were analyzed.
Phylogenetic analysis of partial sequences.
Operational taxonomic units (OTUs) were defined as 18S rRNA gene sequences that shared ≥99% sequence similarity. The obtained sequences of approximately 550 bp were compared to sequences in the National Center for Biotechnology (NCBI) database by BLAST searches and were also imported and aligned into the SILVA 94 SSU Ref database (52), released in April 2008, using the ARB software package (42). A distance matrix (no filter and no corrections) was calculated for all clones. This distance matrix was used as an input file in the DOTUR program (59). OTUs for the purpose of community analysis were defined by a 1% difference in nucleic acid sequences, as determined using the furthest neighbor algorithm in DOTUR. Similarity percentages were determined for complete and partial 18S rRNA gene sequences in the SILVA 94 SSU Ref database for genera and species closely related to the obtained OTUs. The partial sequences used correspond to the fragments amplified with the primers Euk1a-f and Euk516-r. The OTU richness was estimated by the Chao1 estimator (11) and was calculated from randomized data as described by Hughes et al. (27).
The similarity of each OTU to 18S rRNA gene sequences in the SILVA 94 SSU Ref database was analyzed by adding one representative sequence of each OTU to the main phylogenic tree by using parsimony criteria without changing the overall tree topology. The POS_VAR_Eukarya_94 filter (excludes highly variable positions 1 to 7) was used. The obtained sequences were divided into taxa based on the classification system of Cavalier-Smith (10) and the structure in the SILVA 94 SSU Ref database (52). Sequences with similarities to described species of <75% were excluded from further analysis.
Statistical analysis.
The F test, with log transformation of the concentrations, was used to determine the difference between the concentrations of H. vermiformis in the distributed water in the summer and the autumn.
Nucleotide sequence accession numbers.
All partial 18S rRNA gene sequences determined in this study have been deposited in GenBank under accession numbers EU860442 to EU860974.
RESULTS
Active biomass (ATP) and water temperature.
ATP concentrations in treated water and in distributed water at supply A generally were below 1.0 ng liter−1, and the average was 123.4 ± 87.7 pg ATP cm−2 in the biofilm in the pipes. The concentration of active biomass in treated water of supply B was 10.6 ± 4.9 ng ATP liter−1, and that in the distribution system was 4.7 ± 1.2 ng ATP liter−1, with a biofilm concentration of 334.7 ± 226.1 pg ATP cm−2. The temperature of the treated water at both plants was close to 10°C and showed little variation during the seasons (Table 1). For supply A, the average temperature of the water samples collected from the distribution system was 14.8 ± 2.3°C in July and 11.1 ± 0.6°C in November. The average temperature of the distributed water in supply B was 13.6 ± 1.5°C in July and 12.5 ± 0.8°C in November. Hence, the temperature of the water in the distribution system increased during the summer. In summary, the concentration of active biomass in supply B was higher than that in supply A, and both water types were characterized by relatively low temperatures.
T-RFLP analysis of eukaryotic communities.
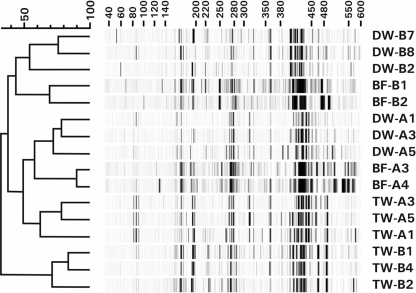
T-RFLP analyses using 18S rRNA gene primers revealed complex eukaryotic communities in the water samples of both supplies (Fig. 1). The fingerprints of the samples of each supply clustered together, indicating that the water type affected the eukaryotic community (Fig. 1). Fingerprints of TW-A analyzed in duplicate showed a minimum similarity of 90%, and those obtained in different seasons showed similarities between 61.8% and 78.7%. Duplicate fingerprints of each TW-B sample showed a minimum similarity of 87%, and the fingerprints of different samples showed similarities between 70.5% and 81.3%. Hence, the eukaryotic communities in treated water showed some variation, but more variation was observed between the fingerprints of the biofilm samples within each supply.
FIG. 1.
Dendrogram, created by unweighted-pair group method using average linkages, of T-RFLP fingerprints of treated water (TW), biofilms (BF) from the distribution systems, and distributed water (DW) of supply A and supply B. Samples of distributed water were taken from different locations in July, and biofilm samples from different locations were taken in September and October.
Diversity in eukaryotic communities in treated water and in biofilms.
Clone libraries were constructed for one treated water sample and three biofilm samples from each supply. In total, 545 partial 18S rRNA gene sequences of 550 bp were analyzed (Table 2). The results for the three biofilm samples were combined for comparison with the results for the treated water samples. All sequences showed the highest similarity to 18S rRNA gene sequences in the NCBI and SILVA databases (release 94; April 2008), confirming the specificity of the primers used for eukaryotic sequences.
TABLE 2.
Classification of clones of eukaryotes retrieved from treated water and biofilms of supply A and supply B
| Kingdom or subkingdom | Water supply Aa
|
Water supply Ba
|
All analyzed samples
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TW-A1
|
BF-A
|
TW-B1
|
BF-B
|
|||||||
| No. (%) of OTUs | % of clones in library | No. (%) of OTUs | % of clones in library | No. (%) of OTUs | % of clones in library | No. (%) of OTUs | % of clones in library | No. (%) of OTUsb | No. (%) of clones in library | |
| Free-living protozoa | 25 (48.1) | 33.4 | 29 (43.3) | 64.5 | 43 (67.2) | 52.2 | 31 (56.4) | 36.6 | 127 (58.0) | 253 (46.4) |
| Fungi | 7 (13.5) | 8.9 | 12 (17.9) | 20.3 | 4 (6.3) | 2.9 | 6 (10.9) | 5.2 | 27 (12.3) | 51 (9.4) |
| Metazoa | 4 (7.7) | 24.4 | 3 (4.4) | 4.3 | 10 (15.6) | 23.5 | 14 (25.5) | 47.8 | 28 (12.8) | 134 (24.6) |
| Protophyta and plants | 3 (5.8) | 4.4 | 2 (3.0) | 1.5 | 0 (0) | 0 | 1 (1.8) | 0.7 | 5 (2.3) | 9 (1.7) |
| Organisms with <75% similarity | 13 (25.0) | 28.9 | 10 (14.9) | 9.4 | 7 (10.9) | 21.4 | 3 (5.5) | 9.7 | 32 (14.6) | 98 (17.9) |
| Total | 52 (100) | 100 | 56 (100) | 100 | 64 (100) | 100 | 55 (100) | 100 | 219 (100) | 545 (100) |
TW, treated water obtained directly from plant; BF, biofilm. The biofilm data are totals for the three analyzed biofilm samples.
The total number of OTUs includes OTUs which were obtained from more than one sample type.
