Abstract
Fusarium oxysporum f. sp. cubense, the causal agent of fusarium wilt of banana (Musa spp.), is one of the most destructive strains of the vascular wilt fungus F. oxysporum. Genetic relatedness among and within vegetative compatibility groups (VCGs) of F. oxysporum f. sp. cubense was studied by sequencing two nuclear and two mitochondrial DNA regions in a collection of 70 F. oxysporum isolates that include representatives of 20 VCGs of F. oxysporum f. sp. cubense, other formae speciales, and nonpathogens. To determine the ability of F. oxysporum f. sp. cubense to sexually recombine, crosses were made between isolates of opposite mating types. Phylogenetic analysis separated the F. oxysporum isolates into two clades and eight lineages. Phylogenetic relationships between F. oxysporum f. sp. cubense and other formae speciales of F. oxysporum and the relationships among VCGs and races of F. oxysporum f. sp. cubense clearly showed that F. oxysporum f. sp. cubense's ability to cause disease on banana has emerged multiple times, independently, and that the ability to cause disease to a specific banana cultivar is also a polyphyletic trait. These analyses further suggest that both coevolution with the host and horizontal gene transfer may have played important roles in the evolutionary history of the pathogen. All examined isolates harbored one of the two mating-type idiomorphs, but never both, which suggests a heterothallic mating system should sexual reproduction occur. Although, no sexual structures were observed, some lineages of F. oxysporum f. sp. cubense harbored MAT-1 and MAT-2 isolates, suggesting a potential that these lineages have a sexual origin that might be more recent than initially anticipated.
Fusarium oxysporum Schlechtendahl emend. Snyder and Hansen is a cosmopolitan species (9) comprised of both pathogenic and nonpathogenic isolates (20). The pathogenic isolates of F. oxysporum cause fusarium wilt of several agricultural crops, and are accordingly subdivided into formae speciales (3, 26, 55). One of the economically more important and destructive formae speciales is the causal agent of fusarium wilt (Panama disease) of banana (Musa spp.), F. oxysporum f. sp. cubense (E. F. Smith) Snyder et Hansen. This disease has been reported in all banana production regions of the world, except those bordering the Mediterranean, Melanesia, Somalia, and some islands in the South Pacific (66, 77).
A range of approaches are typically employed for the characterization of F. oxysporum f. sp. cubense isolates. Based on virulence to specific banana cultivars (66, 67), the pathogen may be classified into one of three races (i.e., races 1, 2, and 4), although this designation may be contingent on environmental conditions. For instance, genetically identical isolates of F. oxysporum f. sp. cubense are classified as race 4 isolates in the subtropics and as race 1 isolates in the tropics because they cause disease to Cavendish bananas under subtropical conditions only (67, 86). Based on vegetative compatibility, F. oxysporum f. sp. cubense isolates have been separated into 24 so-called vegetative compatibility groups (VCGs) (5, 29, 47, 68). Finally, various DNA-based tools have been used to separate F. oxysporum f. sp. cubense into a number of clonal lineages that more or less correspond to their grouping based on VCGs (6, 22, 38, 59).
The evolutionary history of F. oxysporum f. sp. cubense is complex. Based on the results of phylogenetic studies (4-7, 22, 38, 57, 59). F. oxysporum f. sp. cubense represent multiple unrelated lineages, some of which are more closely related to other formae speciales of F. oxysporum than to other F. oxysporum f. sp. cubense lineages (3, 57, 59). This has lead to speculations that new pathogenic forms of F. oxysporum may be derived from other pathogenic and nonpathogenic members of this species (21). Factors such as coevolution with the plant host and the spread of virulence determinants via processes such as parasexuality, heterokaryosis, and sexual recombination also have been implicated in the evolution of this pathogen (11, 36, 37, 39, 64, 65, 69). Although parasexuality and heterokaryosis are known to occur in F. oxysporum (11, 39), sexual fruiting structures have never been observed in the species and only indirect evidence for sexual recombination has been detected (82). Indeed, the organization of the F. oxysporum f. sp. cubense mating type locus (MAT) is similar to those found in the closely related Gibberella fujikuroi (Sawada) Ito in Ito et K. Kimura complex and other heterothallic ascomycetes (2, 90).
Development of appropriate disease management strategies and the selection of F. oxysporum f. sp. cubense-resistant banana cultivars may benefit from a better understanding of the diversity and evolutionary history of the pathogen. Although most previous DNA-based studies provided knowledge regarding the diversity of F. oxysporum f. sp. cubense, the genetic relatedness among the lineages identified in these studies remains uncertain (22). It is also not clear how the different races and VCGs of F. oxysporum f. sp. cubense are related to one another and to other isolates of F. oxysporum. Therefore, the main objective of this study was to resolve the relationships among the F. oxysporum f. sp. cubense VCGs and determine their relationships with other formae speciales and nonpathogenic members of F. oxysporum by using a multigene phylogenetic approach (8, 32, 52, 53, 62, 75, 91). To facilitate the rapid differentiation of the various F. oxysporum f. sp. cubense lineages, we also aimed to develop a diagnostic PCR-restriction fragment length polymorphism (RFLP) procedure. To evaluate the potential of F. oxysporum f. sp. cubense to reproduce sexually, sexual crosses among isolates of opposite mating types were attempted after PCR-based detection of the MAT-1 and MAT-2 idiomorphs (34).
MATERIALS AND METHODS
Fungal isolates.
A global collection of 70 F. oxysporum isolates representing 20 of the 24 VCGs, a new F. oxysporum f. sp. cubense VCG from Vietnam, other formae speciales of F. oxysporum, F. oxysporum isolates from heliconia (Heliconia sp.), and nonpathogenic F. oxysporum isolates from the rhizosphere of banana plants in South Africa (48) were included in this study (Table 1). All cultures are maintained in the culture collection (CAV) of the Forestry Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa. DNA for cultures that were not available was kindly supplied by Suzy Bentley from the Queensland Department of Primary Industries (QDPI) in Brisbane, Australia.
TABLE 1.
Origin, pathogenicity, VCG, and mating type of isolates of Fusarium oxysporum f. sp. cubense, F. oxysporum, and F. circinatum selected for a multigene phylogenetic comparison
| Isolate no.a | Other identification no.a | Species and forma specialis | VCG | Host or cultivar | Origin | Collector(s) | Pathogenicityb | Mating typec |
|---|---|---|---|---|---|---|---|---|
| CAV 009 | F. oxysporum f. sp. cubense | 0120 | Cavendish | South Africa | A. Viljoen | Not tested | MAT-2 | |
| CAV 045 | F. oxysporum f. sp. cubense | 0120 | Cavendish | South Africa | A. Viljoen | Not tested | MAT-2 | |
| CAV 105 | F. oxysporum f. sp. cubense | 0120 | Cavendish | South Africa | A. Viljoen | Not tested | MAT-2 | |
| CAV 293 | IC-1 | F. oxysporum f. sp. cubense | 0120 | Dwarf Cavendish | Canary Islands | J. Hernandez | Pathogenic | MAT-2 |
| CAV 294 | 34661 | F. oxysporum f. sp. cubense | 0120 | Highgate | Honduras | R. Ploetz, American Type Culture Collection | Pathogenic | MAT-2 |
| CAV 296 | STH1 | F. oxysporum f. sp. cubense | 0120 | Highgate | Honduras | R. Ploetz, R. H. Stover | Not tested | MAT-2 |
| CAV 298 | BR18 | F. oxysporum f. sp. cubense | 0120/15 | Banana | Brazil | Unknown | Not tested | MAT-2 |
| CAV 299 | PD14-1 | F. oxysporum f. sp. cubense | 0120/15 | Gros Michel | Nigeria | C. Pausberg-Gauhl | Not tested | MAT-2 |
| CAV 612 | RPCR1-1 | F. oxysporum f. sp. cubense | 01215 | Gros Michel | Costa Rica | Unknown | Pathogenic | MAT-2 |
| CAV 607 | RP13 | F. oxysporum f. sp. cubense | 0122 | Cavendish | Philippines | R. Ploetz | Pathogenic | MAT-1 |
| CAV 605 | RP14 | F. oxysporum f. sp. cubense | 0122 | Cavendish | Philippines | R. Ploetz | Pathogenic | MAT-1 |
| CAV 613 | Phil 7 | F. oxysporum f. sp. cubense | 0126 | Latundan | Philippines | Unknown | Pathogenic | MAT-2 |
