Abstract
The cellulase and hemicellulase genes of the filamentous fungus Trichoderma reesei have been shown to be under carbon catabolite repression mediated by the regulatory gene cre1. In this study, strains were constructed in which the cre1 gene was either completely removed or replaced by a truncated mutant variant, cre1-1, found previously in the Rut-C30 mutant strain with enhanced enzyme production capability. The T. reesei transformants with either deletion or truncation of cre1 had clearly altered colony morphology compared with the parental strains, forming smaller colonies and fewer aerial hyphae and spores. Liquid cultures in a medium with glucose as a carbon source showed that the transformants were derepressed in cellulase and hemicellulase production. Interestingly, they also produced significantly elevated levels of these hydrolytic enzymes in fermentations carried out in a medium inducing the hydrolase genes. This suggests that cre1 acts as a modulator of cellulase and hemicellulase gene expression under both noninducing and inducing conditions. There was no phenotypic difference between the Δcre1 and cre1-1 mutant strains in any of the experiments done, indicating that the cre1-1 gene is practically a null allele. The results of this work indicate that cre1 is a valid target gene in strain engineering for improved enzyme production in T. reesei.
The filamentous fungus Trichoderma reesei (Hypocrea jecorina) produces large amounts of extracellular enzymes. The majority of the secreted proteins are various plant polymer-degrading enzymes; the most abundant of these enzymes are the cellobiohydrolases and endoglucanases that act synergistically to break down cellulose. This fungus has been used as a production host for various industrial enzymes, including products tailored for textile, feed, food, and pulp and paper applications (6, 10). It has been reported that protein production levels in the industrial T. reesei process exceed 100 g/liter (7).
The major cellulase and hemicellulase genes are regulated in a coordinate manner by the carbon source available (2, 9, 14). Cellulose and other plant materials and other substances (for example, lactose) induce the expression of cellulase and hemicellulase genes, while glucose acts as a repressing carbon source. Several genes coding for regulators of cellulase and hemicellulase expression have been isolated. These include CREI mediating carbon catabolite repression, the repressor ACEI, the activator ACEII, the CCAAT binding complex Hap2/3/5 (reviewed in references 2, 17, and 27) and the activator XYRI (29). The CREI protein has sequence similarity with other fungal proteins mediating glucose repression, such as Aspergillus nidulans CREA (8) and Saccharomyces cerevisiae MIG1 and RGR1 (22). In T. reesei, glucose repression has been shown to occur upon binding of CREI to specific sequences in the cbh1 promoter (13). A mutant cre1 gene (cre1-1) encoding a truncated form of CREI has been isolated from the hypercellulolytic T. reesei strain Rut-C30, which is capable of cellulase and hemicellulase production on glucose-containing media. Further evidence for the function of CREI in glucose repression was obtained by complementation of the cre1-1 mutation of Rut-C30 by the wild-type cre1 gene, which restored the glucose-repressed phenotype of the strain (15).
In this paper, we wanted to address three questions. (i) What is the effect of cre1 mutations in the wild-type background? (ii) Is cre1-1 a null mutation? (iii) Can enzyme production be further improved by cre1 deletion in an industrial production strain improved greatly by mutagenesis and screening programs? Therefore, we introduced cre1-1 allele and cre1 deletion to the wild-type strain QM6a and the cre1 deletion into the industrial strain VTT-D-80133 and studied the effects of these mutations on enzyme production.
MATERIALS AND METHODS
Strains.
T. reesei natural isolate strain QM6a (19) and the hypercellulolytic mutant strain VTT-D-80133 (4) originating from strain QM9414 were used as parental strains. The Δcre1 and cre1-1 transformants of these strains are listed in Table 1. The T. reesei strains were grown and sporulated on potato dextrose agar slants (BD Diagnostics, Sparks, MD) at 30°C for 1 week and stored at +4°C.
TABLE 1.
T. reesei strains used in this study
Construction of T. reesei Δcre1 and cre1-1 strains.
For replacement of the wild-type cre1 gene in the genome with either the acetamidase (amdS) marker gene or with the T. reesei cre1-1 mutant gene, two plasmids were constructed. The pTNS34 plasmid, used for replacement of the cre1 gene with the amdS cassette, contains a 1.9-kb cre1 3′ flanking fragment cloned at the EcoRI site and a 2.8-kb EcoRI-EcoRV cre1 5′ flanking fragment cloned at the SalI site of p3SR2 (12). The p3SR2 plasmid carries a 5-kb genomic fragment of Aspergillus nidulans amdS gene. The 1.9-kb cre1 3′ flanking fragment was amplified with PCR using 5′-GGG GAA TTC ATA GAT GGA TAG AAA GAG TTG G-3′ as the sense oligonucleotide and 5′-GGG GAA TTC CTC ACT ATA GGG AGA CCG GCC TCG AGT TAA TTA AGC TT-3′ (EcoRI sites underlined) as the antisense oligonucleotide and using pMI-41 as a template (15), which carries the cre1 genomic fragment of the VTT-D-80133 strain.
The plasmid pTNS36 was constructed for replacement of the cre1 gene with the mutant gene cre1-1. It contains the cre1-1 coding region operably linked to the cre1 promoter, followed by the amdS cassette and cre1 3′ flanking sequence. To construct pTNS36, the following cloning steps were carried out. First, the 3.0-kb EcoRI-BstEII fragment of plasmid pMI-41 (17) was used to replace the EcoRI-BstEII fragment in pMI-42 (15), which resulted in pMI-60. Second, the 1.9-kb cre1 3′ flanking fragment amplified by PCR as described above was cloned at the EcoRI site of the amdS-containing plasmid pToC202. In the last step, an EcoRI-XhoI fragment from pMI-60 was cloned at the SalI site of pToC202.