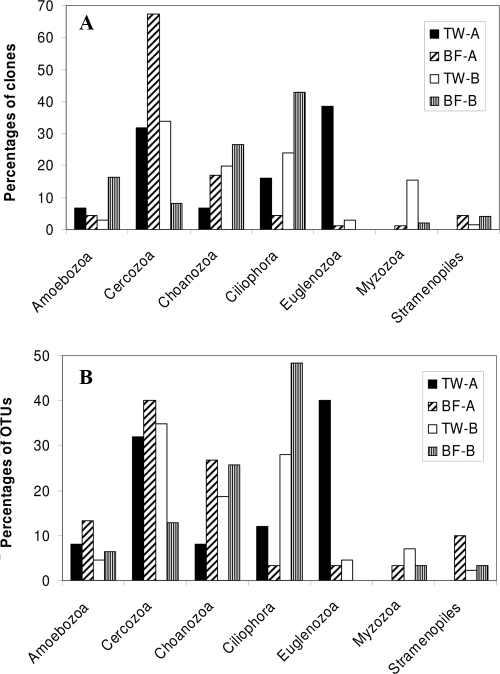
A total of 219 different OTUs (sequence similarity of ≥99%) were distinguished (Table 2). Eight OTUs were observed in more than one sample type, and therefore the sums of the OTUs in Tables 2 and 3 give excess values. The other 211 OTUs were unique for specific samples, demonstrating the high level of diversity in the eukaryotic communities in the two supplies. Table 3 shows that the coverage of the clone libraries for both supplies was similar and also that the estimated total OTU richness values were not significantly different between the two supplies. In treated water of both supplies, the free-living protozoa constituted the largest proportion (>48%) of the obtained OTUs, with fungi having the second largest number in supply A and metazoa having the second largest number in supply B (Table 2). In addition, protophyta and cellular plants were represented. Thirty-two (14.6%) of the obtained OTUs, one of which was detected in TW-A and in BF-A, had similarity percentages below 75% for described sequences in the SILVA database and remained unidentified (Table 2).
TABLE 3.
Diversity of organisms in clone libraries for treated water at the plant and biofilms in the distribution systems of supply A and supply B
| Source and type of organism | No. of clones | No. of OTUs identified | Coverage indexa | Total OTU richness (Chao1 estimation)b
|
||
|---|---|---|---|---|---|---|
| Mean | Lower limit | Upper limit | ||||
| Eukaryotes in clone libraries | ||||||
| Supply A | 272 | 108 | 39.7 | 159 | 136 | 204 |
| Supply B | 273 | 115 | 42.1 | 145 | 91 | 277 |
| Total | 545 | 219c | 40.2 | 390 | 328 | 487 |
| Free-living protozoa in clone libraries | ||||||
| Supply A | 133 | 54c | 40.6 | 113 | 81 | 187 |
| TW-A | 44 | 25 | 56.8 | 34 | 28 | 55 |
| BF-Ad | 89 | 30 | 33.7 | 158 | 70 | 444 |
| Supply B | 120 | 72c | 60.0 | 163 | 112 | 274 |
| TW-B | 71 | 43 | 60.6 | 134 | 75 | 297 |
| BF-Bd | 49 | 31 | 63.3 | 45 | 32 | 88 |
| Total | 253 | 127c | 50.2 | 281 | 212 | 407 |
A total of 27 OTUs (12.3%) showed highest similarity to phyla within the fungi, viz., Chytridiomycota (3 OTUs), Zygomycota (2 OTUs), Ascomycota (20 OTUs), and Basidiomycota (2 OTUs). Two of these OTUs, which showed the highest similarity to Triparticalar arcticum and an uncultured Banisveld eukaryote, were retrieved from both supplies.
A total of 28 OTUs (12.8%) showed highest similarity to metazoan phyla, viz., Porifera (5 OTUs), Cnidaria (8 OTUs), Platyhelminthes (1 OTU), Rotifera (2 OTUs), Gastrotricha (4 OTUs), Nematoda (2 OTUs), Annelida (2 OTU), and Arthropoda (4 OTUs). Two of these OTUs were retrieved from both supplies and showed the highest similarity to Lepadella patella and Rhabdolaimus terrestris. The clone libraries of BF-B5 and BF-B6 are predominated by an OTU with highest similarity (99%) to the metazoan freshwater jellyfish Craspedacusta sowerbyi. Five OTUs (2.3%) clustered with protophyta or cellular plants, and one of these OTUs was obtained from BF-A and BF-B. The other four OTUs were obtained only from supply A (Table 2). Four OTUs (1.8%) clustering with the protophyta showed highest similarity with species within the phylum Cryptophyta, viz., Plagioselmis prolonga (75.8% similarity), Chlorella sp. (76.5% similarity), Staurastrum polymorphum (82.7% similarity), and Goniomonas pacifica (91.3% similarity). One OTU clustered within the family Poaceae (grasses).
Identities of and diversity in free-living protozoa in treated water and in biofilms.
A total of 253 sequences (46.4%) and more than half of the obtained OTUs (127 OTUs) showed highest similarity to free-living protozoa (Table 2). The coverage of the clone libraries for supply A was lower than that for supply B, but the estimated total OTU richness values for these supplies were not significantly different (Table 3). The obtained OTUs had similarities of 57% to 100% with eukaryotic sequences in the SILVA database (release 94; April 2008). Similarity threshold percentages for eukaryotic genera and species at the 18S rRNA gene level have not yet been established. Therefore, similarity percentages for 18S rRNA gene sequences most closely related to the same genera and species included in the SILVA database were derived. Data for nine different genera of free-living protozoa revealed that the minimum similarities ranged from 75% to 92%. For sequences in the SILVA database most closely related to the cluster of Hartmannella (n = 19), Acanthamoeba (n = 211), and Vorticella (n = 7) species, minimum similarities of 75.2%, 78.1%, and 91.9%, respectively, were obtained. Minimum similarities ranging from 86.6% (Bodo saltans; n = 23) to 99.7% (H. vermiformis; n = 15) were calculated for sequences of seven protozoan species (not all data shown) most closely related to those collected in this study. A total of 98 sequences (32 OTUs) had ≤75% similarity to sequences in the database and thus were considered unidentifiable (Table 2). These unidentified OTUs showed a minimum similarity of 44.4% and a maximum similarity of 98.2% to each other. Eleven of these OTUs, including nine OTUs from supply A and two OTUs from supply B, clustered with each other with more than 95% similarity.
In the clone libraries for both supplies, we observed sequences clustering with seven protozoan phyla (Fig. 2 and Table 4). The results show that a few protozoan phyla predominated in the different clone libraries and that the diversity within each phylum varied between the different sample locations. None of the 127 OTUs with highest similarity to free-living protozoa were observed in both supplies, demonstrating that there are highly diverse protozoan communities in each supply (Table 4).