| CAV 793 | Indo 33 | F. oxysporum f. sp. cubense | 0126 | Pisang Rubus | Indonesia | R. Shivas | Pathogenic | MAT-2 |
| CAV 794 | Indo 38 | F. oxysporum f. sp. cubense | 0126 | Pisang Rubus | Indonesia | Unknown | Not tested | MAT-2 |
| CAV 1051 | RP52 | F. oxysporum f. sp. cubense | 01210 | Apple | United States | R. Ploetz | Pathogenic | MAT-1 |
| CAV 632 | RP26 | F. oxysporum f. sp. cubense | 01210 | Highgate | Honduras | R. Ploetz | Pathogenic | MAT-1 |
| CAV 847 | Indo 35 | F. oxysporum f. sp. cubense | 01219 | Pisang Raja Sereh | Indonesia | H. Stover | Pathogenic | MAT-1 |
| CAV 195 | Indo 25 | F. oxysporum f. sp. cubense | 01219 | Pisang Ambon | Indonesia | N. Moore | Not tested | MAT-1 |
| RP7 | F. oxysporum f. sp. cubense | 0121 | Cavendish | Taiwan | R. Ploetz | Pathogenic | MAT-2 | |
| RP8 | F. oxysporum f. sp. cubense | 0121 | Cavendish | Taiwan | R. Ploetz | Not tested | MAT-2 | |
| RP9 | F. oxysporum f. sp. cubense | 0121 | Cavendish | Taiwan | R. Ploetz | Not tested | MAT-2 | |
| CAV 810 | Indo 34 | F. oxysporum f. sp. cubense | 01213 | Pisang Berangan | Indonesia | I. Buddenhagen, J. C. Barlett | Pathogenic | MAT-1 |
| CAV 811 | Indo 30 | F. oxysporum f. sp. cubense | 01213 | Pisang Susu | Indonesia | H. Stover | Pathogenic | MAT-1 |
| CAV 300 | CV-1 | F. oxysporum f. sp. cubense | 01213 | Valery | Indonesia | Jepara Banana Plantation | Not tested | MAT-1 |
| CAV 312 | RPML 25 | F. oxysporum f. sp. cubense | 01213/16 | Pisang udang | Malaysia | R. Ploetz | Pathogenic | MAT-1 |
| CAV 313 | RPML 47 | F. oxysporum f. sp. cubense | 01213/16 | Pisang awak legor | Malaysia | R. Ploetz | Not tested | MAT-1 |
| CAV 814 | Indo 47 | F. oxysporum f. sp. cubense | 01216 | Cavendish | Indonesia | I. Buddenhagen, R. Shivas | Pathogenic | MAT-1 |
| CAV 815 | Indo 56 | F. oxysporum f. sp. cubense | 01216 | Cavendish | Indonesia | I. Buddenhagen, G. P. Salingay | Pathogenic | MAT-1 |
| CAV 604 | Indo 50 | F. oxysporum f. sp. cubense | 01216 | Cavendish | Indonesia | Unknown | Not tested | MAT-1 |
| CAV 602 | 23534 | F. oxysporum f. sp. cubense | 0124 | Lady finger | Australia | Unknown | Pathogenic | MAT-2 |
| CAV 609 | 23538 | F. oxysporum f. sp. cubense | 0124 | Lady finger | Australia | Unknown | Pathogenic | MAT-2 |
| CAV 786 | 23734 | F. oxysporum f. sp. cubense | 0124 | Lady finger | Australia | K. Pegg | Not tested | MAT-2 |
| 8611 | F. oxysporum f. sp. cubense | 0125 | Lady finger | Australia | Unknown | Pathogenic | MAT-2 | |
| 23480 | F. oxysporum f. sp. cubense | 0125 | Lady finger | Australia | Unknown | Not tested | MAT-2 | |
| 23487 | F. oxysporum f. sp. cubense | 0125 | Lady finger | Australia | Unknown | Not tested | MAT-2 | |
| CAV 1097 | 22993 | F. oxysporum f. sp. cubense | 0128 | Blue Java | Australia | Unknown | Pathogenic | MAT-2 |
| CAV 1096 | 22994 | F. oxysporum f. sp. cubense | 0128 | Bluggoe | Australia | Unknown | Pathogenic | MAT-2 |
| 24211 | F. oxysporum f. sp. cubense | 01220 | Cavendish | Australia | Unknown | Not tested | MAT-2 | |
| 24219 | F. oxysporum f. sp. cubense | 01220 | Cavendish | Australia | Unknown | Not tested | MAT-2 | |
| CAV 957 | Thai 37 | F. oxysporum f. sp. cubense | 0123 | Kluai Namwa | Thailand | S. Kooariyakul | Not tested | MAT-2 |
| CAV 929 | PHIL 13 | F. oxysporum f. sp. cubense | 0123 | Latundan | Philippines | L. Magnaye | Pathogenic | MAT-2 |
| CAV 933 | Thai 2-1 | F. oxysporum f. sp. cubense | 0123 | Kluai Namwa | Thailand | N. Singburaudom | Pathogenic | MAT-2 |
| 23510 | F. oxysporum f. sp. cubense | 0129 | Lady finger | Australia | Unknown | Not tested | MAT-2 | |
| CAV 1100 | 23518 | F. oxysporum f. sp. cubense | 0129 | Lady finger | Australia | K. Pegg | Pathogenic | MAT-2 |
| 23631 | F. oxysporum f. sp. cubense | 01211 | SH3142 | Australia | Unknown | Not tested | MAT-2 | |
| RP58 | F. oxysporum f. sp. cubense | 01212 | Ney poovan | Tanzania | Unknown | Not tested | MAT-2 | |
| CAV 189 | RPMW 40 | F. oxysporum f. sp. cubense | 01214 | Harare | Malawi | R. Ploetz | Pathogenic | MAT-1 |
| CAV 871 | MAL 7 | F. oxysporum f. sp. cubense | 01217 | Pisang Rastali | Malaysia | Unknown | Pathogenic | MAT-2 |
| CAV 791 | Indo 5 | F. oxysporum f. sp. cubense | 01218 | Pisang Siem | Indonesia | N. Moore | Pathogenic | MAT-1 |
| CAV 1107 | Viet 6 | F. oxysporum f. sp. cubense | 0129/11 | Chuoi xiem | Vietnam | I. Buddenhagen, N. Moore, S. Bentley | Not tested | MAT-2 |
| CAV 1020 | Viet 19 | F. oxysporum f. sp. cubense | Unknown | Chuoi xiem | Vietnam | I. Buddenhagen, N. Moore, S. Bentley | Pathogenic | |
| CAV 1788 | F. oxysporum | Heliconia sp. | South Africa | S. Tween | Nonpathogenic | |||
| CAV 1787 | F. oxysporum | Heliconia sp. | South Africa | S. Tween | Nonpathogenic | |||
| F. circinatum | ||||||||
| CAV 211 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 273 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 202 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 261 | F. oxysporum nonpathogenic | soil | South Africa | B. Nel | ||||
| CAV 246 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 275 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 231 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 274 | F. oxysporum nonpathogenic | Soil | South Africa | B. Nel | ||||
| CAV 330 | CBS 413.90 | F. oxysporum f. sp. lycopersici | Tomato | Israel | R. Cohn | |||
| CAV 337 | CBS 411.90 | F. oxysporum f. sp. vasinfectum | Cotton | Israel | J. Katan | |||
| CAV 342 | CBS 101.97 | F. oxysporum f. sp. lupine | Lupinus | The Netherlands | M. Guranowska | |||
| CAV 343 | CBS 424.90 | F. oxysporum f. sp. melonis | Melon | Israel | J. Katan | |||
| CAV 336 | CBS 488.76 | F. oxysporum f. sp. raphanai | Radish | Germany | W. Gerlach | |||
| CAV 341 | CBS 794.70 | F. oxysporum f. sp. perniciosum | Silktree | Iran | W. Gerlach | |||
| CAV 329 | CBS 259.51 | F. oxysporum f. sp. lini | Flax | Canada | J.W. Groves | |||
| CAV 328 | CBS 137.97 | F. oxysporum f. sp. gladioli | Freesia | The Netherlands | E. J. A. Roebroeck, L. B. O. Lisse | |||
| CAV 335 | CBS 101587 | F. oxysporum f. sp. radicis-lycopersici | Tomato | G. V. Bloemberg |
CAV, Culture Collection at FABI, University of Pretoria, South Africa; numerals only, Culture Collection of the Queensland Department of Primary Industries, Brisbane, Australia; RP, Culture Collection of Randy Ploetz at the University of Florida, Homestead. DNA was supplied by Suzy Bentley from the Queensland Department of Primary Industries, Brisbane, Australia.
Pathogenicity tests.
To verify that the isolates included in this study are indeed specific pathogens of banana, 27 isolates representing 17 known VCGs of F. oxysporum f. sp. cubense were selected for pathogenicity tests on banana plants (Table 1). All cultures were grown on 20 g/liter potato dextrose agar (PDA) (Biolab Diagnostics, Wadeville, South Africa) for 10 days. A spore suspension was prepared by washing spores from mycelia with sterile distilled water, followed by filtration through cheese cloth and adjustment of the spore concentration to 1 × 106 spores/ml. Pathogenicity tests were performed with all isolates on Gros Michel tissue culture banana plantlets or Gros Michel and Bluggoe tissue culture banana plantlets for the new VCG from Vietnam and the F. oxysporum isolates from heliconia. Five tissue culture banana plantlets were inoculated for each isolate tested. The tests were conducted in a hydroponics system (49), and disease severity was measured after 6 weeks using a disease rating scale developed previously (12).
Morphological characterization.
To confirm that the isolates included in this study represent F. oxysporum, their cultural and morphological characteristics were studied using the procedures described by Nelson et al. (50) and Leslie and Summerell (41). All F. oxysporum f. sp. cubense isolates were cultured on PDA (40 g/liter) and carnation leaf agar (41) and incubated at 25°C under white and near-UV fluorescent light for 12 days. Morphological features such as the presence and abundance of micro- and macroconidia, chlamydospores, and the size and shape of the macroconidia produced on carnation leaf agar were examined using light microscopy. Colony color and colony diameter were recorded after 3, 7, and 10 days of growth on PDA, and the presence of sclerotia and sporodochia was documented after 12 days.