Fungal transformations.
The linearized pTNS34 (Δcre1) plasmid and the expression cassette from pTNS36 (cre1-1) were transformed into the QM6a and VTT-D-80133 strains using acetamide selection by the method of Penttilä et al. (25). A selection of AmdS+ transformants were purified through single conidia on plates containing acetamide, and then they were screened for correct replacement of the cre1 gene using Southern analysis.
Southern and PCR analyses of cre1 loci.
Fungal DNA was isolated using the Easy DNA kit of Invitrogen (Carlsbad, CA) according to the manufacturer's instructions. Five to 10 μg of DNA was digested with PvuII, which had known recognition sequences within the coding and flanking regions of the cre1 sequence. The cleaved DNAs were run on an agarose gel and transferred to a Hybond N nylon filter (Amersham Biosciences, Uppsala, Sweden). Southern hybridizations with T. reesei cre1- and A. nidulans amdS-specific probes labeled with [α-32P]dCTP by using the Random Primed DNA labeling kit (Roche Applied Science, Indianapolis, IN) were carried out under stringent conditions in a hybridization mix with 50% formamide at 42°C (26). The filters were washed under stringent conditions in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.6])-0.1% sodium dodecyl sulfate for 15 min at room temperature and twice in 0.1× SSC-0.1% sodium dodecyl sulfate at 65°C for 45 min. The washed filters were exposed on Kodak X-Omat film (Kodak Industrie, Chalon-sur-Saone, France). For the strains chosen for further cultivations and analysis (Table 1), additional restriction digestions and Southern blotting were performed to confirm the single-copy replacement event.
Plate assays.
To study the morphology of the mutant strains, transformants and their host strains were cultivated on three different types of minimal medium (25) agar plates containing either (i) potato dextrose (BD Diagnostics, Sparks, MD), (ii) 2% glucose and 0.2% peptone (BD Diagnostics), or (iii) 2% Solka floc cellulose (James River Corporation, Berlin, NH) and 0.2% peptone as carbon sources. The experiment was done on plates containing 0.1% Triton X-100 to restrict the growth of the fungal colonies and on plates with no Triton X-100, and both plates gave similar results (data in Fig. 1 shown for the Triton X-100-containing plates). The plates were grown at 28°C for 5 days.
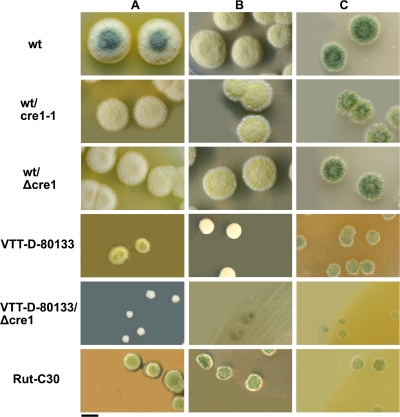
FIG. 1.
Colonies of T. reesei cre1-1 and Δcre1 transformants and their parental strains and strain Rut-C30 on potato dextrose (A), glucose-containing (B), and cellulose (C) agar plates. Bar, 1 cm.
Bioreactor cultivations.
The bioreactor cultivations were carried out at 28°C and pH 4.2 in Braun Biostat B bioreactors with a working volume of 2 liters (B. Braun, Melsungen, Germany). Cultivations were performed on glucose-containing medium and on two types of inducing media, Solka floc cellulose-based medium with solid cellulose and lactose-based liquid medium. The media in glucose-based cultivations contained the following (all ingredients shown in g liter−1): glucose, 30; (NH4)2SO4, 5; KH2PO4, 5; MgSO4·7H2O, 0.6; CaCl2, 0.6; FeSO4·7H2O, 0.005; MnSO4·H2O, 0.0016; ZnSO4·7H2O, 0.0014; and CoCl2, 0.002. The cellulose-based medium contained the following (all ingredients shown in g liter−1): Solka Floc cellulose, 30; (NH4)2SO4, 5; KH2PO4, 5; and a complex nitrogen source, 15. The lactose-based medium contained the following (all ingredients shown in g liter−1): lactose, 30; (NH4)2SO4, 5; KH2PO4, 5; and a complex nitrogen source, 30. An extract from this nitrogen source for the lactose-based medium was prepared by heating and decanting by the method of Paloheimo et al. (24). In the beginning of the cultivations, 1 ml liter−1 of structol (Schill & Seilacher Structol AG, Hamburg, Germany), an antifoam agent, was added to all the fermentations.
The bioreactors were inoculated either with spores (QM6a/QM6a-based transformants in glucose and lactose cultivations) or with 200-ml precultures grown in Erlenmeyer flasks for 48 h at 28°C and 250 rpm (QM6a/QM6a-based transformants in Solka floc cellulose cultivations and cultivations of VTT-D-80133/VTT-D-80133-based transformant). Spores from potato dextrose agar slant were suspended in 5 ml of 0.9% NaCl-0.01% Tween 80 solution, and the solution was filtered through a glass wool filter and used for inoculation. The spore concentrations were determined using a Bürker chamber (HBG Henneberg-Sander GmbH, Giessen-Luetzellinden, Germany). The precultures and lactose-based bioreactor cultivations were inoculated with spores from one well-grown confluent potato dextrose agar slant each. The glucose-based bioreactor cultivations were inoculated by 1.2 × 108 spores each.
To measure mass (dry weight), 10 ml of the culture broth from different time points was filtered through 3.0-μm and 0.45-μm-pore-size Millipore membrane filters (Millipore Oy, Espoo, Finland), washed with an equal volume of 0.7% NaCl solution, dried at 80°C for 24 h, and weighed.
Lactose, galactose, and glucose concentrations were monitored during cultivation using commercial kits (R-Biopharm AG, Darmstadt, Germany).