FIG. 2.
(A) Taxonomic distribution of free-living protozoa, based on 18S rRNA gene clones retrieved from treated water (TW) and biofilms (BF) of supply A and supply B. Biofilm data are totals for the three analyzed biofilm samples per supply. (B) Taxonomic distribution of OTUs with highest similarity to free-living protozoa retrieved from treated water (TW) and from biofilms (BF) of supply A and supply B. Biofilm data are totals for the three analyzed biofilm samples per supply.
TABLE 4.
Classification of OTUs clustering with free-living protozoa obtained from treated water and from biofilms in the distribution systems of water supply A and water supply Ba
| Organism(s) with highest similarity (GenBank accession no.)b | Similarity (%) | No. of OTUs | No. of clones
|
|||
|---|---|---|---|---|---|---|
| TW-A | BF-Ac | TW-B | BF-Bc | |||
| Amoebozoa | 10 | 3 | 4 | 2 | 8 | |
| Acanthamoeba polyphaga (AF260725) | 89.3 | 1 | 1 | |||
| Echinamoeba thermarum (AJ489264) | 85.7 | 1 | 1 | |||
| Eimeriidae environmental sample clone (EF024503)/Acanthamoeba sp. (AY173000) | 96.8/85.9 | 1 | 2 | |||
| Uncultured endolithic amoeba (AB257667)/Hartmannellidae sp. strain LO57N/1 (AY145442) | 79.2/77.9 | 1 | 7 | |||
| Neoparamoeba aestuarina (AY121851) | 89.3 | 1 | 1 | |||
| Pterocystis tropica (AY749612) | 93.4 | 1 | 1 | |||
| Raineriophrys erinaceoides (AY749633) | 94.7, 93.3 | 2 | 2 | |||
| Uncultured eukaryote clone (AY749523)/Raineriophrys sp. (AY749606) | 89.0/88.4 | 1 | 1 | |||
| Uncultured Sarcosomataceae clone (EF023269)/Amastigomonas mutalitis (AY050182) | 78.3/77.1 | 1 | 1 | |||
| Cercozoa | 39 | 14 | 60 | 24 | 4 | |
| Athalamea environmental sample clone (EF024169)/soil flagellate AND25 (AY965868) | 91.1/88.5 | 1 | 2 | |||
| Athalamea environmental sample clone (EF024169)/Hedriocystis reticulata (AY305010) | 90.3/85.0 | 1 | 1 | |||
| Athalamea environmental sample clone (EF024169)/Exuviaella pusilla (DQ388459) | 81.4/81.0 | 1 | 1 | |||
| Cercomonas longicauda (AY496047, AF411270, AY496047) | 96.3, 94.6, 91.9 | 3 | 4 | 1 | ||
| Cercomonas metabolicus (DQ211597) | 97.5, 95.6, 94.8 | 3 | 6 | |||
| Cercomonas sp. (AF534712) | 95.2, 95.0, 80.6 | 3 | 3 | 5 | ||
| Cercomonadida environmental sample clone (EF024293 and EF024163)/soil flagellate AND24 (AY965867) | 98.1, 93.4/94.1, 92.6 | 2 | 1 | 1 | ||
| Cercomonadidae environmental sample clone (EF024692)/Cercomonas sp. (AF411266) | 95.1/95.0 | 1 | 1 | |||
| Cercomonadida environmental sample clone (EF024163)/Masisteria marina strain DFS1 (AF174371) | 87.4/84.6 | 1 | 1 | |||
| Dimorpha-like sp. strain ATCC 50522 (AF411283) | 93.4 | 1 | 10 | |||
| Dodomorpha sp. strain HFCC57 (DQ211596) | 99.4 | 1 | 24 | |||
| Ebria triparrtita (DQ303922) | 88.2 | 1 | 3 | |||
| Pseudodiffllugia cf. gracilis (AJ418794) | 88.7 | 1 | 1 | |||
| Trachelocorythion pulchellum (AJ418789) | 76.5 | 1 | 13 | |||
| Uncultured Banisveld eukaryote clone (EU091827)/soil flagellate AND21 (AY965866) | 99.8/99.1 | 1 | 1 | |||
| Uncultured cercozoan sample clone (EF023523)/Cercomonadida environmental sample clone (EF024163) | 92.6/91.9 | 1 | 2 | |||
| Uncultured cercozoan clone (AY620301)/Cercomonas sp. (AF411266) | 98.0/91.4 | 1 | 1 | |||
| Uncultured cercozoan clone (AY620269)/Massisteria marina (AF174373) | 89.1/87.4 | 1 | 1 | |||
| Uncultured cercozoan clone (AY821946)/Pseudodiffllugia cf. gracilis (AJ418794) | 90.0/87.5 | 1 | 2 | |||
| Uncultured cercozoan partial 18S rRNA gene (AM114807)/soil flagellate AND24 (AY965867) | 99.0/93.0 | 1 | 3 | |||
| Uncultured cercozoan clone (AY620268)/Cercomonas sp. (AF411266) | 97.9/89.1 | 1 | 1 | |||
| Uncultured eukaryote clone (AY082981)/Ebria tripartita (DQ303922) | 96.1/88.0 | 1 | 1 | |||
| Uncultured eukaryote clone (EF024996)/Protaspis grandis (DQ303924) | 96.1/92.6 | 1 | 1 | |||
| Uncultured eukaryote clone (AY082993)/uncultured freshwater cercozoan (DQ243993) | 95.1/94.0 | 1 | 1 | |||
| Uncultured eukaryote clone (EF023764)/Protaspis grandis (DQ303924) | 99.4/94.0 | 1 | 1 | |||
| Uncultured freshwater cercozoan clone (DQ244000)/Cercomonadidae environmental sample clone (EF024294) | 83.3/81.4 | 1 | 1 | |||
| Uncultured freshwater cercozoan clone (DQ243992 and (DQ243993)/Cercomonas sp. (AF411271 and AF411266) | 92.6, 89.0/91.4, 88.5 | 2 | 2 | 1 | ||
| Uncultured freshwater cercozoan clone (DQ243993)/Rigidomastix-like sp. (AF411279) | 93.5/92.9 | 1 | 1 | |||
| Uncultured rhizosphere cercozoan (AJ506007)/soil flagellate AND21 (AY905866) | 78.9/74.5 | 1 | 1 | |||
| Unidentified eukaryote (AJ130856)/Lecythium sp. (AJ514867) | 97.0, 95.9/95.9, 94.4 | 2 | 3 | |||
| Choanozoa | 26 | 3 | 15 | 14 | 13 | |
| Amoebidium parasiticum strain ATCC 32708 (Y19155) | 90.5 | 1 | 1 | |||
| Codonosigidae evironmental sample clone (EF024012)/Monosiga ovata (AF271999) | 97.1/93.5 | 1 | 1 | |||