DNA isolation, PCR amplification, and sequencing.
Isolates of F. oxysporum f. sp. cubense and F. oxysporum were grown on 20 g/liter PDA medium for 7 days. DNA was isolated from the isolates as described previously (22). For phylogenetic analyses, we targeted the translation elongation factor-1α (TEF) and the mitochondrial small subunit (MtSSU) rRNA genes, as well as the rRNA intergenic spacer (IGS) region and a repeat region encoded in the mitochondrial genome (MtR) (T. Gordon, unpublished data). For this purpose, we used the primer sets EF1 and EF2 (59), MS1 and MS2 (88), and PNFo and PnF22 (17) to amplify regions of TEF, MtSSU, and IGS, respectively. To target the MtR region, primers R117 (5′-GTCAACCAGGAGCAGACTG-3′) and U9 (5′-GTAACCTCTGACTCACCG-3′) were used. Each amplification reaction mixture contained ∼5 ng/μl DNA, 0.3 μM of each primer, 250 μM deoxynucleoside triphosphates (dNTPs; Fermantas, Nunningen, Switzerland), 0.04 U/μl Taq DNA polymerase (Roche Molecular Biochemicals, Manheim, Germany), and PCR buffer with MgCl2 (Roche). Cycling conditions consisted of 35 cycles at 94°C for 45 s, 60°C (TEF), 53°C (MtSSU), 50°C (IGS), or 59°C (MtR) for 45 s and 72°C for 90 s. Each PCR was preceded by an initial denaturation step at 94°C for 2 min and concluded with a final extension step at 72°C for 5 min. PCR products were purified using the High Pure PCR product purification kit (Roche Applied Biochemicals) and sequenced using the Big Dye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI 377 automated sequencer (Applied Biosystems). In the case of IGS, we also designed and used an internal reverse primer, IGS2 (5′-GCCGGATTTGCTCCCTTCT-3′), for sequencing of the entire 1,500-bp fragment.
Phylogenetic analysis.
Multiple sequence alignments were constructed using MAFFT, version 5.85 (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/), with the L-INS-i option effective (30, 33). This option utilizes an iterative refinement method with various algorithms for optimization of local and pairwise alignments (30). Four data sets were constructed for the sequenced gene regions, where three of these comprised the MtR, IGS, and combined TEF-plus-MtSSU sequences of F. oxysporum f. sp. cubense generated in this study. The fourth data set represented an extended TEF-MtSSU data set that included F. oxysporum f. sp. cubense sequences, as well as sequences for other formae speciales of F. oxysporum, nonpathogenic F. oxysporum from South Africa, F. oxysporum isolates from heliconia, and sequences for F. oxysporum isolates that were obtained from GenBank. All ambiguously aligned sites, including a 148- or 156-bp insertion/deletion (indel) within MtR, were excluded from these analyses.
To test for the combinability of data sets, the partition homogeneity test (18) implemented in PAUP*, version 10b (79), was used on parsimony informative sites only (14, 16, 40). These tests were based on 1,000 repartitions and heuristic searches using 100 rounds of random sequence additions with tree bisection reconnection branch swapping. Phylogenies based on maximum parsimony (MP), Bayesian inference (BI), and maximum likelihood (ML) methods were inferred for the different data sets using PAUP*, version 10b, MrBayes, version 3.b.4 (27), and PhyML, version 2.4.3 (23), respectively. For these analyses, the best-fit models of evolution, as indicated by MrModeltest 2.2 (54) and Modeltest 3.7 (71), were used. For the F. oxysporum f. sp. cubense TEF-MtSSU data set, the BI analysis used separate parameters for each gene (27), where the Hasegawa, Kishino, and Yano (HKY) model (25) plus proportion invariable sites (I) were used for TEF and Felsenstein's model (19) for MtSSU. For ML analysis of the F. oxysporum f. sp. cubense TEF-MtSSU data set, the transitional model with equal base frequencies (TIMef) (81) was used. The IGS data set used the general time reversible (GTR) model (81) plus I and gamma correction for among-site variation (G) for BI and Kimura's (35) three-parameter model for the ML analysis. For the MtR data set, the BI analysis used Felsenstein's model (19), while ML used Tumara and Nei's (TN) (84) model with equal base frequencies (TNef). BI of the extended TEF-MtSSU data set utilized HKY plus I for TEF and HKY plus I plus G for MtSSU, while ML analysis of this data set utilized TN plus G. BI trees were constructed using the Metropolis-coupled Monte Carlo Markov chain with 2,000,000 generations, after which Bayesian posterior probabilities were calculated. MP and ML bootstrap confidence values were based on 1,000 replications and the same parameters described above.
DNA-based diagnosis of the F. oxysporum f. sp. cubense lineages.
All sequences were screened for VCG- or lineage-specific polymorphisms in BioEdit, version 6.0.7 (24). As the IGS region contained polymorphisms for the different F. oxysporum f. sp. cubense lineages, five restriction enzymes were used for diagnostic PCR-RFLP purposes. These enzymes included AvaI (New England BioLabs, Hitchin, England), BbvI (New England BioLabs), BceAI (New England BioLabs), BsrDI (New England BioLabs), and Csp6I (Fermentas). All enzymes were used separately in PCR-RFLP digestion reactions and consisted of 5 μl IGS PCR product, 2 U of the restriction enzyme, and 2 μl of the supplied restriction buffer. After incubation at 37°C for 3 h, the restricted fragments were separated by agarose (3% [wt/vol]) gel electrophoresis (72).
Mating type diagnoses and mating studies.
Mating types of the various F. oxysporum f. sp. cubense isolates were determined by PCR using the primer set Falpha 1 and Falpha 2 for MAT-1 (2), and the primers GFmat2c (76) and FF1 (87) for MAT-2. PCR conditions were similar to those described above, apart from the use of annealing temperatures of 55°C for MAT-1 and 54°C for MAT-2. Selected MAT-1 and MAT-2 products were also sequenced in both directions with the original PCR primers, as described above and compared to those in GenBank (http://www.ncbi.nlm.nih.gov/) using BLASTN. Once the mating type of the isolates was known, two F. oxysporum f. sp. cubense isolates in each VCG were crossed in all possible combinations with isolates of the opposite mating type in other VCGs by procedures described by Leslie and Summerell (41). For the F. oxysporum f. sp. cubense lineages containing both mating types, all isolates of opposite mating type were crossed with each other. For all of the crosses, isolates were treated as males and females. F. oxysporum f. sp. cubense isolates were also crossed with the mating type tester of Fusarium circinatum (MRC 6213 or MRC 7488) (10) for control purposes, and crosses between the F. circinatum tester isolates were included as positive controls.
Nucleotide sequence accession numbers.
Sequences determined in this study have been submitted to Genbank under accession no. FJ664901 to FJ66531.
RESULTS
Pathogenicity tests.
All isolates designated F. oxysporum f. sp. cubense caused disease symptoms typical of fusarium wilt on Gros Michel and/or Bluggoe plantlets. After symptom development, the inoculated pathogens were reisolated from randomly selected plants to confirm Koch's postulates. The two F. oxysporum isolates obtained from heliconia, which were previously designated as F. oxysporum f. sp. cubense race 3 (68, 78), did not cause any symptoms on the respective banana hosts.
Morphological characterization.
Isolates of F. oxysporum f. sp. cubense developed cultural and morphological traits typical of those described for F. oxysporum (41, 50). No significant differences were found in growth rate between isolates representing different VCGs, lineages, or clades of F. oxysporum f. sp. cubense. No sclerotium-like structures or sporodochia were produced by any of the isolates after 12 days. Microconidia were produced in false heads on short monophialides and were mostly single celled and kidney shaped. All isolates produced microconidia in abundance, with the exception of isolates associated with F. oxysporum f. sp. cubense VCGs 0126, 01210, and 01219, which produced few microconidia. Thin, sickle-shaped macroconidia were sparse or absent in most isolates, except for one isolate representing each of F. oxysporum f. sp. cubense VCGs 01210, 0126, and 0123. Chlamydospores were produced by F. oxysporum isolates from heliconia after 12 days, and for the F. oxysporum f. sp. cubense isolates only after 4 weeks, and in some cases after 6 weeks. Chlamydospores were formed singly and sometimes in pairs with a coarse protective wall.
Sequence and phylogenetic analysis.