Analysis of proteins in culture media.
Protein and activity assays were performed from the culture supernatants after removing the mycelia by centrifugation. Soluble protein from glucose cultivations was assayed by the method of Lowry (16) using bovine serum albumin (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) as the standard. Analysis of proteins from other cultivations was performed by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA) using bovine gamma globulin (Bio-Rad) as the standard.
Xylanase activity was measured by the method described by Bailey et al. (3) using birch xylan (Carl Roth GmbH, Karlsruhe, Germany) as a substrate. The reaction time for the samples taken from the glucose cultivations was increased from 5 to 60 min. Cellulase activity was measured using two different substrates. β-1,4-Endoglucanase activity against hydroxylethylcellulose (middle viscosity 1; Fluka Chemie AG, Buchs, Switzerland) was measured by the method of Bailey and Nevalainen (4). The reaction time for the samples taken from the glucose cultivations was increased from 10 to 180 min and 0.1% bovine serum albumin was added to the reaction mixture. The combined action of two main cellulase components, cellobiohydrolase I (CBHI) and endoglucanase I (EGI), was assayed based on hydrolysis of the low-molecular-weight substrate 4-methylumbelliferyl-β-d-lactoside (MUL) (Sigma-Aldrich) and by detection of the fluorescent hydrolysis product methylumbelliferone (5).
Protease activity was measured using 2% hemoglobin (Sigma-Aldrich) as a substrate. The enzymatic reaction was performed at 40°C and pH 4.7 using 30-min reaction time. The nonhydrolyzed hemoglobin was removed from the reaction mixture by precipitation with 14% trichloroacetic acid (for 1 h) and filtering through Whatman no. 42 filter (Whatman International Ltd., Maidstone, United Kingdom). The absorbance of the filtrate was measured at 275 nm against a tyrosine standard curve. One unit of protease activity is the amount of enzyme that hydrolyzes hemoglobin in 1 minute in such an amount that the absorbance of the filtrate is similar to that of a tyrosine solution with a concentration of 1.10 μg ml−1.
Northern analysis.
For isolation of RNA, mycelia were harvested from culture media by filtration through GF/B glass microfiber filters (Whatman), washed with sterile water, and stored at −70°C. Total RNA was isolated with the Trizol reagent (Life Technologies Inc., Gaithersburg, MD). Five micrograms of RNA was glyoxylated and electrophoresed on a 1% agarose gel and blotted onto a Hybond N nylon filter (Amersham) (26). The probes used for Northern analysis were cbh1 (GenBank accession number P62694), cbh2 (P07987), egl1 (AAX28897), and xyn1 (P36218) gene fragments released from vectors by digestion. For an internal loading control, the membranes were hybridized with glyceraldehyde-3-phosphate dehydrogenase gene gpd1 (protein. ID 21248 in JGI T. reesei genome database http://genome.jgi-psf.org/Trire2/Trire2.home.html). The probes were labeled with [α-32P]dCTP by using the random primed DNA labeling kit (Roche Applied Science). Hybridizations in a mix with 50% formamide at 42°C and subsequent washing steps were performed by the method of Sambrook et al. (26). Hybridization signals were detected on phosphor screen autoradiographs by using PhosphorImager SI, quantified by using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA), and normalized by using gpd1 as a loading control.
RESULTS
Construction of T. reesei Δcre1 and cre1-1 strains.
To modify carbon catabolite repression in T. reesei, two replacement cassettes were constructed, pTNS34 for deleting the wild-type glucose repressor gene cre1 by replacing it with the amdS marker gene and pTNS36 for replacing the cre1 gene with the mutant gene cre1-1. The pTNS34 (Δcre1) and pTNS36 (cre1-1) constructions were transformed to the wild-type strain QM6a and to the hypercellulolytic mutant strain VTT-D-80133, shown to carry the wild-type cre1 gene (data not shown). Transformants were purified through single spore cultures, and Southern analysis was performed to verify that the cre1 gene was correctly replaced in the genomes with one copy of either the pTNS34 or pTNS36 deletion/expression cassette. Several QM6a- and VTT-D-80133-based transformants were obtained where the cre1 gene had been correctly deleted. Also, a number of cre1-1 transformants were obtained for strain QM6a. However, despite numerous trials, no VTT-D-80133 transformants were obtained where the wild-type cre1 gene would have been correctly replaced by the cre1-1 gene. The reason for this is not known. From the transformants obtained, two single-copy Δcre1 and cre1-1 strains were chosen for further analysis (Table 1).
Morphology of Δcre1 and cre1-1 transformants.
During the screening for correct Δcre1 and cre1-1 transformants, it was noticed that some of the transformants formed fewer aerial hyphae and spores than the parental strain did on potato dextrose agar plates that were used to obtain sporulating cultures. Such transformants were later shown to have cre1 replaced with the amdS marker (pTNS34) or cre1-1 (pTNS36) on the basis of Southern analysis. In order to study the morphology of the mutant strains in more detail, Δcre1 and cre1-1 transformants and their host strains QM6a and VTT-D-80133, and as a comparison, the natural cre1-1 mutant strain Rut-C30, were cultivated on agar plates with three different carbon sources, either potato dextrose, glucose, or cellulose.
Clear morphological differences between the strains were detected (Fig. 1). On potato dextrose agar plates (Fig. 1A), T. reesei QM6a colonies were approximately 1.5 times larger than the colonies of its Δcre1 and cre1-1 transformants. Also, the wild-type strain colonies were pigmented and sporulated, unlike the Δcre1 and cre1-1 transformants, which further indicates that the wild-type strain grows faster on this carbon source. The T. reesei VTT-D-80133 Δcre1 strain formed very small and dense colonies. These colonies (ca. 2 mm in diameter) were about 1/4 of the size of the colonies of the parental strain VTT-D-80133 and approximately 1/10 of the size of the QM6a colonies on the same carbon source. Like QM6a, VTT-D-80133 sporulated on potato dextrose agar, whereas its Δcre1 transformant did not.