| Corallochytrium limacisporum (L42528) | 79.5 | 1 | 1 | |||
| Diaphanoeca grandis (DQ103820, AY753614, AF084234) | 92.9, 82.8, 75.7 | 3 | 3 | 3 | ||
| Eimeriidae environmental sample clone (EF024885)/Diaphanoeca grandis (AF084234) | 92.3/90.1 | 1 | 1 | |||
| Eimeriidae environmental sample clone (EF024885)/Endochytrium sp. (AY635844) | 91.9/90.2 | 1 | 1 | |||
| Eimeriidae environmental sample clone (EF023936)/Monosiga ovata (AF084230) | 94.9, 94.1, 90.2/92.1, 91.4, 88.9 | 3 | 2 | 1 | ||
| Eimeriidae environmental sample clone (EF024885)/Stephanoeca diplocostata (AF084235 and AY149899) | 91.5, 93.5/90.2, 92.8 | 2 | 7 | 1 | ||
| Ichthyophonus irregularis (AF232303 and AF232303) | 92.4, 79.4 | 2 | 2 | 1 | ||
| Nuclearia moebiusi (AF484686) | 91.8 | 1 | 1 | |||
| Rhinosporidium seeberi (AF118851) | 90.1 | 1 | 1 | |||
| Uncultured eukaryote (AB275066)/Diaphanoeca grandis (AF684234) | 90.7/89.6 | 1 | 1 | |||
| Uncultured eukaryotic picoplankton clone (AY642728)/Monosiga ovata (AF084230) | 94.8, 98.5, 90.7/94.4, 90.4 | 3 | 5 | 1 | ||
| Uncultured eukaryotic picoplankton clone (AY642707)/Stephanoeca diplocostata (AY149899) | 92.2/92.0 | 1 | 1 | |||
| Uncultured marine eukaryote clone (EF526879)/Corallochytrium limacisporum (L42528) | 90.4/89.9 | 1 | 2 | |||
| Uncultured marine eukaryote clone (EF526803 and DQ103820)/Diaphanoeca grandis (AF084234 and L10824) | 94.0, 88.7/92.4, 88.4 | 2 | 1 | 4 | ||
| Uncultured marine eukaryote clone (EF526803)/Stephanoeca diplocostata (AY149899) | 93.4/92.0 | 1 | 3 | |||
| Ciliophora | 29 | 7 | 4 | 17 | 21 | |
| Dexitrichides pangi (AY212805) | 91.2 | 1 | 2 | |||
| Heliophrya erhardi (AY007445) | 86.3, 85.6 | 2 | 5 | |||
| Hemiophrys macrostoma (AY102173) | 82.6 | 1 | 1 | |||
| Hemiophrys procera (AY102175) | 98.1, 96.8 | 2 | 2 | |||
| Holosticha diademata (DQ059583) | 98.7, 96.7 | 2 | 2 | |||
| Oxytrichidae environmental sample clone (EF024903)/Gonostomum namibiense (AY498655) | 97.5/96.9 | 1 | 1 | |||
| Parabirojimia similis (DQ503584) | 97.3 | 1 | 1 | |||
| Tokophrya lemnarum clone (AY332720) | 87.6 | 1 | 4 | |||
| Unidentified eukaryote (AJ130855)/Carchesium polypinum (AF401522) | 90.4/90.2 | 1 | 1 | |||
| Uncultured eukaryote clone (EF024996)/Dexitrichides pangi (AY212805) | 98.9, 87.1 | 1 | 1 | |||
| Unidentified eukaryote (AJ130851)/Ophrydium versatile (AF401526) | 85.9, 85.3 | 1 | 1 | |||
| Unidentified eukaryote (AJ130851)/Vorticella campanula (AF335518 and DQ662849) | 94.8, 92.4/94.6, 92.1 | 2 | 4 | |||
| Uncultured hypotrichid ciliate clone (AY821937)/Aspidisca steini (AF305625) | 88.9/87.7 | 1 | 2 | |||
| Uncultured marine eukaryote clone (DQ103847)/uncultured ciliate (AM114813) | 77.0/76.2 | 1 | 3 | |||
| Uncultured marine eukaryote clone (EF526916)/Plagiopyliella pacifica (AY541685) | 94.5/94.5 | 1 | 2 | |||
| Uroleptus gallina (AF164130) | 87.6 | 1 | 1 | |||
| Vorticella campanula (DQ662849 and AF335518) | 99.2, 98.4, 93.6, 95.2, 93.4, 91.6 | 6 | 1 | 7 | ||
| Vorticella fusca (DQ190468) | 99.3 | 1 | 1 | |||
| Vorticella sp. strain JCC-2006-4 (DQ868349) | 96.6 | 1 | 5 | 1 | ||
| Zoothamnium niveum (DQ868350) | 94.8 | 1 | 1 | |||
| Euglenozoa | 13 | 17 | 1 | 2 | 0 | |
| Bodo saltans (DQ207571); (AY490229) | 95.6, 77.2 | 2 | 2 | |||
| Neobodo designis (AY753616, AY753616, DQ207583) | 97.4, 94.3, 93.0, 91.1, 88.3, 87.7 | 6 | 10 | 1 | ||
| Petalomonas cantuscygni CCAP 1259/1 (AF386635) | 85.8 | 1 | 1 | |||
| Rhynchomonas nasuta (DQ207595 and AY425023) | 98.1; 85.4 | 2 | 4 | |||
| Uncultured eukaryote clone (AY753980)/Bodo saltans (AY490232) | 94.5/93.6 | 1 | 1 | |||
| Uncultured eukaryote clone (EF100316)/Petalomonas cantuscygni (U84731) | 80.5/78.5 | 1 | 1 | |||
| Myzozoa | 5 | 0 | 1 | 11 | 1 | |
| Pseudoperkinsus tapetis (AB300505) | 86.8 | 1 | 1 | |||
| Uncultured alveolate clone (AF372776)/Corallochytrium limacisporum (L42528) | 87.8/78.9 | 1 | 1 | |||
| Uncultured eukaryote clone (EF100258)/uncultured alveolate clone (AF372776) | 97.3/91.0 | 1 | 9 | |||
| Uncultured eukaryote clone (EF100258)/Colpodella pontica (AY078092) | 98.1/77.3 | 1 | 1 | |||
| Uncultured marine eukaryote clone (DQ103862)/Peridinium wierzejskii (AY443018) | 97.1/96.7 | 1 | 1 | |||
| Stramenopiles | 5 | 0 | 4 | 1 | 2 | |
| Aphanomyces invadans (DQ403202 and AF396684) | 98.7, 98.4 | 2 | 3 | |||
| Hyphochytrium catenoides (AF163294) | 97.3 | 1 | 2 | |||
| Paraphysomonas foraminifera (AB022864) | 98.5 | 1 | 1 | |||
| Rhizidiomyces apophysatus (AF163295) | 98.3 | 1 | 1 | |||
| Total | 127 | 44 | 89 | 71 | 49 | |
Most closely related sequence/most closely related genus or species. More than one accession number indicates that more than one OTU had the highest similarity to the same genus or species, but to different sequence types.