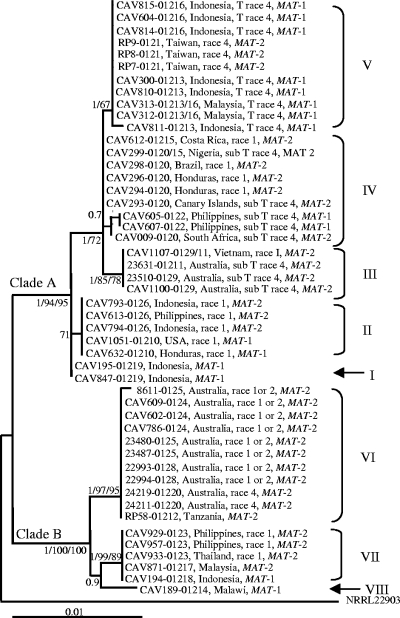
In this study, we sequenced ∼650 bp, 700 bp, and 1,500 bp of the regions encoding TEF, MtSSU, and IGS, respectively. The length of our F. oxysporum f. sp. cubense MtR sequences ranged between 1,100 and 1,250 bp. Isolates associated with the same VCG had identical sequences for all of these regions. The partition homogeneity test supported the combination of the TEF and MtSSU datasets (P = 0.9) but rejected all of the possible combinations of the other regions (P ≤ 0.01). Phylogenetic analyses of the combined F. oxysporum f. sp. cubense TEF-MtSSU data set separated the 48 isolates into two distinct clades (A and B) and eight lineages (I to VIII) (Fig. 1). Clade A included lineage I (VCG 01219), lineage II (VCGs 0126 and 01210), lineage III (VCGs 0129, 0129/11, and 01211), lineage IV (VCGs 0120, 01215, 0120/15, and 0122), and lineage V (VCGs 0121, 01216, 01213, and 01213/16). Clade B included lineage VI (VCGs 0123, 01217, and 01218), lineage VII (VCGs 0124, 0125, 0128, 01220, and 01212), and lineage VIII (VCG 01214).
FIG. 1.
ML phylogenetic tree of F. oxysporum f. sp. cubense inferred from combined TEF and MtSSU rRNA sequence data. A tree with a similar topology was generated using BI and MP (tree scores, confidence interval = 0.882 and retention index = 0.988). The two major clades are indicated at their respective branches with A and B, while the various F. oxysporum f. sp. cubense lineages (I to VIII) are indicated to the right of the tree. For each taxon, VCG and race designation, geographic origin, and mating type are indicated. Bayesian posterior probabilities (>0.7) and bootstrap values (>70%) for the ML analyses and MP are indicated in that order at the internodes. The tree is rooted with Fusarium sp. strain NRRL 22903.
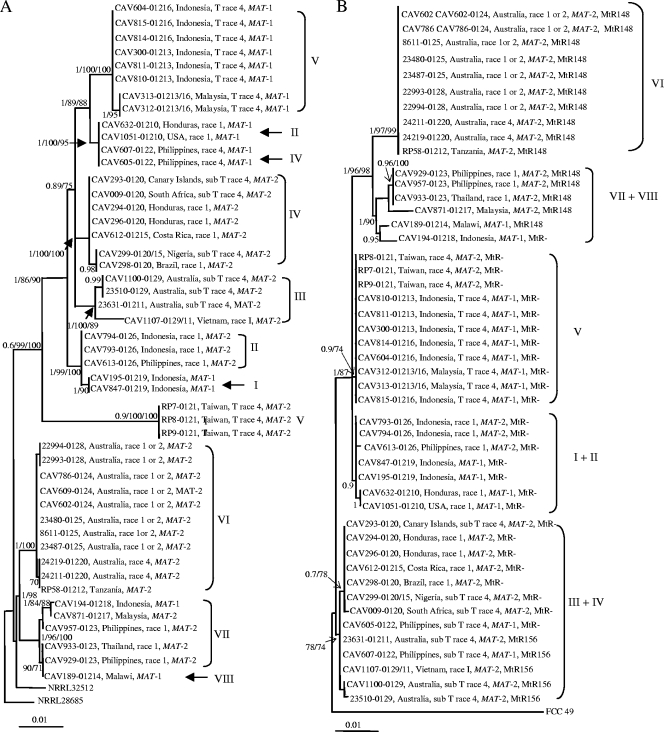
The IGS and MtR (Fig. 2) data did not support the two main clades revealed by the TEF-MtSSU data. The MtR data did, however, cluster the 48 F. oxysporum f. sp. cubense isolates into groups that match the TEF-MtSSU-based lineages (Fig. 1). The IGS sequences also allowed separation of the isolates into groups that broadly match those based on the TEF-MtSSU and MtR data sets. The only exceptions were the divergent placement of F. oxysporum f. sp. cubense VCGs 0121, 0122, 01210, and 01214 in the IGS phylogeny. F. oxysporum f. sp. cubense VCG 0121 formed part of lineage V, based on the TEF-MtSSU data set, whereas it is associated with an exceptionally long branch in the IGS tree and was not related to other lineage V members. In the TEF-MtSSU tree, F. oxysporum f. sp. cubense VCGs 0122 and 01210 formed part of lineages IV and II, respectively, but grouped together in the IGS tree separate from isolates representing TEF-MtSSU-based lineages IV and II. In the IGS and MtR trees, the lineage VIII taxon, F. oxysporum f. sp. cubense VCG 01214, was nested in a clade of lineage VI isolates but displayed a sister group relationship with this lineage based on the TEF-MtSSU data.
FIG. 2.
ML phylogenetic tree of F. oxysporum f. sp. cubense inferred from sequences for the IGS region of the rRNA operon data (A) and the MtR (B). A tree with a similar topology was generated using BI and MP (tree scores, confidence interval [CI] = 0.779 and retention index [RI] = 0.956 for IGS and CI = 0.85 and RI = 0.98 for MtR). The TEF-MtSSU-based F. oxysporum f. sp. cubense lineages (I to VIII) identified in Fig. 1 are indicated to the right of the tree. For each taxon, VCG and race designation, geographic origin, and mating type are indicated. Isolates haboring the 156-bp and 148-bp MtR insertions are indicated by “MtR156” and “MtR148,” respectively, and those lacking an insertion are indicated by “MtR−”. Bayesian posterior probabilities (>0.7) and bootstrap values (>70%) for the ML analyses and MP are indicated in that order at the internodes. The tree is rooted with Fusarium sp. strain NRRL 28687 for IGS and F. circinatum for MtR.
The presence of a 148- or 156-bp indel at nucleotide position 901 relative to the R117 primer position in the IGS sequences separated the F. oxysporum f. sp. cubense isolates into three groups. One group of isolates representing F. oxysporum f. sp. cubense VCGs 0120, 0120/15, 01215, 0126, 01210, 01219, 0121, 01213, 01213/16, 01216, and 01218 lacked an insertion at this position and mostly corresponded to those included in clade A (Fig. 1). Isolates representing F. oxysporum f. sp. cubense VCGs 0124, 0125, 0128, 01220, 01212, 0123, and 01214 harbored a 148-bp insertion at this position, while the third group of isolates harbored a 156-bp insertion and included isolates representing VCGs 01211, 0122, 0129, and 0129/11 and corresponded to those in clade B. The only exceptions are the clade B taxon, VCG 01218, which lacked an insertion, and the clade A VCGs 0122, 01211, 0129/11, and 0129, which contained the 156-bp insert.
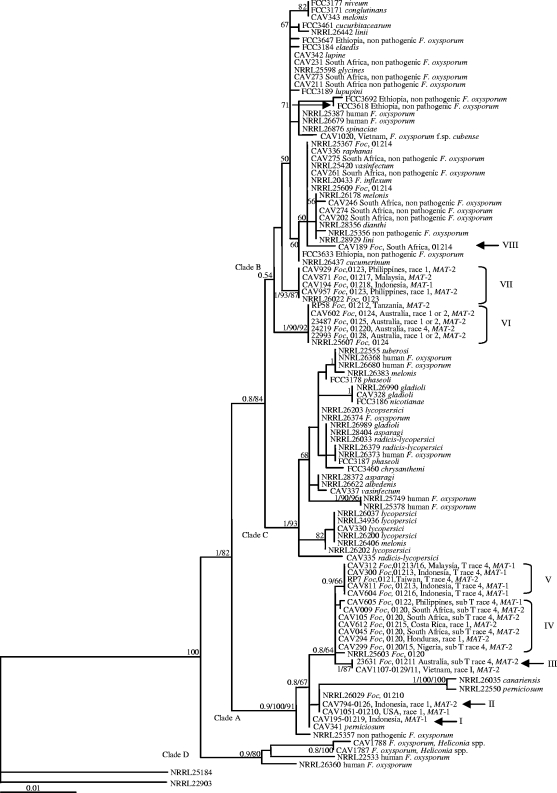
The extended TEF-MtSSU data set that included the F. oxysporum f. sp. cubense sequences, as well as those for other F. oxysporum isolates determined in this study and those obtained from GenBank, separated the isolates into four well-supported clades (Fig. 3). Two of these clades (A and B) correspond with those identified using the smaller TEF-MtSSU data set (Fig. 1) and included a representative set of all of the F. oxysporum f. sp. cubense isolates examined here. We also included isolates from the phylogenetic analysis by O'Donnell et al. (59), which allowed for a direct comparison between the two studies. Lineages C1, C2, C3, C4, and C5 identified in the previous study (59) correspond to our lineages VII, I, VI, II, and VIII, respectively. However, our results show that F. oxysporum f. sp. cubense is even more diverse than initially thought as it includes at least three additional lineages (lineages I, III, and V). Clade C included human pathogens, as well as formae speciales of F. oxysporum other than F. oxysporum f. sp. cubense. Clade D included the F. oxysporum isolates from heliconia and a single F. oxysporum isolate from human tissue. Although clade A consisted predominantly of F. oxysporum f. sp. cubense isolates, it also included isolates of F. oxysporum f. sp. canariensis and F. oxysporum f. sp. perniciosum. Clade B included a number of nonpathogenic F. oxysporum isolates, two F. oxysporum isolates from human tissue, Fusarium inflexum, and several formae speciales of F. oxysporum. Within clade B, an F. oxysporum f. sp. cubense isolate typed as VCG 01214 appeared to be more closely related to nonpathogenic F. oxysporum isolates; other formae speciales of F. oxysporum such as raphanai, vasinfectum, melonis, and dianthi; and F. inflexum than to F. oxysporum f. sp. cubense. Isolate CAV 1020, representing a novel F. oxysporum f. sp. cubense VCG from Vietnam, was included in clade B but was also more closely related to nonpathogenic isolates than to known F. oxysporum f. sp. cubense VCGs.