On plates containing glucose (Fig. 1B) and cellulose (Fig. 1C), no substantial differences in colony size, pigmentation, or sporulation was observed between wild-type strain QM6a and its Δcre1 and cre1-1 transformants. The colonies sporulated on plates containing cellulose, but not on plates containing glucose. Strains originating from strain QM6a formed colonies two to three times larger than those of strain VTT-D-80133, its Δcre1 transformant, and Rut-C30. The colonies of the VTT-D-80133 Δcre1 transformant were about two times smaller than those of its parental strain both on glucose- and cellulose-containing plates. The Rut-C30 strain (cre1-1 mutant) sporulated on glucose-containing and potato dextrose agar plates, unlike the QM6a transformant with cre1 replaced with cre1-1. Microscopic examination did not reveal clear differences in the mycelial fine structure of the strains.
Effects of cre1 deletion and truncation on cellulase production in glucose-based cultivations.
Glucose represses the expression of cellulases and other plant material-hydrolyzing enzymes of T. reesei. To compare the effects of the two different cre1 replacements on cellulase expression and on protein production more generally, bioreactor cultivations with the wild-type strain QM6a and its Δcre1 and cre1-1 transformants were performed in glucose-containing medium.
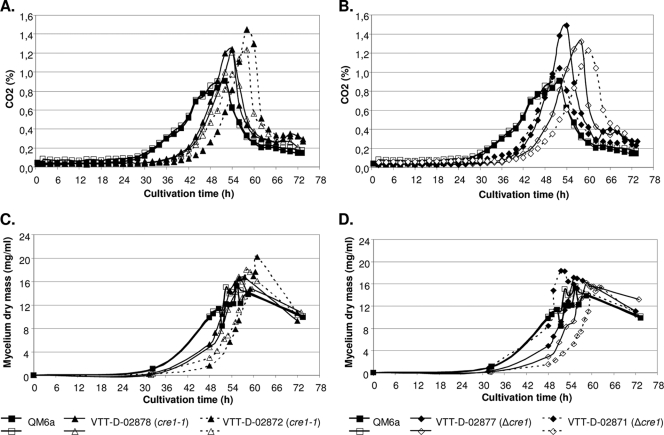
All the growth parameters indicated that the Δcre1 and cre1-1 transformants grew similarly (Fig. 2A to D). However, the growth of both types of transformants started about 10 h later than that of the host (Fig. 2A and B). The maximum weight (dry weight) of the mycelia of transformants was similar to that of the host. As expected, both Δcre1 and cre1-1 mutations resulted in derepression of cellulase and hemicellulase expression. The amounts of secreted proteins in the cultivations were somewhat variable between the parallel cultivations, but on average, the transformants produced about fourfold-higher amount of proteins into their culture supernatants than the wild-type strain QM6a did (Table 2). The cellulase activities were analyzed using MUL and hydroxyethylcellulose as substrates. MUL is specific for two major cellulases of T. reesei, the CBHI cellobiohydrolase and EGI endoglucanase, whereas activity against hydroxyethylcellulose shows the total endoglucanase activity in the samples. The parental strain QM6a produced only a very minute level of MUL activity, while an approximately 10-fold increase in the MUL activity was detected from the culture supernatants of both the Δcre1 and cre1-1 transformants. Accordingly, the total endoglucanase and xylanase activities were clearly higher for the transformants than for the parental strain. There was no difference between the Δcre1 and cre1-1 transformants in the amounts of total protein or the enzyme activities measured from the culture supernatants.
FIG. 2.
Growth of T. reesei strain QM6a and its cre1-1 and Δcre1 transformants in glucose-based medium. The concentration of CO2 (A and B) and weight of the mycelium (dry weight) (mycelium dry mass) (C and D) in each cultivation are shown. Two parallel cultivations in bioreactors were performed for each transformant and the parental strain.
TABLE 2.
Effects of cre1-1 and Δcre1 on protein production in glucose-based bioreactor cultivations at 72 ha
| Strain | Amt of secreted protein (mg/ml) | Enzyme activity (nkat/ml)
|
||
|---|---|---|---|---|
| CBHI and EGI | Endoglucanase | Xylanase | ||
| QM6a | 0.26 ± 0.04 | 0.36 | Ndb | 0.24 ± 0.01 |
| VTT-D-02878 (cre1-1) | 0.92 ± 0.11 | 3.73 | 0.42 | 0.91 |
| VTT-D-02872 (cre1-1) | 1.14 ± 0.25 | 3.32 | 0.32 ± 0.02 | 0.86 ± 0.29 |
| VTT-D-02877 (Δcre1) | 1.09 ± 0.33 | 4.36 | 0.48 | 1.03 |
| VTT-D-02871 (Δcre1) | 0.99 ± 0.38 | 3.15 | 0.34 ± 0.01 | 0.69 ± 0.28 |
Results are from one or two parallel cultivations (with standard deviation) of each strain.
Nd, not detected (a value which is below the detection level of the measurement).