Data are totals for the three analyzed biofilm samples.
Occurrence of protozoan hosts and pathogenic free-living protozoa in treated water and biofilms.
All samples of treated water, water from distribution systems, and biofilms for both supplies were analyzed for the presence of the L. pneumophila host Hartmannella vermiformis, using quantitative PCR (38). Inhibition of PCR amplification was not observed in any of the samples. H. vermiformis was detected in four of the seven samples of TW-A, at concentrations between 0.49 and 29.3 cells liter−1 (median, 1 cell liter−1), but was not detected in any of the DW-A samples or in the BF-A samples. Two of the seven samples of TW-B were positive for H. vermiformis, both at a concentration of 1.5 cells liter−1, and one (of eight) biofilm sample was positive for H. vermiformis, at a concentration of 4.3 cells per 10 cm2. The organism was detected in all DW-B samples, at concentrations between 2.3 and 815 cells liter−1, and concentrations in July (median, 70 cells liter−1) were significantly (P < 0.05) higher than the concentrations in November (median, 4 cells liter−1). H. vermiformis was not detected in the clone libraries. These observations demonstrated that this organism was commonly present but constituted a minor fraction of the protozoan community. One OTU of the clone library of BF-B2 showed the highest similarity (77.9%) to a sequence belonging to the family of Hartmannellidae (Table 4). A total of 2.2% of the sequences representing free-living protozoa obtained from supply A showed highest similarity (85.9% to 89.3%) to species within the genus Acanthamoeba. Several Acanthamoeba spp. can serve as hosts for L. pneumophila and other undesired bacteria (24). One OTU (0.8%) obtained from TW-B showed the highest similarity to Echinamoeba thermarum (85.7%), a potential host for L. pneumophila. One OTU had the highest similarity to Acanthamoeba polyphaga, a potential pathogen (Table 4).
DISCUSSION
Analytical procedures.
To our knowledge, primers for the amplification of all free-living protozoa included in public databases are not available. Therefore, we selected 18S rRNA gene primers amplifying most, but not all, eukaryotic organisms represented in public databases. Two genera serving as hosts for L. pneumophila, viz., Naegleria spp. and Vahlkampfia spp., were not amplified with these primers. Recently, a primer set for vahlkampfiid amoeba has been developed for direct detection of Acanthamoeba spp., Naegleria spp., and Vahlkampfia spp. (13).
The variation in the T-RFLP fingerprints of treated water and biofilms exceeded the reproducibility of the T-RFLP method, demonstrating differences in the involved eukaryotic communities. However, a limitation of the T-RFLP method is that similar fragment lengths may represent different sequences, implying that the diversity in the sample may be higher than the number of observed fragments. The use of clone libraries to study the diversity in eukaryotic communities and the estimation of diversity with the Chao1 index (11) also have a few limitations. In most eukaryotes, 18S rRNA genes are organized in tandem repeat units (41), and the copy number differs significantly by genus, e.g., H. vermiformis has about 1,330 copies per cell (38) and Acanthamoeba spp. have about 600 copies per cell (9). The clone libraries from the treated water and biofilm samples were constructed using a fraction (5% to 6.7%) of the isolated DNA, and therefore only organisms with more than 15 to 20 copies of the 18S rRNA gene per cell could be represented in the clone libraries. Hence, the composition of the clone libraries does not exactly reflect the composition of the involved eukaryotic communities. The effect of copy number is most pronounced with multicellular eukaryotes, containing more DNA (copies) than unicellular eukaryotes, as demonstrated in the clone libraries of BF-B5 and BF-B6, with an OTU with >50% of the clone sequences representing metazoa (Table 2). Furthermore, only the predominating sequences were analyzed (Table 3).
Identification of obtained partial 18S rRNA gene sequences.
Similarity percentages at the 18S rRNA gene level have not been published for members of eukaryotic genera and species. For the genera of free-living protozoa most closely related to those observed in this study, similarities between 76% and 92% were derived from the 18S rRNA gene sequences in the SILVA database. On the species level, similarities between 86.6% and 99.7% were calculated for sequences of a number of protozoan species most closely related to those collected in this study. Morphologically well-defined ciliate species vary highly at the small-subunit rRNA sequence level (30, 67). Hence, genera and species of many free-living protozoa may show relatively high levels of sequence diversity in the 18S rRNA gene. Therefore, with the division of sequences into OTUs with 99% similarity, almost all different species can be distinguished. The large proportion of sequences with relatively low similarity percentages to sequences included in the databases further indicates that many eukaryotic organisms in freshwater and marine environments are not yet described (5, 15, 46, 67, 70, 79). A total of 32 OTUs showed the highest similarity to a specific eukaryote but clustered in the phylogenic tree with another group of eukaryotic organisms. We used the information on highest similarity (by BLAST search) for these OTUs for identification. These observations demonstrate that identification of freshwater protozoa is limited by the currently available database, but the large variety of sequences retrieved in the present study will facilitate further investigations of free-living protozoan communities in water supplies.
Host protozoa for Legionella spp. and pathogenic free-living protozoa.
H. vermiformis, a commonly observed protozoan host for L. pneumophila (17, 39, 80), was detected in both supplies. This protozoan has also been observed in treated groundwater in Germany (34, 47), using culture methods, in drinking water supplies (47, 70), in warm water supplies (54, 69), and in surface water (38, 47, 70), demonstrating its ubiquitous presence in the freshwater environment. However, H. vermiformis was not a predominant protozoan in the eukaryotic communities in any of the samples in this study for which clone libraries were prepared. In the distributed water, H. vermiformis was detected only in supply B, with higher concentrations in the summer than in the autumn. The presence of H. vermiformis in supply B is associated with an elevated level of active biomass and a high level of NOM.
A total of 2.3% of the protozoan sequences retrieved from supply A and 6.7% of the protozoan sequences of supply B had highest similarities to genera with one or more protozoan species described as hosts for L. pneumophila. Water temperatures in supplies A and B were below 20°C and thus were too low for growth of L. pneumophila (32), but uncultured Legionella species, including Legionella-like amoeba pathogens (82), can multiply in this temperature range in water supplies. A number of the detected free-living protozoa can probably serve as hosts for these uncultured Legionella bacteria. At elevated temperatures in warm water installations, H. vermiformis and Acanthamoeba spp. are available to promote the growth of L. pneumophila and other undesired bacteria (24, 56, 80). Acanthamoeba spp. (12, 31, 48) and H. vermiformis (1) have also been identified as opportunistic human pathogens, but it is unclear whether the sequences related to such species represent organisms with pathogenic characteristics.