FIG. 3.
ML phylogenetic tree of F. oxysporum f. sp. cubense (Foc) and other isolates in the F. oxysporum complex inferred from combined TEF and MtSSU rRNA sequence data. A tree with a similar topology was generated using BI and MP (tree scores, confidence interval = 0.73 and retention index = 0.95). The three main clades are indicated at their respective branches with A, B, C, and D to the right of the tree. For each F. oxysporum f. sp. cubense taxon, VCG and race designation, geographic origin, and mating type are indicated. Taxa representing other F. oxysporum isolates are indicated as a human pathogen or nonpathogenic or with the specific forma specialis. Bayesian posterior probabilities (>0.7) and bootstrap values (>70%) for the ML analyses and MP are indicated in that order at the internodes. The tree is rooted with Fusarium sp. strains NRRL 22903 and NRRL 25184.
IGS PCR-RFLP.
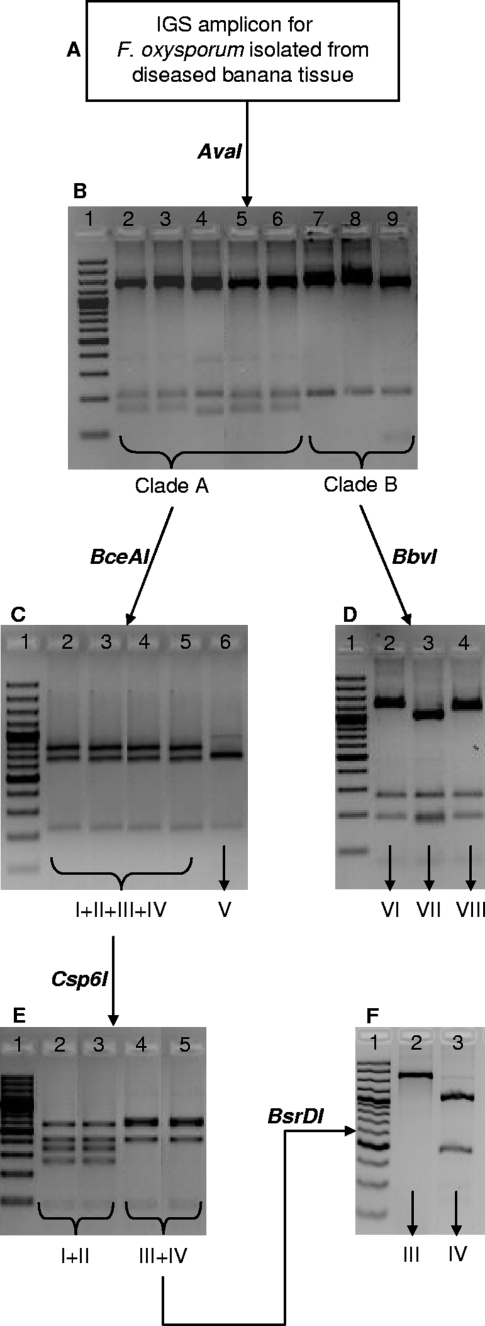
Of the four regions sequenced, IGS was most useful for identifying the different TEF-MtSSU-based lineages of F. oxysporum f. sp. cubense, and we selected five restriction enzymes to apply for diagnostic purposes (Fig. 4). Enzyme AvaI allowed separation of clades A and B (Fig. 4A). Within clade B, BbvI separates lineage VII from lineages VI and VIII (Fig. 4B). Among the clade A lineages, BceAI separates lineage V from lineages I, II, III, and IV (Fig. 4C), while Csp6I separates lineages I and II and lineages III and IV (Fig. 4D), and BsrDI separates lineages III and IV (Fig. 4E). No restriction enzyme was able to separate isolates of lineages VI and VIII from one another as well as from lineages I and II. However, isolates from lineage VIII (VCG 01214) harbor a 94-bp deletion within the MtR gene region at position 747 with respect to the forward primer and can therefore be separated from lineage VI by means of conventional agarose gel electrophoresis.
FIG. 4.
PCR-RFLP analysis of the rRNA IGS region for differentiating the lineages of F. oxysporum f. sp. cubense. For this procedure, DNA obtained from a putative F. oxysporum f. sp. cubense isolate is used as a template for amplication of the IGS region (A), after which the amplicon is sequentially subjected to digestion with restriction enzymes AvaI (B), BceAI (C), BbvI (D), Csp6I (E), and BsrDI. (F) (B) Lane 1, 100-bp marker; lane 2, CAV 847 (lineage I); lane 3, CAV 613 (lineage II); lane 4, CAV 1100 (lineage III); lane 5, CAV 009 (lineage IV); lane 6, CAV 810 (lineage V); lane 7, CAV 957 (lineage VI); lane 8, CAV 608 (lineage VII); lane 8, CAV 189 (lineage VIII). (C) Lane 1, 100-bp marker; lane 2, CAV 794 (lineage I); lane 3, CAV 815 (lineage II); lane 4, CAV 294 (lineage III); lane 5, CAV 933 (lineage IV); lane 6, CAV 1098 (lineage V); lane 7, CAV 189 (lineage VI); lane 8, CAV (lineage VII); lane 8, CAV (lineage VIII). (D) Lane 1, 100-bp marker; lane 2, CAV 957 (lineage VI); lane 3, CAV 608 (lineage VII); lane 4, CAV 189 (lineage VIII). (E) Lane 1, 100-bp marker; lane 2, CAV 847 (lineage I); lane 3, CAV 613 (lineage II); lane 4, CAV 1100 (lineage III); lane 5, CAV 009 (lineage IV); lane 6, CAV 810 (lineage V). (F) Lane 1, 100-bp marker; lane 2, CAV 794 (lineage I); lane 3, CAV 815 (lineage II); lane 4, CAV 294 (lineage III); lane 5, CAV 933 (lineage IV).
Mating type diagnoses and mating studies.
The mating types of the F. oxysporum f. sp. cubense isolates were identified as MAT-1 and MAT-2 based on the presence of 370- and 700-bp fragments, respectively. Only one MAT amplicon was present per F. oxysporum f. sp. cubense isolate. MAT-1 was present in F. oxysporum f. sp. cubense VCGs 0122, 01210, 01219, 01213, 01213/16, 01216, 01214, and 01218, and MAT-2 was present in F. oxysporum f. sp. cubense VCGs 0120, 01215, 0120/15, 0126, 0121, 0124, 0124/5, 0125, 0128, 01220, 0123, 0129, 01211, 01212, 01217, and 0129/11 (Table 1 and Fig. 1 to 3). Both MAT amplicons were present within clades A and B, as well as within lineages II, IV, V, and VII.
No sexual fruiting structures were produced in any of the crosses between F. oxysporum f. sp. cubense isolates 8 weeks after incubation. Protoperithecium-like structures were, however, formed in some crossing combinations. These protoperithecial structures were dark purple to black and superficially resembled the perithecia (9) that were produced by crosses between the F. circinatum tester strains. Structures that were too small to be protoperithecia were also observed. Protoperithecia were abundantly produced when individuals from the opposite mating type within the same lineage were crossed with one another. No protoperithecium-like structures were produced when individuals from clade A were mated with those in clade B and vice versa. One or two protoperithecium-like structures were observed when F. oxysporum f. sp. cubense isolates were crossed with the two F. circinatum tester strains. The tester strains, when crossed with each other, produced abundant perithecia with fertile ascospores.
DISCUSSION
This study considered the evolution of the causal agent of fusarium wilt of banana, F. oxysporum f. sp. cubense, and its various VCGs and races. Based on the DNA sequence information of two nuclear (TEF and IGS) and two mitochondrial (MtSSU and MtR) regions, we demonstrate that F. oxysporum f. sp. cubense's ability to cause disease on banana has emerged multiple times, independently in the F. oxysporum complex. Within the F. oxysporum phylogenetic framework, relationships between the VCGs and races of F. oxysporum f. sp. cubense are complex, which is consistent with pathogenicity to a specific banana cultivar being a polyphyletic trait. Also, as described more fully below, our data suggest that factors such as coevolution with the banana host, horizontal gene transfer events, and sexual reproduction may have played important roles in shaping the evolutionary history of the causal agent of fusarium wilt of banana.
Species concepts applicable to filamentous fungi (74) may be roughly divided into two broad categories: tree based, such as the phylogenetic species concept, and non-tree based, such as the biological and morphological species concepts (74). Among the taxa under study, the morphological species concept has no utility because the F. oxysporum f. sp. cubense isolates examined could not be differentiated based on morphological characters. Likewise, the biological species concept is not applicable because no fertility was observed in any crosses between isolates of opposite mating type, and a teleomorph stage for F. oxysporum has never been reported (41). On the other hand, a multigene phylogeny separated F. oxysporum f. sp. cubense isolates into eight distinct and mostly unrelated lineages (Fig. 1 and 3), which is consistent with results from previous studies (8, 37, 55, 59-61). These findings suggest that various F. oxysporum f. sp. cubense and other F. oxysporum lineages probably constitute distinct species for which discriminatory morphological properties may never be identified. The taxonomy of this group therefore requires extensive reevaluation using DNA-based measures of relationships.
