Abstract
Fungal and oomycete populations and their dynamics were investigated following the introduction of the biocontrol agent Pythium oligandrum into the rhizosphere of tomato plants grown in soilless culture. Three strains of P. oligandrum were selected on the basis of their ability to form oospores (resting structures) and to produce tryptamine (an auxin-like compound) and oligandrin (a glycoprotein elicitor). Real-time PCR and plate counting demonstrated the persistence of large amounts of the antagonistic oomycete in the rhizosphere throughout the cropping season (April to September). Inter-simple-sequence-repeat analysis of the P. oligandrum strains collected from root samples at the end of the cropping season showed that among the three strains used for inoculation, the one producing the smallest amount of oospores was detected at 90%. Single-strand conformational polymorphism analysis revealed increases in the number of members and the complexity of the fungal community over time. There were no significant differences between the microbial ecosystems inoculated with P. oligandrum and those that were not treated, except for a reduction of Pythium dissotocum (ubiquitous tomato root minor pathogen) populations in inoculated systems during the last 3 months of culture. These findings raise interesting issues concerning the use of P. oligandrum strains producing elicitor and auxin molecules for plant protection and the development of biocontrol.
In soilless cultures, the recycling of drainage water within a system is the consequence of new laws concerning water saving and limitation of pollution. Such closed systems minimize costs by conserving water and reducing fertilizer input; however, they may favor the dissemination of pathogens (13). When pathogens manage to enter recirculation systems, they are rapidly disseminated and may cause disease epidemics, particularly during periods of stress, e.g., stress due to high temperatures and/or to low levels of dissolved oxygen in the nutrient solution. Thus, numerous facultative pathogens commonly found in conventional cultures may become economically significant (53). Several of them, e.g., Pythium spp. and Phytophthora spp., are well adapted to the aquatic environment of hydroponic systems: they produce flagellate zoospores which enable them to swim in the nutrient solution, facilitating the spread of infection (18, 21, 36, 54, 61).
Various methods are used to reduce the risks to plant health. Over recent years, the disinfection of nutrient solutions by physical or chemical treatments, e.g., ozonization, UV irradiation, chlorination, and thermo-disinfection, has been developed (13, 38). These methods effectively destroy pathogenic microorganisms but are harmful to species liable to benefit the plant, to be used as biocontrol agents, or both. Indeed, recirculation of nutrient solutions in closed hydroponic systems favors the establishment of a potentially suppressive microflora besides the pathogenic microflora (16, 28, 39, 41). The development of a beneficial microflora may thus be impeded by treatments used to destroy pathogenic microorganisms. Consequently, interest has been focused on the management of microorganisms in soilless cultures (12). Postma and coworkers (40) found that the extent of root disease is increased by the use of autoclaved rock wool. Tu and coworkers (59) observed that root rot disease was less severe in closed hydroponic systems than in open cultures and suggested that the difference was due to a higher density of bacteria in the closed systems. According to Paulitz (34), the diversity of microorganisms in soilless cultures is more limited than that in conventional soil cultures, such that conditions are more suitable for beneficial microorganisms, and consequently for effective biological control, in soilless than in conventional soil cultures.
Biocontrol strategies are promising (7, 35). However, both biotic and abiotic factors may affect the performance of biocontrol methods. Relevant biotic factors include interactions with nontarget microorganisms (6), poor implantation of the biocontrol agent due to nonadaptation to the hydroponic system or resistance from the native microflora, shelf life and formulation, and host plant species and cultivar effects. Abiotic factors include climatic, chemical, and physical conditions of the soil or rhizosphere.
Despite the limitations, various studies report evidence of the suppression of disease following the inoculation of hydroponic systems with antagonistic microorganisms. In particular, Pythium oligandrum is an effective biocontrol agent (2, 14, 49, 64). This oomycete colonizes roots without damaging the host plant cells (24, 45) and survives in the rhizosphere, where it exerts its biocontrol (57). P. oligandrum acts through both direct effects (mycoparasitism, antibiosis, and competition for nutrients and space) and indirect effects (stimulation of plant defense reactions and plant growth promotion) (49). The operating effects seem to depend on the type of pathogenic fungi being controlled (3, 48, 49). Le Floch and coworkers suggested that mycoparasitism is not the main mode of action (23). Root colonization by P. oligandrum may induce systemic resistance associated with the synthesis of elicitors protecting the plant from its aggressors (4, 17, 31, 37, 56). Several studies have investigated formulations of P. oligandrum oospores applied to soil or seeds, and their production and use, to optimize the efficacy of biocontrol (9, 30).