Fungi, metazoa, protophyta, and cellular plants.
Fungi, metazoa, protophyta, and cellular plants were detected in the clone libraries of nearly all samples (Table 2). Fungi (25) and metazoa (75) can multiply in water treatment and distribution systems (4). Some of the OTUs had highest similarities to fungi, e.g., Aspergillus spp., Fusarium spp., and Cladosporium spp., which have also been observed in drinking water in Slovakia (21), Norway (25), and Germany (23). A few OTUs had highest similarities (>99%) to pathogenic fungi, e.g., Candida albicans (14), but it is not possible to determine whether the obtained partial sequences represent pathogenic organisms.
Metazoa such as nematodes and cnidarians (e.g., freshwater jellyfish [74]) are common inhabitants of treated water in distribution systems and play a role in the food chain (4, 45, 46, 67, 75). None of the sequences obtained in the present study were related to pathogenic metazoa.
Four OTUs (1.8%) clustered with the Cryptophyta, which contains a large number of mixotrophic species (51), but identification of these protophyta is limited by the currently available database. The OTU clustering with the family of grasses probably originated from a contamination with pollen via the air during sampling or sample treatment, although it was not observed in the negative control.
Eukaryotic diversity in supplies A and B.
The concentration of NOM in treated water of supply A (<0.5 ppm of C) was much lower than that in supply B (7.9 ppm of C) (Table 1). This difference is reflected in the concentrations of active biomass measured as ATP in these water types, viz., < 1 ng liter−1 for TW-A and 10.6 g liter−1 for TW-B, confirming the ultraoligotrophic nature of water type A. PCR-based identification methods can detect more variation at a low DNA concentration than at a high concentration (60). Indeed, the fingerprints of TW-A showed more variation than the fingerprints of TW-B (Fig. 1), but overall, the total number of OTUs observed in the clone libraries of supply A was not significantly different from the number observed in the clone libraries of supply B (Table 3).
The coverage index of the clone libraries for all eukaryotes was 40% based on 99% similarity between the sequences within one OTU and 37% when OTUs were based on 97% similarity (Tables 3 and 4). These coverage indexes are low in comparison with the values derived for the communities of small eukaryotes in an anaerobic aquifer (66%) (8) and in a mesotrophic lake (91%) (40) but higher than the value (22%) reported for eukaryotes in a suboxic and an oxid lake in France (67).
Only eight OTUs were observed in more than one biofilm sample from supply A, and only two OTUs were obtained from more than one biofilm sample from supply B. Obviously, differences in environmental conditions for biofilms sampled at different locations within the distribution system promoted the growth of different types of eukaryotes. Still, the T-RFLP fingerprints of the communities of eukaryotes clustered within each supply (Fig. 1).
Diversity in free-living protozoa in supplies A and B.
Free-living protozoa feed on bacteria, other protozoa, and detritus and play an important role in the transfer of energy through the trophic levels (7, 50, 76). Due to their rapid response to environmental changes, free-living protozoa have been used as water quality indicators, and the diversity in free-living protozoa generally increases with improved water quality (29, 34-36, 45). Consequently, differences in the protozoan communities in the two supplies can be attributed to differences in raw water composition, treatment processes, and conditions in the distribution system (hydraulics, materials, and residence time). However, the estimated diversity in the free-living protozoa was not significantly different between the two supplies (Table 3).
Based on morphological studies, free-living protozoa have been divided into flagellates, ciliates, and amoebae (50, 76). Table 4 shows that representatives of these groups were identified in the different samples by molecular techniques. Microscopic studies have shown that sand filters operating under similar conditions in water treatment systems harbor different numbers and types of ciliates and amoebae (43, 70). Microscopic analysis also showed that flagellates predominated (93%) in drinking water in an experimental distribution system with pipes of concrete and polyvinyl chloride supplied for 4 months with treated water with a dissolved organic carbon concentration of 2.3 mg C liter−1 (64). However, in the biofilm, no flagellates were detected, but ciliates (52%) and amoebae (48%) were observed. In the present study, many OTUs observed in the biofilms had highest similarities to flagellates, including Cercomonas spp., Bodo saltans, and Rhynchomans nasuta (Table 4) (51).
Twelve OTUs of the clone libraries for supply A and 26 OTUs of the clone libraries for supply B had highest similarities to genera that have been used as indicator organisms in the saprobic index for organic pollution (20, 66). A total of 87% of these organisms belong to genera that indicate moderate pollution at a high dissolved oxygen content, e.g., Hemiophrys, Rhynchomonas, and Vorticella (Table 4). However, elucidation of the relationship between environmental conditions in water treatment and distribution systems, e.g., water composition, and the occurrence of free-living protozoa is not possible because (i) the communities are highly diverse, (ii) species and genus boundaries of eukaryotes are still unclear, (iii) little information is available about the growth conditions of free-living protozoa, and (iv) the diversity in the clone libraries is not proportional to the diversity in the protozoa in the samples.
In conclusion, in two groundwater supplies with a large difference in the concentration of NOM, highly diverse communities of free-living protozoa were observed. These communities differed between locations within the distribution system. Hence, a large variety of microhabitats, defined by as yet unknown environmental conditions, exist within water supplies and affect the eukaryotic composition. Furthermore, high levels of NOM and active biomass in treated water corresponded with elevated concentrations of H. vermiformis. Consequently, quantitative detection of selected protozoa by molecular techniques may be promising in elucidating the relationship between drinking water quality and the presence of specific organisms.
Acknowledgments
This study was financed by Delft Cluster project CT 06.10 and by the water supply companies in The Netherlands in the framework of the Joint Research Program.
We thank Leo Heijnen, Jörg Peplies, and Paul Baggelaar for helping with phylogenic and statistical analysis. We thank Wim Hoogenboezem and Johannes Hackstein for valuable discussions and the staff of the microbiologic laboratory of KWR, Watercycle Research Institute, for skillful assistance with the experiments.
Footnotes
Published ahead of print on 22 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aitken, D., J. Hay, F. B. Kinnear, C. M. Kirkness, W. R. Lee, and D. V. Seal. 1996. Amebic keratitis in a wearer of disposable contact lenses due to a mixed Vahlkampfia and Hartmannella infection. Ophthalmology 103:485-494. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amblard, C., G. Bourdier, J.-F. Carrias, N. Maurin, and C. Quiblier. 1996. Seasonal evolution of microbial community structure in a drinking water reservoir. Water Res. 30:613-624. [Google Scholar]
- 4.Anonymous. 1989. Problem organisms in water: identification and treatment. American Water Works Association, Denver, CO.