The separation of F. oxysporum f. sp. cubense VCGs into distinct phylogenetic lineages consisting of clusters of related VCGs (Fig. 1, 2, and 3) correlates well with earlier studies using DNA fingerprinting techniques such as RFLPs (38), randomly amplified polymorphic DNAs and DNA amplification fingerprints (4-6), and amplified fragment length polymorphisms (22). This is also true for previous DNA-based phylogenetic studies (59). For all three of the F. oxysporum f. sp. cubense data sets (TEF-MtSSU, IGS, and MtR) used in the current study, isolates associated with the same VCG had identical sequences and clustered together irrespective of their geographic origin. All three of these data sets also consistently clustered the same sets of VCGs into each of the F. oxysporum f. sp. cubense lineages, with the notable exceptions of F. oxysporum f. sp. cubense VCGs 0121, 0122, and 01210 in the IGS tree (Fig. 2). These discrepancies may potentially be associated with the specific nature of the IGS region as a relatively quickly evolving region with the potential for more than one sequence to reside within a single genome (1). This could preclude the inference of the true phylogenetic history from relationships based solely on IGS. A similar topological conflict between TEF and IGS was reported in a recent study of F. oxysporum isolates (58). In some Fusarium species, the inference of the true phylogenetic relationships using nucleus-encoded rRNA regions may also be complicated by the presence of multiple nonorthologous copies (56). Nevertheless, despite these potential limitations, the IGS region has proven to be an excellent marker for diagnoses of Fusarium spp. (31, 32, 73), as was apparent from this study in which lineage-specific PCR-RFLP fingerprints could be developed for F. oxysporum f. sp. cubense (Fig. 4).
The IGS PCR-RFLP fingerprinting method developed in this study presents a quick, easy, and accurate method to identify the lineages of F. oxysporum f. sp. cubense. These fingerprints also allow separation of F. oxysporum f. sp. cubense from nonpathogenic isolates of F. oxysporum (47). They could, therefore, be used for the early detection and characterization of F. oxysporum f. sp. cubense in infected planting material, whether symptomatic or not, in water, and in the soil. In laboratories without sequencing facilities and where VCG testers of F. oxysporum f. sp. cubense cannot be used because of national quarantine regulations, these fingerprints could be of great value in the characterization of the fusarium wilt pathogen of banana. It is, specifically, its ability to rapidly and accurately detect F. oxysporum f. sp. cubense “tropical” race 4 VCG 01213, 01216, and 01213/16 isolates from lineage V in new regions where this pathogen is introduced that could be invaluable in the isolation and management of this most devastating form of F. oxysporum f. sp. cubense.
Our results show that isolates representing F. oxysporum f. sp. cubense races 1 and 2 are scattered among the lineages in clades A and B, while isolates representing F. oxysporum f. sp. cubense race 4 are restricted to clade A (lineages IV and V), with the exception of VCG 01220 (clade B, lineage IV) from Australia, which caused disease in stressed Cavendish bananas (Fig. 1 and 3) (63). Our phylogeny therefore does not reflect the current race designation within F. oxysporum f. sp. cubense. Race designations in F. oxysporum f. sp. cubense and other F. oxysporum formae speciales are based on field evaluation and are generally known not to produce stable classifications (13, 15, 46). The classification of F. oxysporum f. sp. cubense into races in the greenhouse is even more difficult, as virulence is influenced by variables such as temperature, host age, and method of inoculation (13), and different pathogenicity tests used in different laboratories around the world could easily generate discordant results (15). Once universally acceptable greenhouse inoculation techniques have been developed and new potentially differential banana cultivars have been selected for race designation in F. oxysporum f. sp. cubense, the lineages in this study could serve as candidates for developing a new race structure.
In general, the formae speciales of F. oxysporum are not monophyletic (3, 22, 38, 59, 61) (Fig. 3). This is evident from phylogenetic trees where isolates representing different formae speciales grouped together, rather than with representatives from the same forma specialis. In our study, for example, F. oxysporum f. sp. cubense lineages I and II form part of clade A, where they are more closely related to F. oxysporum f. sp. canariense and F. oxysporum f. sp. perniciosum than to other isolates of F. oxysporum f. sp. cubense (Fig. 3). Also, within clade B, lineage VIII forms part of a group containing formae speciales such as lini, dianthi, melonis, vasinfectum, and rhaphania. In contrast, isolates of formae speciales such as F. oxysporum f. sp. albedinis (80), F. oxysporum f. sp. ciceris (28), or F. oxyposrum f. sp. canariensis (70) always represent each others' closest relatives, thus representing some of the few known instances of monophyletic formae speciales. However, from a genetic point of view, the polyphyletic nature of formae speciales is not surprising. This informal taxonomic rank is based on pathogenicity toward a specific plant host (36) and is largely dependent on the products of the avirulence genes harbored by the fungus (51). Recent studies have shown that these genes are generally subject to strong selection and horizontal gene transfer (83, 85, 89). As a result, the grouping of isolates based solely on host pathogenicity would commonly hide genetic diversity and biological differences (36) and may also artificially cluster unrelated isolates together, as has been demonstrated here and elsewhere (3, 22, 38, 59, 61). This highlights the importance of knowledge regarding pathogen diversity for development of reliable/durable plant resistance.
The occurrence of both mating types in F. oxysporum f. sp. cubense is reported for the first time in this study. Our results therefore confirm that F. oxysporum f. sp. cubense would be heterothallic should sexual reproduction take place, as either MAT-1 or MAT-2 sequences (never both) were detected in each of the isolates examined. The fact that in some cases, both MAT-1 and MAT-2 individuals were detected in a single closely related group of isolates implies that different lineages of the fusarium wilt pathogen have sexual origins that could be more recent than initially anticipated. These results, therefore, support the hypothesis that all fungi were originally sexual (42, 43, 82) and that sexual recombination may be followed by phases of clonal propagation of opportunistic varieties (44).
The results of all previous phylogenetic studies (22, 38, 59) demonstrate multiple origins for the evolution of F. oxysporum f. sp. cubense as a pathogen of bananas. However, the results presented here suggest that coevolution with the plant host in its center of origin in Wallace's Indo-Malayan region in Southeast Asia (69) has played an important role during this process. For example, the majority of F. oxysporum f. sp. cubense isolates in clade B originate from banana cultivars that represent Musa balbisiana × Musa acuminata hybrids with at least one chromosome derived from M. balbisiana (e.g., Lady finger and Bluggoe), while those in clade A mostly originate from banana cultivars with pure “A” genomes (i.e., all chromosomes derived from M. acuminata; e.g., Cavendish and Gros Michel) (7). It is therefore possible that F. oxysporum f. sp. cubense lineages I to V derived their ability to cause disease on banana, specifically on M. acuminata, from the ancestor of clade A. The ancestor of clade B, on the other hand, appears to have potentially imparted to its descendants the ability to cause disease to banana cultivars of pure and hybrid background, as well as to plants in the related genus Ensete. This is because one member of clade B lineage VIII (VCG 01214) has the capacity to cause disease not only to enset, but also banana cultivars with pure A and mixed A-B genomes (Fig. 1).
In addition to coevolution with the banana host in its center of origin, the evolution of F. oxysporum f. sp. cubense might also have been influenced by other factors. Although F. oxysporum is considered to be strictly mitotic (20, 82), previous research has suggested that genetic exchange among and within individual lineages might occur more frequently than originally thought (82). This possibility is further emphasized by the results of the current study showing that one of the mitochondrial and nuclear regions examined (MtR and IGS, respectively) supported phylogenies that were highly incongruent with the F. oxysporum f. sp. cubense TEF-MtSSU tree. Therefore, the fact that some F. oxysporum f. sp. cubense VCGs cluster together in the IGS tree and separate in the TEF-MtSSU tree potentially reflects ancient recombination or genetic exchange beween F. oxysporum f. sp. cubense lineages. Such genetic exchange or recombination could be due to parasexuality, a nonsexual mode of genetic exchange, or heterokaryosis, a process that is initiated by fusion of vegetative hyphae (anastomosis) between individuals with very similar genomes and that has been shown to occur in F. oxysporum (11, 39). Taylor et al. (82) also demonstrated the possibility of recombination within some of the F. oxysporum f. sp. cubense clonal lineages. In their study, they reanalyzed previous RFLP data (38) and showed that recombination within some of the clonal lineages may exist. They also concluded that the lack of association between DNA amplification fragment-based DNA fingerprint groups (6) and VCGs is further evidence for recombination. The findings presented in the current study also support this notion as both mating types were detected in some F. oxysporum f. sp. cubense lineages, and crosses between many pairs of isolates of the opposite mating type resulted in the production of structures resembling immature perithecia.