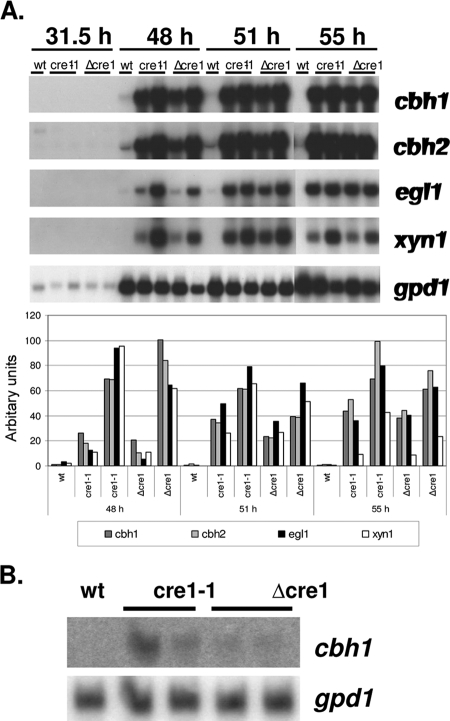
According to Northern analysis (see Fig. 4B), the major cellulase gene, cbh1, which is under the regulation of cre1, is expressed in cultures of Δcre1 and cre1-1 transformants grown on glucose. However, the expression level is very low and not comparable to its expression level under inducing conditions. In the parental strain QM6a, cbh1 remained repressed as expected.
FIG. 4.
Expression of the major cellulases and hemicellulases in wild-type strain QM6a (wt) and its cre1-1 and Δcre tranformants during bioreactor cultivations on lactose at different time points during cultivation (A) and on glucose after 55 h of cultivation (B). Genes encoding cellobiohydrolase 1 (cbh1), cellobiohydrolase 2 (cbh2), endoglucanase 1 (egl1), and xylanase 1 (xyn1) are shown. Hybridization to a glyceraldehyde-3-phosphate dehydrogenase (gpd1) probe is shown as a control. The signals from the lactose cultivation were normalized against those obtained with the gpd1 probe.
The QM6a Δcre1 and cre1-1 transformants produce increased amounts of cellulases under inducing conditions.
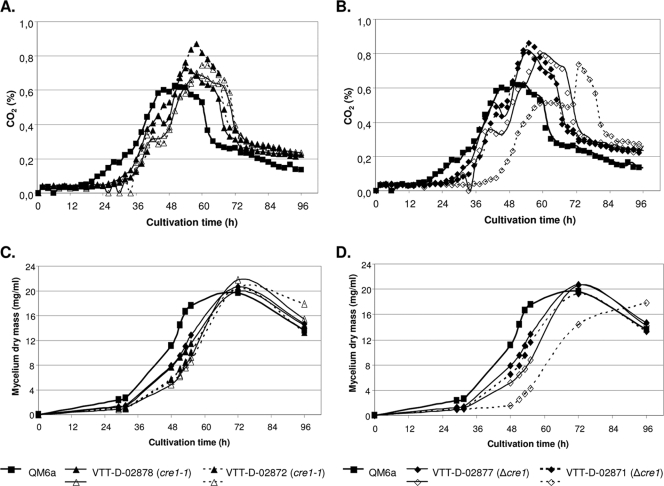
The effects of cre1 deletion and truncation on cellulase and hemicellulase production were also studied under conditions where the expression of the corresponding genes is naturally induced. The wild-type strain QM6a and its two Δcre1 and cre1-1 transformants were cultivated in lactose-containing medium in a bioreactor. Each of the cultivations was inoculated with spores harvested from one potato dextrose agar slant. It was estimated that about three to four times more QM6a spores compared to the transformants were used for inoculation due to more efficient sporulation of QM6a. The growth of the strains was monitored by online determination of CO2 (Fig. 3A and B). Also, the weight of the mycelium (dry weight) (Fig. 3C and D) and the amount of sugars reacting to 3,5-dinitrosalisylic acid (not shown) as well as protein and enzyme activities (Table 3) were measured, and Northern analysis (Fig. 4A) was performed on culture supernatant/mycelium samples taken at different time points of the cultivation. As in the glucose cultivations, the growth of both types of transformants was similar and started later than the parental strain QM6a (Fig. 3A to D). Also, as in the glucose cultivations, the maximum weight (dry weight) of mycelia of the transformants were similar to that of the host.
FIG. 3.
Growth of strain QM6a and its cre1-1 and Δcre1 transformants in lactose-based medium. The concentration of CO2 (A and B) and weight of the mycelium (dry weight) (mycelium dry mass) (C and D) in each cultivation are shown. Two parallel cultivations in bioreactors were performed for each transformant and one with the parental strain.
TABLE 3.
Protein production of T. reesei QM6a and QM6a-based cre1-1 and Δcre1 transformants in lactose-induced bioreactor cultivationsa
| Strain | Total amt of secreted protein (mg/ml) | Enzyme activity
|
|||
|---|---|---|---|---|---|
| CBHI + EGI (nkat/ml) | Endoglucanase (nkat/ml) | Xylanase (nkat/ml) | Protease (U/ml) | ||
| QM6a | 0.45 ± 0.05 | 0.62 ± 0.09 | 20 ± 0 | 25 ± 0 | 18.9 ± 0.9 |
| VTT-D-02878 (cre1-1) | 1.00 ± 0.14 | 12.05 ± 0.35 | 90 ± 14 | 195 ± 21 | 22.4 ± 0.6 |
| VTT-D-02872 (cre1-1) | 1.00 ± 0.20 | 14.70 ± 2.30 | 110 ± 28 | 227 ± 109 | 25.1 ± 0.6 |
| VTT-D-02877 (Δcre1) | 0.90 ± 0.00 | 10.55 ± 0.45 | 75 ± 6 | 185 ± 3 | 22.3 ± 3.3 |
| VTT-D-02871 (Δcre1) | 1.25 ± 0.05 | 18.6 ± 2.60 | 127 ± 23 | 313 ± 110 | 25.5 ± 3.9 |
Samples were collected after 96 h of cultivation. Averages from two parallel cultivations of each strain and standard deviations of the results are shown.