- 5.Bass, D., and T. Cavalier-Smith. 2004. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int. J. Syst. Evol. Microbiol. 54:2393-2404. [DOI] [PubMed] [Google Scholar]
- 6.Block, J. C., K. Haudidier, J. L. Paquin, J. Miazga, and Y. Lévi. 1993. Biofilm accumulation in drinking water distribution systems. Biofouling 6:333-343. [Google Scholar]
- 7.Bloem, J., M. B. Bar-Gilissen, and T. E. Cappenberg. 1986. Fixation, counting, and manipulation of heterotrophic nanoflagellates. Adv. Appl. Microbiol. 52:1266-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brad, T., M. Braster, B. M. van Breukelen, N. M. van Straalen, and W. F. Roling. 2008. Eukaryotic diversity in an anaerobic aquifer polluted with landfill leachate. Appl. Environ. Microbiol. 74:3959-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byers, T. J., E. R. Hugo, and V. J. Stewart. 1990. Genes of Acanthamoeba: DNA, RNA and protein sequences. J. Protozool. 37:17S-25S. [DOI] [PubMed] [Google Scholar]
- 10.Cavalier-Smith, T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52:297-354. [DOI] [PubMed] [Google Scholar]
- 11.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 12.Culbertson, C. G. 1961. Pathogenic Acanthamoeba (Hartmannella). Am. J. Clin. Pathol. 35:195-202. [DOI] [PubMed] [Google Scholar]
- 13.De Jonckheere, J. F., and S. Brown. 2005. The identification of vahlkampfiid amoebae by ITS sequencing. Protist 156:89-96. [DOI] [PubMed] [Google Scholar]
- 14.Dixon, D. M., M. M. McNeil, M. L. Cohen, B. G. Gellin, and J. R. La Montagne. 1996. Fungal infections: a growing threat. Public Health Rep. 111:226-235. [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein, S., and P. López-García. 2008. “Missing” protists: a molecular prospective. Biodivers. Conserv. 17:261-276. [Google Scholar]
- 16.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 17.Fields, B. S., G. N. Sanden, J. M. Barbaree, W. E. Morrill, R. M. Wadowsky, E. H. White, and J. C. Feeley. 1989. Intracellular multiplication of Legionella pneumophila in amoebae isolated from hospital hot water tanks. Curr. Microbiol. 18:131-137. [Google Scholar]
- 18.Fields, B. S., E. B. Shotts, Jr., J. C. Feeley, G. W. Gorman, and W. T. Martin. 1984. Proliferation of Legionella pneumophila as an intracellular parasite of the ciliated protozoan Tetrahymena pyriformis. Appl. Environ. Microbiol. 47:467-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foissner, W. 2008. Protist diversity and distribution: some basic considerations. Biodivers. Conserv. 17:235-242. [Google Scholar]
- 20.Foissner, W. 1988. Taxonomic and nomenclatural revision of Sládeček's list of ciliates (Protozoa: Ciliophora) as indicators of water quality. Hydrobiologia 166:1-64. [Google Scholar]
- 21.Franková, E., and M. Horecká. 1995. Filamentous soil fungi and unidentified bacteria in drinking water from wells and water mains near Bratislava. Microbiol. Res. 150:311-313. [DOI] [PubMed] [Google Scholar]
- 22.Good, I. J. 1953. The population frequencies of species and the estimation to the population parameters. Biometrika 40:237-264. [Google Scholar]
- 23.Gottlich, E., W. van der Lubbe, B. Lange, S. Fiedler, I. Melchert, M. Reifenrath, H. C. Flemming, and S. de Hoog. 2002. Fungal flora in groundwater-derived public drinking water. Int. J. Hyg. Environ. Health 205:269-279. [DOI] [PubMed] [Google Scholar]
- 24.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hageskal, G., A. K. Knutsen, P. Gaustad, G. S. de Hoog, and I. Skaar. 2006. Diversity and significance of mold species in Norwegian drinking water. Appl. Environ. Microbiol. 72:7586-7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, M. W., and M. G. Hofle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inglis, T. J., P. Rigby, T. A. Robertson, N. S. Dutton, M. Henderson, and B. J. Chang. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect. Immun. 68:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, J. G., and Y. F. Shen. 2007. Development of the microbial communities in Lake Donghu in relation to water quality. Environ. Monit. Assess. 127:227-236. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, M. D., T. Tengs, D. W. Oldach, C. F. Delwiche, and D. K. Stoecker. 2004. Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist 155:347-359. [DOI] [PubMed] [Google Scholar]
- 31.Jones, D. B., G. S. Visvesvara, and N. M. Robinson. 1975. Acanthamoeba polyphaga keratitis and Acanthamoeba uveitis associated with fatal meningoencephalitis. Trans. Ophthalmol. Soc. U. K. 95:221-232. [PubMed] [Google Scholar]
- 32.Katz, S. M., and J. M. Hammel. 1987. The effect of drying, heat, and pH on the survival of Legionella pneumophila. Ann. Clin. Lab. Sci. 17:150-156. [PubMed] [Google Scholar]
- 33.Kikuhara, H., M. Ogawa, H. Miyamoto, Y. Nikaido, and S. Yoshida. 1994. Intracellular multiplication of Legionella pneumophila in Tetrahymena thermophila. J. UOEH 16:263-275. [DOI] [PubMed] [Google Scholar]
- 34.Kolkwitz, R., and M. Marsson. 1902. Grundsätze für die biologische Beurteilung des wassers nach seiner Flora und Fauna. Mitt. Prüfungsanst. Wasserversorg. Abwasserreinig. 1:33-72. [Google Scholar]
- 35.Kolkwitz, R., and M. Marsson. 1908. Ökologie der pflanzlichen Saprobien. Ber. Bot. Ges. 26A:505-519. [Google Scholar]
- 36.Kolkwitz, R., and M. Marsson. 1909. Ökologie der tierischen saprobien. Int. Rev. Hydrobiol. 2:126-152. [Google Scholar]
- 37.Krishna-Prasad, B., and S. K. Gupta. 1978. Preliminary report on engulfment and retention of mycobacteria by trophozoites of axenically grown Acanthamoeba casterllanii Douglas. Curr. Sci. 47:245-247. [Google Scholar]
- 38.Kuiper, M. W., R. M. Valster, B. A. Wullings, H. Boonstra, H. Smidt, and D. van der Kooij. 2006. Quantitative detection of the free-living amoeba Hartmannella vermiformis in surface water by using real-time PCR. Appl. Environ. Microbiol. 72:5750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuiper, M. W., B. A. Wullings, A. D. Akkermans, R. R. Beumer, and D. van der Kooij. 2004. Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl. Environ. Microbiol. 70:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lèpere, C., I. Domaizon, and D. Debroas. 2008. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl. Environ. Microbiol. 74:2940-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, E. O., and I. B. Dawid. 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49:727-764. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madoni, P., D. Davoli, G. Cavagnoli, A. Cucchi, M. Pedroni, and F. Rossi. 2000. Microfauna and filamentous microflora in biological filters for tap water production. Water Res. 34:3561-3572. [Google Scholar]
- 44.Magic-Knezev, A., and D. van der Kooij. 2004. Optimisation and significance of ATP analysis for measuring active biomass in granular activated carbon filters used in water treatment. Water Res. 38:3971-3979. [DOI] [PubMed] [Google Scholar]
- 45.Margalef, R. 1969. Diversity and stability: a practical proposal and a model of interdependence. Brookhaven Symp. Biol. 22:25-37. [PubMed] [Google Scholar]
- 46.Massana, R., J. Castresana, V. Balague, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedros-Alio. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel, R., R. Hoffmann, A. Giese, and K. D. Muller. 1995. Untersuchung von drei Grundwasserwerken auf Vorkommen von Acanthamoeben, Naeglerien und anderen freilebenden Amöben. Acta Hydrochim. Hydrobiol. 23:202-211. [Google Scholar]
- 48.Nagington, J., P. G. Watson, T. J. Playfair, J. McGill, B. R. Jones, and A. D. M. G. Steel. 1974. Amoebic infection of the eye. Lancet ii:1537-1540. [DOI] [PubMed] [Google Scholar]
- 49.Newsome, A. L., R. L. Baker, R. D. Miller, and R. R. Arnold. 1985. Interactions between Naegleria fowleri and Legionella pneumophila. Infect. Immun. 50:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parry, J. D. 2004. Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54:167-196. [DOI] [PubMed] [Google Scholar]
- 51.Patterson, D. J. 1992. Free-living freshwater protozoa. A colour guide. Mansons Publishing Ltd., London, United Kingdom.