Inclusion of isolates representing formae speciales other than F. oxysporum f. sp. cubense, nonpathogenic F. oxysporum isolates, and F. oxysporum isolates from human tissue in our phylogenetic analyses illustrates the great diversity that exists within the F. oxysporum complex. In this study, a single isolate from Vietnam (CAV 1020) that was confirmed as pathogenic to banana and shown to be associated with a novel VCG of F. oxysporum f. sp. cubense (Fig. 4) (6) grouped separately from all other F. oxysporum f. sp. cubense isolates (Fig. 3.). This result suggests that many distinct lineages of F. oxysporum f. sp. cubense may remain to be discovered and that additional surveys and research are needed for the full appreciation of the evolution of this pathogen. It also demonstrates that focusing on a single agricultural crop may lead to an overestimation of the clonality (45). Therefore, in order to pinpoint potential species boundaries within F. oxysporum and to elucidate the true relationships among the VCGs and lineages of F. oxysporum f. sp. cubense, the diversity of the F. oxysporum complex needs to be fully characterized.
Acknowledgments
We thank the Banana Growers Association of South Africa, the Technology and Human Resources of Industry Programme, and the University of Pretoria for financial support.
We also thank The Queensland Department of Primary Industries, Brisbane Australia; Randy Ploetz, Tropical Research and Education Centre, University of Florida, Homestead; and the Tree Pathology Cooperative Programme, Pretoria, South Africa, for the use of some of the isolates in this study and for the F. oxysporum f. sp. cubense VCG determinations. Finally, we thank Grieta Mahlangu from the Tissue Culture Facility at FABI for the tissue culture-derived Gros Michel and Bluggoe banana plantlets that were used in pathogenicity trials.
Footnotes
Published ahead of print on 29 May 2009.
REFERENCES
- 1.Appel, D. J., and T. R. Gordon. 1996. Relationships among pathogenic and non-pathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Mol. Plant-Microbe Interact. 9:125-138. [DOI] [PubMed] [Google Scholar]
- 2.Arie, T., I. Kaneko, T. Yoshida, M. Noguchi, Y. Nomura, and I. Yamaguchi. 2000. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant-Microbe Interact. 13:1330-1339. [DOI] [PubMed] [Google Scholar]
- 3.Baayen, R. P., K. O'Donnell, P. J. M. Bonants, E. Cigelnik, L. P. N. M. Kroon, J. A. Roebroeck, and C. Waalwijk. 2000. Gene genealogies and AFLP analysis in the Fusarium oxysporum complex identify monophyletic and non-monophyletic formae speciales causing wilt and rot disease. Phytopathology 90:891-900. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S., and B. J. Bassam. 1996. A robust DNA amplification fingerprinting system applied to analysis of genetic variation within Fusarium oxysporum f.sp. cubense. J. Phytopathol. 144:207-213. [Google Scholar]
- 5.Bentley, S., and J. L. Dale. 1995. Genetic variation among a world-wide collection of isolates of Fusarium oxysporum f.sp. cubense analysed by RAPD-PCR fingerprinting. Mycol. Res. 99:1278-1384. [Google Scholar]
- 6.Bentley, S., K. G. Pegg, N. Y. Moore, R. D. Davis, and I. W. Buddenhagen. 1998. Genetic variation among vegetative compatibility groups of Fusarium oxysporum f.sp. cubense analysed by DNA fingerprinting. Phytopathology 88:1283-1293. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, E. W. A., R. C. Ploetz, and H. C. Kistler. 1994. Statistical analysis of electropheric karyotype variation among vegetative compatibility groups of Fusarium oxysporum f.sp. cubense. Mol. Plant-Microbe Interact. 7:196-207. [Google Scholar]
- 8.Bogale, M., B. D. Wingfield, M. J. Wingfield, and E. T. Steenkamp. 2006. Characterization of Fusarium oxysporum isolates from Ethiopia using AFLP, SSR and DNA sequencing. Fungal Divers. 23:51-66. [Google Scholar]
- 9.Booth, C. 1971. The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, United Kingdom.
- 10.Britz, H., T. A. Coutinho, M. J. Wingfield, W. F. O. Marasas, T. R. Gordon, and J. F. Leslie. 1999. Fusarium subglutinans f. sp. pini represents a distinct mating population in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:1198-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buxton, E. W. 1962. Parasexual recombination in the banana-wilt Fusarium. Trans. Br. Mycol. Soc. 45:133-139. [Google Scholar]
- 12.Carlier, J., D. De Waele, and J. V. Escelant. 2002. Global evaluation of Musa germplasm for resistance to Fusarium wilt, Mycosphaerella leaf spot diseases and nematodes, vol. 6. INIBAP Technical Guidelines. INIBAP, Montpellier, France.
- 13.Correll, J. C. 1991. The relationship between formae speciales, races and vegetative compatibility groups in Fusarium oxysporum. Phytopathology 81:1061-1064. [Google Scholar]
- 14.Darlu, P., and G. Lecointre. 2002. When does the incongruence length difference test fail? Mol. Biol. Evol. 19:432-437. [DOI] [PubMed] [Google Scholar]
- 15.Davis, R. D., N. Y. Moore, and J. K. Kochman. 1996. Characterisation of a population of Fusarium oxysporum f.sp. vasinfectum causing wilt of cotton. Aust. J. Agric. Res. 47:1143-1156. [Google Scholar]
- 16.Dolphin, K., R. Belshaw, C. D. L. Orme, and D. L. J. Quicke. 2000. Noise and incongruence: interpreting results of the incongruence length difference test. Mol. Phylogenet. Evol. 17:401-406. [DOI] [PubMed] [Google Scholar]
- 17.Edel, V., C. Steinberg, I. Avelange, G. Laguerre, and C. Alabouvette. 1995. Comparison of three molecular methods for the characterization of Fusarium oxysporum strains. Phytopathology 85:579-585. [Google Scholar]
- 18.Farris, J. S., M. Kallersjö, A. G. Kluge, and C. Bult. 1994. Testing significance of incongruence. Cladistics 10:315-319. [Google Scholar]
- 19.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, T. R., and R. D. Martyn. 1997. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35:111-128. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, T. R., and D. Okamoto. 1992. Population structure and the relationship between pathogenic and non-pathogenic strains of Fusarium oxysporum. Phytopathology 82:73-77. [Google Scholar]
- 22.Groenewald, S., N. Van Den Berg, W. F. O. Marasas, and A. Viljoen. 2006. The application of high-throughput AFLPs in assessing genetic diversity in Fusarium oxysporum f.sp. cubense. Mycol. Res. 110:297-305. [DOI] [PubMed] [Google Scholar]
- 23.Guidon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 24.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 25.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 26.Hawksworth, D. L., P. M. Kirk, B. C. Sutton, and D. N. Pegler. 1995. Dictionary of the fungi. CAB International, Wallingford, United Kingdom.
- 27.Heulsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollack. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez-Gasco, M. M., M. G. Milgroom, and R. M. Jiménez-Díaz. 2002. Gene genealogies support Fusarium oxysporum f.sp. ciceris as a monophyletic group. Plant Pathol. 51:72-77. [Google Scholar]
- 29.Katan, T. 1999. Current status of vegetative compatibility groups in Fusarium oxysporum. Phytoparasitica 27:51-64. [Google Scholar]
- 30.Katoh, K., K.-I. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawabe, M., K. Katsube, T. Yoshida, T. Arie, and K. Tsuchiya. 2007. Genetic diversity of Fusarium oxysporum f. sp. spinaciae in Japan based on phylogenetic analysis of rDNA-IGS and MAT1 sequences. J. Gen. Plant Pathol. 73:353-359. [Google Scholar]
- 32.Kawabe, M., Y. Kobayashi, G. Okada, I. Yamaguchi, T. Teraoka, and T. Arie. 2005. Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f.sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J. Gen. Plant Pathol. 71:263-272. [Google Scholar]
- 33.Kazutaka, K., M. Kazuharu, K. Kei-ichi, and M. Takashi. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerényi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:4071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura, M. 1981. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci USA 78:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kistler, H. C. 2000. Evolution of host specificity in Fusarium oxysporum, p. 70-82. In B. A. Summerell, J. F. Leslie, D. Backhouse, W. L. Bryden, and L. W. Burgess (ed.), Fusarium: Paul E. Nelson Memorial Symposium. APS Press, Saint Paul, MN.
- 37.Kistler, H. C. 1997. Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 87:474-479. [DOI] [PubMed] [Google Scholar]
- 38.Koenig, R. L., R. C. Ploetz, and H. C. Kistler. 1997. Fusarium oxysporum f.sp. cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathology 87:915-923. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn, D. N., B. Cortes, T. Pinto, and J. Weaver. 1995. Parasexuality and heterokaryosis in Fusarium oxysporum f.sp. cubense. Phytopathology 85:1119. [Google Scholar]
- 40.Lee, M. S. Y. 2001. Uninformative characters and apparent conflict between molecules and morphology. Mol. Biol. Evol. 18:676-680. [DOI] [PubMed] [Google Scholar]
- 41.Leslie, J. F., and B. A. Summerell. 2006. The Fusarium laboratory manual. Blackwell Publishing-Wiley, Hoboken, NJ.