The difference in the production levels between the parental strain QM6a and the Δcre1 and cre1-1 transformants was clear. Larger amounts of total proteins (on average, 1.8- to 2.4-fold-higher amounts) were secreted into the culture medium by the transformants (Table 3). The differences in the enzyme activity levels between the transformants and the parental strain were even more pronounced. The cellulase activity determined using MUL as a substrate was slightly more than 20-fold higher and the xylanase activity was 10-fold higher in the culture supernatants of the transformants than in the culture supernatant of the parent strain. The increase was similar in the Δcre1 and cre1-1 transformants. However, the production of protease activity was not greatly affected by either the deletion or truncation of the cre1 gene. Similar results were also obtained from bioreactor cultivations with complex cellulose medium. In these cultivations, 2.5-fold-higher secreted total protein levels and 6-fold-higher cellulase and 2.5-fold-higher xylanase levels were obtained with both the Δcre1 and cre1-1 transformants compared to strain QM6a (data not shown).
The expression patterns of the major cellulase genes (cbh1, cbh2, and egl1) and one of the xylanases (xyn1) are shown in Fig. 4A. The results of Northern analysis clearly show that expression of all of the above genes is higher in the Δcre1 and cre1-1 transformants than in their parental strain. The increase varies between 10- and 100-fold, depending on the gene and time point of cultivation. These results are in accordance with the enzyme activity results. It is noteworthy that in the QM6a strain, these mRNA levels are quite low and barely detectable with Northern hybridization.
Effect of the Δcre1 mutation on protein production of the hypercellulolytic VTT-D-80133 strain.
The VTT-D-80133 strain and its Δcre1 transformant, VTT-D-02874, were cultivated in cellulose- and lactose-containing media in a bioreactor (one cultivation per strain and medium). The parental strain and the Δcre1 transformant grew similarly, according to the data obtained from the cultivations and including online determination of base consumption, CO2 concentration, dissolved oxygen concentration, and, in the case of the lactose cultivation, biomass accumulation and sugar consumption (data not shown). The protein and activity results from the fermentations of VTT-D-80133 and its Δcre1 transformant are shown in Fig. 5. In both the cellulose and lactose cultivations, the Δcre1 transformant produced larger amounts of proteins than the parent strain did (1.4- and 1.2-fold, respectively). Also, higher cellulase and xylanase activities were obtained from cultures of the Δcre1 strain than from cultures of the parental strain (cellulase activity as measured with MUL, 1.1- to 1.3-fold higher and xylanase activity 1.5- to 2.3-fold, respectively, depending on the cultivation/carbon source). However, protease activity was not increased in the Δcre1 transformant compared to the parental strain in either of the cultivations.
FIG. 5.
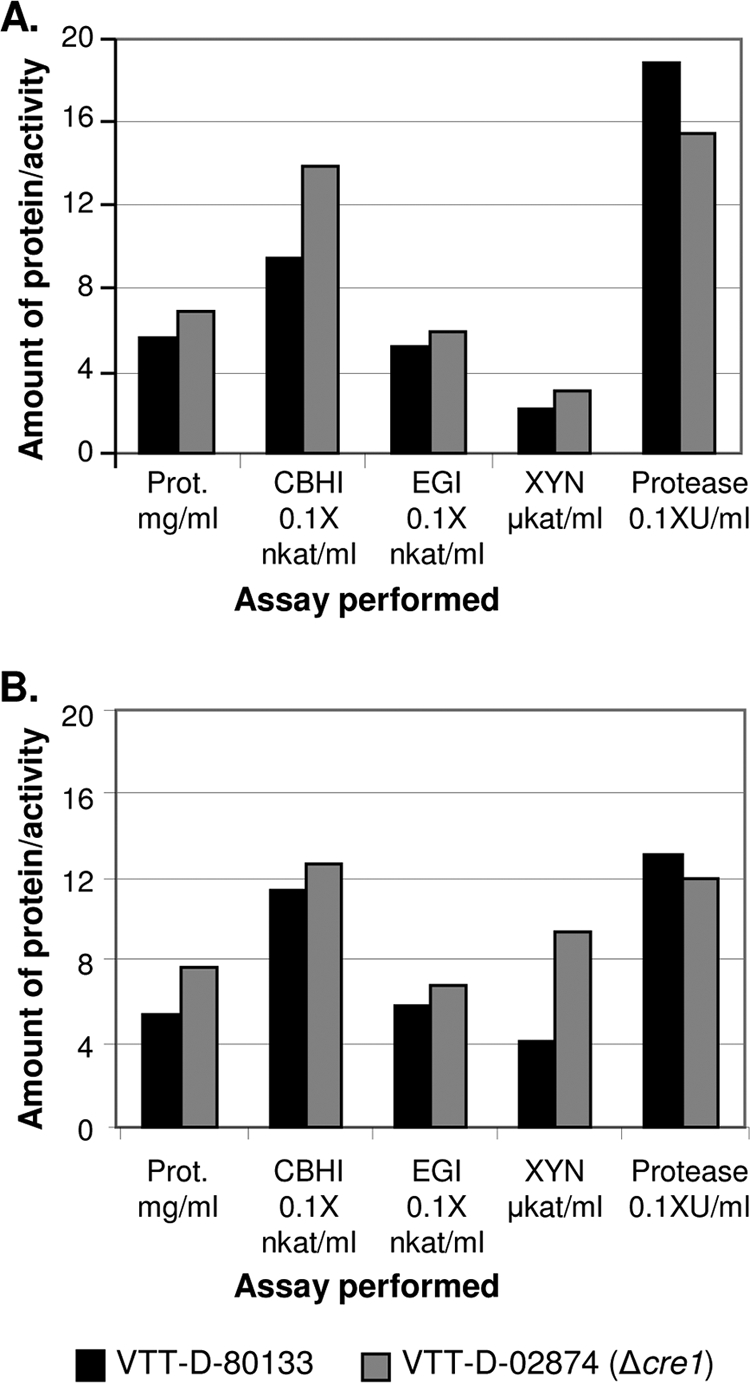
Effect of the cre1 deletion on protein production of the hypercellulolytic VTT-D-80133 strain in cellulose-based (A) and lactose-based (B) bioreactor cultivations. Samples were collected after 70 and 96 h of cultivation in panels A and B, respectively. Prot., protein.