Effective biocontrol by P. oligandrum may be limited by its heterogeneous implantation in the rhizosphere (46). Therefore, enhanced implantation and persistence of P. oligandrum in the rhizosphere should improve plant protection. We report an investigation of the persistence of P. oligandrum and its impact on the native fungal microflora of the roots. Three strains with characteristic traits were selected to constitute an inoculum applied to tomato plant roots. The characteristics of the strains were the production of oospores to allow root colonization and favor persistence, the synthesis of tryptamine, a plant growth enhancer (22), and the production of oligandrin, a plant-protective elicitor (37). The inoculated rhizospheres were monitored to evaluate the persistence of the strains and their effects on the microflora. The populations of the common tomato root pathogen P. dissotocum (endemic in the studied systems) and of P. oligandrum were both assessed by plate counting and real-time PCR. The strain(s) of P. oligandrum responsible for the colonization of the rhizosphere was identified by inter-simple-sequence-repeat (ISSR) methodology. Single-strand conformational polymorphism (SSCP) investigations were used to study the effects of P. oligandrum on the fungal populations colonizing the rhizosphere and the fungal dynamics throughout the cropping season.
MATERIALS AND METHODS
Fungal culture.
The strains of Pythium oligandrum Dreschler used in this study were obtained from the CBS collection (CBS 149.84, CBS 382.34, CBS 530.74, CBS 109980, and CBS 109981) and the Souchotèque de Bretagne (Université Européenne de Bretagne and Université de Brest, ESMISAB) (LMSA 1.01.631, LMSA 1.03.024, and LMSA 1.03.025). Other Pythium mycoparasitic and pathogenic species (P. periplocum LMSA 1.01.640, P. dissotocum LMSA 1.01.536, P. ultimum LMSA 1.01.653, P. aphanidermatum LMSA 1.01.476, and P. violae LMSA 1.01.639) were obtained from the Souchotèque de Bretagne.
All strains were grown at 24°C in the dark on corn meal agar supplemented with antibiotics (19) and monitored regularly.
Molecular characterization of strains by sequencing.
DNA was extracted from 5-day-old cultures by use of a FastDNA spin kit (MP Biomedicals), with slight changes to the manufacturer's instructions. Approximately 0.02 g of mycelium was scraped from the margin of a colony and added to a Fast-Prep tube which contained garnet matrix, ceramic spheres, and 1 ml of CLS-Y buffer. The internal transcribed spacer 1 (ITS-1; complete sequence), ITS-2 (partial sequence), and 5.8S gene (complete sequence) were sequenced to confirm the identities of the strains used. Pythium sequences were determined with the primers described by Schurko and coworkers (51) and the protocol described by Allain-Boulé and coworkers (1), with no preliminary purification of the PCR products. The Pythium sequences obtained were compared to those belonging to clades C and D of the Pythium taxonomy reported by Lévesque and De Cock (26) and to Pythium oligandrum sequences recently added to GenBank. The sequences were aligned with Megalign software (version 5.06; DNAStar, Madison, WI), and PAUP (version 4.0b10) (55) was used with default parameters and 100 bootstrap replicates for heuristic searches.
Evaluation of oospore production for inocula.
To stimulate the production of oospores, the Pythium strains were cultured in liquid medium containing molasses as described by Le Floch and coworkers (23). Briefly, 100-ml aliquots of culture medium in flasks were each inoculated with 10 agar plugs of P. oligandrum and incubated at 25°C in the dark for 7 days. Each sample was cultured in triplicate. The mycelial mats were removed and fragmented in distilled water with a blender. Oospore counts in these preparations were determined three times with a Malassez cell.
Evaluation of oligandrin production and immunodetection by Western blotting.
Liquid cultures of P. oligandrum were obtained by growing oomycetes in a medium stimulating the production of elicitin (5). The flasks were incubated at 24°C in the dark for 14 days. The mycelium was removed and the concentrated filtrates of P. oligandrum were resuspended in sodium dodecyl sulfate (SDS) sample buffer. A Mini-Protean II system (Bio-Rad) was used for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (15% acrylamide gel). The proteins were electrotransferred onto 0.2-μm-pore-size nitrocellulose filters, and the filters were incubated with anti-oligandrin rabbit antiserum. Bound antibodies were detected with a second antibody (phosphatase-conjugated goat anti-rabbit antibody). The specificity of the anti-oligandrin antiserum was assessed by using two other elicitins (cryptogein and parasiticein) as negative controls (data not shown). Pythium intermedium culture filtrate was also used as a control.