- 42.Lobuglio, K. F., J. I. Pitt, and J. W. Taylor. 1993. Phylogenetic analysis of two independent ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 85:592-604. [Google Scholar]
- 43.Lobuglio, K. F., and J. W. Taylor. 2002. Recombination and genetic differentiation in the mycorrhizal fungus Cenococcum geophilum Fr. Mycologia 94:772-780. [PubMed] [Google Scholar]
- 44.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald, B. A. 1997. The population genetics of fungi: tools and techniques. Phytopathology 87:448-453. [DOI] [PubMed] [Google Scholar]
- 46.Migheli, Q., T. Berio, M. L. Gullino, and A. Garibaldi. 1995. Electrophoretic karyotype variation among pathotypes of Fusarium oxysporum f.sp. dianthi. Plant Pathol. 44:308-315. [Google Scholar]
- 47.Moore, N. Y., K. G. Pegg, R. N. Allen, and J. A. G. Irwin. 1993. Vegetative compatibility and distribution of Fusarium oxysporum f.sp. cubense in Australia. Aust. J. Exp. Agric. 33:797-802. [Google Scholar]
- 48.Nel, B., C. Steinberg, N. Labuschagne, and A. Viljoen. 2006. Isolation and characterization of nonpathogenic Fusarium oxysporum isolates from the rhizosphere of healthy banana plants. Plant Pathol. 55:207-216. [Google Scholar]
- 49.Nel, B., C. Steinberg, N. Labuschagne, and A. Viljoen. 2006. The potential of non-pathogenic Fusarium oxysporum and other biological control organisms for suppressing fusarium wilt of banana. Plant Pathol. 55:217-223. [Google Scholar]
- 50.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species: an illustrated manual for identification. Pennsylvania State University, University Park, PA.
- 51.Nimchuk, Z., T. Eulgem, B. F. Holt, and J. L. Dangi. 2003. Recognition and response in the plant immune system. Annu. Rev. Genet. 37:579-609. [DOI] [PubMed] [Google Scholar]
- 52.Nirenberg, H. I., and K. O'Donnell. 1997. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90:434-458. [Google Scholar]
- 53.Nirenberg, H. I., and K. O'Donnell. 1998. Two new species of Fusarium: Fusarium brevicatenulatum from the noxious weed Striga asiatica in Madagascar and Fusarium pseudoanthophilum from Zea mays in Zimbabwe. Mycologia 90:459-464. [Google Scholar]
- 54.Nylander, J. A. A. 2004. MrModeltest ver. 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 55.O'Donnell, K., and E. Cigelnik. 1999. A DNA sequenced-based phylogenetic structure for the Fusarium oxysporum complex. Phytoparasitica 27:69. [Google Scholar]
- 56.O'Donnell, K., and E. Cigelnik. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7:103-116. [DOI] [PubMed] [Google Scholar]
- 57.O'Donnell, K., E. Cigelnik, and H. I. Nirenberg. 1998. Molecular systematic and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465-493. [Google Scholar]
- 58.O'Donnell, K., C. Gueidan, P. Johnston, P. Crous, P. Colyer, A. Glenn, R. Riley, N. Zitomer, C. Waalwijk, D. Geiser, S. Kang, J. Juba, D. A. Sutton, R. Ploetz, M. Eliot, H. C. Kistler, M. Davis, S. Sink, and B. Sarver. 2008. A two-locus DNA sequence database for identifying host-specific pathogens and phylogenetic diversity within the Fusarium oxysporum species complex, s3.20. In K. O'Donnell and K. Seifert (ed.), X International Fusarium Workshop and Fusarium Genomics Workshop. Alghero, Sardinia, Italy.
- 59.O'Donnell, K., H. C. Kistler, E. Cigelnik, and R. C. Ploetz. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from the nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 95:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Donnell, K., H. C. Kistler, B. K. Tacke, and H. H. Casper. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 97:7905-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Donnell, K., D. A. Sutton, M. G. Rinaldi, K. C. Magnon, P. A. Cox, S. G. Revankar, S. Sanche, D. M. Geiser, J. H. Juba, J. H. Van Burik, A. Padhye, E. J. Anaissie, A. Francesconi, T. J. Walsh, and J. S. Robinson. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequences data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 42:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Donnell, K., J. W. Todd, D. M. Geiser, H. C. Kistler, and T. Aoki. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41:600-623. [DOI] [PubMed] [Google Scholar]
- 63.Pegg, K. G., R. G. Shivas, N. Y. Moore, and S. Bentley. 1995. Characterization of a unique population of Fusarium oxysporum f.sp. cubense causing Fusarium wilt in Cavendish bananas at Carnarvon, Western Australia. Aust. J. Agric. Res. 46:167-178. [Google Scholar]
- 64.Ploetz, R. C. 2005. Panama disease: an old nemesis rears its ugly head. Part 1. The beginnings of the banana export trades. APSnet. http://www.apsnet.org/online/feature/panama/.
- 65.Ploetz, R. C. 2005. Panama disease: an old nemesis rears its ugly head. Part 2. The Cavendish era and beyond. APSnet. http://www.apsnet.org/online/feature/panama/.
- 66.Ploetz, R. C. 1994. Panama disease: return of the banana menace. Int. J. Pest Manag. 40:326-336. [Google Scholar]
- 67.Ploetz, R. C. 1990. Population biology of Fusarium oxysporum f.sp. cubense, p. 63-67. In R. C. Ploetz (ed.), Fusarium wilt of banana. APS Press, St. Paul, MN.
- 68.Ploetz, R. C., and J. C. Correll. 1988. Vegetative compatibility among races of Fusarium oxysporum f.sp. cubense. Plant Dis. 72:325-328. [Google Scholar]
- 69.Ploetz, R. C., and K. G. Pegg. 1997. Fusarium wilt of banana and Wallace's line: was the disease originally restricted to his Indo-Malayan region? Australas. Plant Pathol. 26:239-249. [Google Scholar]
- 70.Plyler, T. R., G. W. Simone, D. Fernandez, and H. C. Kistler. 2000. Genetic diversity among isolates of Fusarium oxysporum f.sp. canariensis. Plant Pathol. 49:155-164. [Google Scholar]
- 71.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 72.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 73.Schweigkofler, W., K. O'Donnell, and M. Garbelotto. 2004. Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Appl. Environ. Microbiol. 70:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sites, J. W., and J. C. Marshall. 2004. Operational criteria for delimiting species. Annu. Rev. Phytopathol. 35:199-277. [Google Scholar]
- 75.Skovgaard, K., L. Bodker, and S. Rosendahl. 2002. Population structure and pathogenicity of members of Fusarium oxysporum complex isolated from soil and root necrosis of pea. FEMS Microbiol. Ecol. 42:367-374. [DOI] [PubMed] [Google Scholar]
- 76.Steenkamp, E. T., B. D. Wingfield, T. A. Coutinho, K. A. Zeller, M. J. Wingfield, W. F. O. Marasas, and J. F. Leslie. 2000. PCR-based identification of MAT-1 and MAT-2 in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 66:4378-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stover, R. H., and N. W. Simmonds. 1987. Bananas, 3rd ed. Longmans, London, United Kingdom.
- 78.Su, H. J., T. Y. Chuang, and W. S. Kong. 1977. Physiological race of fusarial wilt fungus attacking Cavendish banana of Taiwan. Taiwan Banana Res. Inst. Spec. Publ. 2:22. [Google Scholar]
- 79.Swofford, D. L. 1998. PAUP. Phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associated, Inc., Sunderland, MA.
- 80.Tantaoui, A., M. Quiten, J. P. Geiser, and D. Fernandez. 1996. Characterization of a single clonal lineage of Fusarium oxysporum f.sp. albedinis causing Bayoud disease of date palm in Morocco Phytopathology 86:787-792. [Google Scholar]
- 81.Tavare, S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17:57-86. [Google Scholar]
- 82.Taylor, J. W., D. J. Jacobson, and M. C. Fisher. 1999. The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 37:197-246. [DOI] [PubMed] [Google Scholar]
- 83.Temporini, E. D., and H. D. VanEtten. 2004. An analysis of the phylogenetic distribution of the pea pathogenicity genes of Nectria haematococca MPVI supports the hypothesis of their origin by horizontal transfer and uncovers a potentially new pathogen of garden pea: Noecosmospora boniensis. Curr. Genet. 46:29-36. [DOI] [PubMed] [Google Scholar]
- 84.Tumara, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 85.Van der Does, H. C., and M. Rep. 2007. Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Mol. Plant-Microbe Interact. 20:1175-1182. [DOI] [PubMed] [Google Scholar]
- 86.Viljoen, A. 2002. The status of Fusarium wilt (Panama disease) of banana in South Africa. S. Afr J. Sci. 98:1-4. [Google Scholar]
- 87.Visser, M. 2003. Molecular biological studies of the Fusarium wilt pathogen of banana in South Africa. Ph.D. dissertation. University of Pretoria, Pretoria, South Africa.
- 88.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 313-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 89.Yoon, S. H., Y.-K. Park, S. Lee, D. Choi, T. K. Oh, C.-G. Hur, and J. F. Kim. 2007. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic Acids Res. 35:D395-D500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yun, S.-H., T. Arie, I. Kaneko, O. C. Yoder, and B. G. Turgeon. 2000. Molecular organization of mating type loci in heterothallic, homothallic and asexual Gibberella/Fusarium species. Fungal Genet. Biol. 31:7-20. [DOI] [PubMed] [Google Scholar]
- 91.Zeller, K. A., B. A. Summerell, S. Bullock, and J. F. Leslie. 2003. Gibberella konza (Fusarium konzum) sp. nov., a new biological species within the Gibberella fujikuroi species complex from prairie grasses. Mycologia 95:943-954. [PubMed] [Google Scholar]