DISCUSSION
The cre1 gene encoding a carbon catabolite repressor of T. reesei has been cloned previously, and the truncated mutant gene cre1-1 has been identified and found in strain Rut-C30 (15), which was developed as an enzyme production strain by mutagenesis and screening (21). It was shown by Ilmen et al. (15) that by introducing the wild-type cre1 gene into strain Rut-C30, it was possible to retain glucose repression in the strain. Loss of the repression mechanism in strain Rut-C30 can be considered one likely mechanism that improved the productivity of this strain. Therefore, we wanted to determine the effect of cre1 mutations on the wild-type background and on the other hand to find out whether there is a difference between the cre1-1 mutation and total disruption of the cre1 gene. Furthermore, we wanted to test whether enzyme production can be improved by deleting the cre1 gene from a mutant producing high levels of cellulase, strain VTT-D-80133, shown to contain the wild-type cre1 gene in its genome.
It was observed in this work that cre1 mutation or deletion has clear effects on colony morphology of T. reesei both in glucose-containing medium causing catabolite repression and media inducing the hydrolytic enzyme genes. The colonies of the Δcre1 and cre1-1 transformants were smaller and produced fewer aerial hyphae and spores than colonies of the parental strain. The same effect could be detected in both QM6a and VTT-D-80133 backgrounds. The cre1-1 mutant strain Rut-C30 also produced clearly smaller colonies than the wild-type strain did, and it produced even smaller colonies than did the cre1-1 transformants constructed from the wild-type strain in this work, perhaps owing to other mutations generated in the genome of strain Rut-C30 during mutagenesis. The null mutation or disruption of creA in Aspergillus nidulans has been shown to have drastic effects on colony morphology (28) and hyphal morphology in liquid cultures (1). Also, a reduced maximal growth rate for these Aspergillus mutants was observed (1). We detected no drastic changes in the growth of T. reesei Δcre1 or cre1-1 transformants in our study, even though the QM6a-based transformants showed somewhat slower growth in the glucose and lactose cultivations compared to the host (Fig. 2 and 3). However, no such difference in growth was observed between VTT-D-80133 and its Δcre1 transformant.
Increases in the cellulase and xylanase activities (Tables 2 and 3) and in the amounts of corresponding mRNAs (Fig. 4) were detected in the laboratory scale fermentations of the QM6a-based Δcre1 and cre1-1 transformants compared with the parental strain. The increase was observed both for a repressing medium with glucose as a carbon source and for a complex inducing medium. In the glucose-containing medium, this was an expected result, since loss of cre1 releases the strain from carbon catabolite repression. However, the large effect on protein production detected in the complex medium inducing the hydrolase genes was surprising. This suggests that the cre1 gene also has a role in modulating the levels of expression of the hydrolase genes under inducing conditions. This is consistent with the observation of Ilmén et al. (15) that the cre1 mRNA level is clearly higher when the fungus is grown on an inducing (cellulose) or neutral (sorbitol) carbon source than when grown on glucose. Interestingly, in a recent article by Mach-Aigner et al. (18), it was reported that the xylanase and cellulose activator gene xyr1 is under the control of the CREI repressor. This could provide an explanation for our finding that the cre1 gene mutation also has a strong effect in a medium inducing the (hemi)cellulase genes. There were no clear changes in secreted protease activity between the wild-type strain QM6a and its Δcre1 and cre1-1 transformants. This shows that production of all secreted proteins is not affected by the cre1 modifications and is consistent with the expectation that the protease genes are not controlled by carbon catabolite repression. It is also noteworthy that even though the deletion of cre1 leads to increased production of proteins, the production levels of cellulases and xylanase in glucose-based medium were still clearly lower than the levels obtained from strain QM6a in an inducing medium (Tables 2 and 3). This suggests that induction mechanisms are also needed for full production, such as the direct action of the XYRI activator on the cellulose and hemicellulase promoters (29). The differences in the cellulase and xylanase mRNA levels of the Δcre1 and cre1-1 transformants and QM6a were much larger than the differences in the amounts of the secreted enzymes. This suggests that factors different from gene expression level become limiting for production when the CREI repressor is removed.
An increase in enzyme production was also obtained when the cre1 gene was deleted from VTT-D-80133 (Fig. 5), a strain that has undergone several rounds of mutagenesis and screening for better cellulase production. The relative effect of cre1 deletion on protein production was much milder in this strain than in the QM6a-based transformants. This result is perhaps expected, since VTT-D-80133 probably already has a number of other mutations increasing the productivity, and their effects may mask part of the effects of cre1 deletion. In any case, this result shows that disrupting the cre1 gene is a relevant strategy for improving even highly developed industrial strains which do not already have a mutation in cre1, since in the streamlined industrial process producing low-cost products, even small improvements in lowering the cost or increasing production can be meaningful.
Our results strongly support the suggestions on the truncation of cre1 to cre1-1 being one of the mutations that have elevated the level of cellulase and hemicellulase production in the Rut-C30 strain. In addition to cre1-1, another mutation leading to elevated cellulase expression levels has been identified in strain Rut-C30 by Geysens et al. (11); they found it in a gene encoding the glucosidase II alpha subunit involved in quality control of secreted proteins in the endoplasmic reticulum. The authors showed that when they transformed the wild-type gene into strain Rut-C30, enzyme productivity was reduced. Results for strains where the glucosidase II mutation would have been made in strain QM6a have not been published. It is interesting that the two mutations known to enhance enzyme production in T. reesei are in genes involved in completely different cellular functions. This indicates that both the level of transcription and the level of secretion, among other functions, are important for efficient enzyme production.