- 52.Prüsse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. 2007. SILVA; a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers, J., A. Dowsett, P. Dennis, J. Lee, and C. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohr, U., S. Weber, R. Michel, F. Selenka, and M. Wilhelm. 1998. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rønn, R., A. E. McCaig, B. S. Griffiths, and J. I. Prosser. 2002. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl. Environ. Microbiol. 68:6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 57.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanden, G. N., W. E. Morrill, B. S. Fields, R. F. Breiman, and J. M. Barbaree. 1992. Incubation of water samples containing amoebae improves detection of legionellae by the culture method. Appl. Environ. Microbiol. 58:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwarzenbach, K., J. Enkerli, and F. Widmer. 2007. Objective criteria to assess representativity of soil fungal community profiles. J. Microbiol. Methods 68:358-366. [DOI] [PubMed] [Google Scholar]
- 61.Servais, P., P. Laurent, and G. Randon. 1995. Comparison of the bacterial dynamics in various French distribution systems. J. Water Supply Res. Technol. Aqua 44:10-17. [Google Scholar]
- 62.Shadrach, W. S., K. Rydzewski, U. Laube, G. Holland, M. Ozel, A. F. Kiderlen, and A. Flieger. 2005. Balamuthia mandrillaris, free-living ameba and opportunistic agent of encephalitis, is a potential host for Legionella pneumophila bacteria. Appl. Environ. Microbiol. 71:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherr, E. B., and B. F. Sherr. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie van Leeuwenhoek 81:293-308. [DOI] [PubMed] [Google Scholar]
- 64.Sibille, I., T. Sime-Ngando, L. Mathieu, and J. C. Block. 1998. Protozoan bacterivory and Escherichia coli survival in drinking water distribution systems. Appl. Environ. Microbiol. 64:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Šimek, K., and T. H. Chrzanowski. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl. Environ. Microbiol. 58:3715-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sládeček, V. 1973. System of water quality from the biological point of view. Arch. Hydrobiol. Beith. Ergebn. Limnol. 7:1-218. [Google Scholar]
- 67.Šlapeta, J., D. Moreira, and P. López-Garcia. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. Biol. Sci. 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sogin, M. L., and J. H. Gunderson. 1987. Structural diversity of eukaryotic small subunit ribosomal RNAs. Evolutionary implications. Ann. N. Y. Acad. Sci. 503:125-139. [DOI] [PubMed] [Google Scholar]
- 69.Thomas, V., K. Herrera-Rimann, D. S. Blanc, and G. Greub. 2006. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas, V., J. F. Loret, M. Jousset, and G. Greub. 2008. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environ. Microbiol. 10:2728-2745. [DOI] [PubMed] [Google Scholar]
- 71.Valster, R., B. Wullings, S. Voost, G. Bakker, H. Smidt, and D. van der Kooij. 2006. Detection and identification of free-living protozoa present in drinking water, p. 427-430. In N. P. Cianciotto et al. (ed.), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC.
- 72.Van der Kooij, D. 1999. Potential for biofilm development in drinking water distribution systems. J. Appl. Microbiol. Symp. 85:39S-44S. [DOI] [PubMed] [Google Scholar]
- 73.Van der Kooij, D., J. H. M. van Lieverloo, J. Schellart, and P. Hiemstra. 1999. Maintaining quality without a disinfectant residual. J. Am. Water Works Assoc. 91:55-64. [Google Scholar]
- 74.Van der Land, J., and R. Ates. 1999. Kwallen uit de kraan. Natura 6:166-168. [Google Scholar]
- 75.Van Lieverloo, J. H. M., D. van der Kooij, and W. Hoogenboezem. 2002. Invertebrates and protozoa (free-living) in drinking water distribution systems, p. 1718-1733. In G. Bitton (ed.), Encyclopedia of environmental microbiology. Wiley, New York, NY.
- 76.Vickerman, K. 1992. The diversity and ecological significance of protozoa. Biodivers. Conserv. 1:334-341. [Google Scholar]
- 77.Visvesvara, G. S., F. L. Schuster, and A. J. Martinez. 1993. Balamuthia mandrillaris, n. g., n. sp., agent of amebic meningoencephalitis in humans and other animals. J. Eukaryot. Microbiol. 40:504-514. [DOI] [PubMed] [Google Scholar]
- 78.Volk, C. J., and M. W. LeChevallier. 1999. Impacts of the reduction of nutrient levels on bacterial water quality in distribution systems. Appl. Environ. Microbiol. 65:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Von der Heyden, S., E. E. Chao, K. Vickerman, and T. Cavalier-Smith. 2004. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. J. Eukaryot. Microbiol. 51:402-416. [DOI] [PubMed] [Google Scholar]
- 80.Wadowsky, R. M., L. J. Butler, M. K. Cook, S. M. Verma, M. A. Paul, B. S. Fields, G. Keleti, J. L. Sykora, and R. B. Yee. 1988. Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl. Environ. Microbiol. 54:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willaert, E. 1974. Primary amoebic meningo-encephalitis. A selected bibliography and tabular survey of cases. Ann. Soc. Belg. Med. Trop. 54:429-440. [PubMed] [Google Scholar]
- 82.Wullings, B. A., and D. van der Kooij. 2006. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15 degrees C. Appl. Environ. Microbiol. 72:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]