Acknowledgments
We thank Seija Nordberg, Riitta Nurmi, Sanna Hiljanen-Berg, Sirpa Okko, Kirsti Leskinen, Marko Sivenus, and Anne Halonen for excellent technical assistance and Jussi Joensuu for help with photography.
Footnotes
Published ahead of print on 15 May 2009.
REFERENCES
- 1.Agger, T., J. Petersen, S. O'Connor, R. Murphy, J. Kelly, and J. Nielsen. 2002. Physiological characterisation of recombinant Aspergillus nidulans strains with different creA genotypes expressing A. oryzae alpha-amylase. J. Biotechnol. 92:279-285. [DOI] [PubMed] [Google Scholar]
- 2.Aro, N., T. Pakula, and M. Penttilä. 2005. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29:719-739. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J., P. Biely, and K. Poutanen. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257-270. [Google Scholar]
- 4.Bailey, M. J., and K. M. H. Nevalainen. 1981. Induction, isolation and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulose. Enzyme Microb. Technol. 3:153-157. [Google Scholar]
- 5.Bailey, M. J., and J. Tähtiharju. 2003. Efficient cellulase production by Trichoderma reesei in continuous cultivation on lactose medium with a computer-controlled feeding strategy. Appl. Microbiol. Biotechnol. 62:156-162. [DOI] [PubMed] [Google Scholar]
- 6.Buchert, J., T. Oksanen, J. Pere, M. Siika-aho, A. Suurnäkki, and L. Viikari. 1998. Applications of Trichoderma reesei enzymes in the pulp and paper industry, p. 343-363. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Enzymes, biological control and commercial applications. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 7.Cherry, J. R., and A. L. Fidantsef. 2003. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14:438-443. [DOI] [PubMed] [Google Scholar]
- 8.Dowzer, C., and J. Kelly. 1989. Cloning of the creA gene from Aspergillus nidulans: a gene involved in carbon catabolite repression. Curr. Genet. 15:457-459. [DOI] [PubMed] [Google Scholar]
- 9.Foreman, P. K., D. Brown, L. Dankmeyer, R. Dean, S. Diener, N. S. Dunn-Coleman, F. Goedegebuur, T. D. Houfek, G. J. England, A. S. Kelley, H. J. Meerman, T. Mitchell, C. Mitchinson, H. A. Olivares, P. J. Teunissen, J. Yao, and M. Ward. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278:31988-31997. [DOI] [PubMed] [Google Scholar]
- 10.Galante, Y. M., A. De Conti, and R. Monteverdi. 1998. Applications of Trichoderma enzymes in the food and feed industries, p. 327-342. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Enzymes, biological control and commercial applications. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 11.Geysens, S., T. Pakula, J. Uusitalo, I. Dewerte, M. Penttilä, and R. Contreras. 2005. Cloning and characterisation of the glucosidase II alpha subunit gene of Trichoderma reesei: a frameshift mutation results in the aberrant glycosylation profile of the hypercellulolytic strain Rut-C30. Appl. Environ. Microbiol. 71:2910-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes, M. J., C. M. Corrick, and J. A. King. 1983. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol. Cell. Biol. 3:1430-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilmen, M., M. L. Onnela, S. Klemsdal, S. Keranen, and M. Penttilä. 1996. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 253:303-314. [DOI] [PubMed] [Google Scholar]
- 14.Ilmén, M., A. Saloheimo, M. L. Onnela, and M. E. Penttilä. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilmen, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 17.Mach, R. L., and S. Zeilinger. 2003. Regulation of gene expression in industrial fungi: Trichoderma. Appl. Microbiol. Biotechnol. 60:515-522. [DOI] [PubMed] [Google Scholar]
- 18.Mach-Aigner, A., M. Puchner, S. Steiger, G. Bauer, S. Preis, and R. Mach. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74:6554-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandels, M., and E. T. Reese. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 73:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mäntylä, A., M. Paloheimo, and P. Suominen. 1998. Industrial mutants and recombinant strains of Trichoderma reesei, p. 291-309. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Enzymes, biological control and commercial applications. Taylor & Francis Ltd., London, United Kingdom. [Google Scholar]
- 21.Montenecourt, B. S., and D. E. Eveleigh. 1979. Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 181:283-301. [Google Scholar]
- 22.Nehlin, J., and H. Ronne. 1990. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 9:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Paloheimo, M., A. Mäntylä, J. Kallio, T. Puranen, and P. Suominen. 2007. Increased production of xylanase by expression of a truncated version of the xyn11A gene from Nonomuraea flexuosa in Trichoderma reesei. Appl. Environ. Microbiol. 73:3215-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penttilä, M., H. Nevalainen, M. Ratto, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schmoll, M., and C. P. Kubicek. 2003. Regulation of Trichoderma cellulase formation: lessons in molecular biology from an industrial fungus. A review. Acta Microbiol. Immunol. Hung. 50:125-145. [DOI] [PubMed] [Google Scholar]
- 28.Shroff, R., S. O'Connor, M. Hynes, R. Lockington, and J. Kelly. 1997. Null alleles of creA, the regulator of carbon catabolite repression in Aspergillus nidulans. Fungal Genet. Biol. 22:28-38. [DOI] [PubMed] [Google Scholar]
- 29.Stricker, A., K. Grosstessner-Hein, E. Wurleitner, and R. Mach. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]