Detection of tryptamine in P. oligandrum culture medium by capillary electrophoresis.
To test for the synthesis of auxin compounds by P. oligandrum, oomycetes were grown in a potato dextrose broth medium containing 0.25 mM tryptophan. After growth, indole derivatives were extracted from the P. oligandrum culture medium and assayed by the capillary electrophoresis method described by Rey and coworkers (47).
Measurements of plant material and yield.
Seeds of the tomato plant Lycopersicon esculentum Mill cv. Durinta (Western Seed, France) were sown in rock wool cubes and then transferred (27 January 2006) to coco-fiber slabs (four plants per slab) in two experimental greenhouses in Comité d'Action Technique et Economique precincts (St. Pol de Léon, Brittany, France). Each slab was wrapped in a plastic bag to isolate it from the others. The temperature in the greenhouses was regularly monitored and was between 18°C (±2°C; night) and 21°C (±2°C; day). The nutrient solution (Kemira, France) was delivered to each plant through a capillary system set at the crown. The pH was regularly monitored and was between 5.5 and 6.2. The culture conditions were similar in the two greenhouses, except that the temperature difference between night and day was about 2°C greater in greenhouse 2 than in greenhouse 1. The tomato fruits were collected twice a week from April to September, and the yield per m2 was measured. The Newman-Keuls test from Statbox Pro (version 6.6; Grimmersoft, Paris, France) was used for statistical analysis.
In each greenhouse, three rows (136 plants) of plants were used as controls, and two other rows (136 plants) were inoculated with strains CBS 530.74, CBS 109981, and LMSA 1.01.631 of Pythium oligandrum.
Pythium oligandrum inoculations.
Plants were inoculated with a mixture of P. oligandrum strains on 21 March and 11 April 2006: oospore-mycelium homogenates were used as previously described (23). Briefly, equal volumes of inoculum from each of the three strains were mixed and then diluted in water to the required final concentration of oospores. Aliquots of 100 ml of mixed oospore suspensions at 2.22 × 105 and 3.38 × 105 oospores ml−1 were deposited at the collar and on the roots of each plant, respectively.
Sampling and DNA extraction.
Root samples were collected twice a month throughout the cropping season (March to October 2006). Two coco-fiber slabs (samples) per experimental condition (inoculated and control plants) and per greenhouse were randomly selected and opened for root collection. DNA was extracted from 200 mg of fresh root tissue by use of a Fast DNA spin kit (MP Biomedicals) according to the manufacturer's instructions.
Assessment of root colonization by Pythium spp. by plate counting.
Root colonization by Pythium spp. was monitored from March to September under each condition (inoculated and control plants) by direct plating of about 400 nondisinfected root fragments on selective medium and incubation at 25°C in the dark (50 root segments per sample). After 48 h, P. dissotocum and P. oligandrum thalli were counted, and the ratio of the number of colonized fragments to the number of analyzed fragments was determined for each experimental condition. The differences in Pythium species root colonization between control and inoculated plants throughout the cropping season were assessed by a signed rank test (11) in which the means of the two repetitions per date and per treatment were used as paired data.
Real-time PCR to assess root colonization by Pythium spp.
The primers and probes listed in Table 1 were used for quantitative PCR. To normalize the data, the amount of tomato DNA in each sample was quantified as reported by Le Floch et al. (25) and by using specific primers and a probe for the tomato Lat52 gene (65). A MiniOpticon thermocycler (Bio-Rad) was used for PCR; the reaction volume was 15 μl and consisted of 7.5 μl of 2× Quantitect probe mix (Qiagen) and a 0.4 μM concentration of each primer and probe. The cycling parameters were 95°C for 15 min followed by 40 cycles of 94°C for 30 s (denaturation), 60°C for 30 s (annealing), and 72°C for 30 s (extension). Fluorescence was monitored during every PCR cycle, after the extension step.
TABLE 1.
Primers and fluorogenic probes used for quantitative PCR
| Gene | Primer or probe | Sequence (5′-3′)e | Amplicon size (bp) |
|---|---|---|---|
| Tomato LAT52 gene | up_Lat1a | AGACCACGAGAACGATATTTGC | 92 |
| lo_Lat2a | TTCTTGCCTTTTCATATCCAGACA | ||
| Lpa | FAM-CTCTTTGCAGTCCTCCCTTGGGCT-BHQ1 | ||
| P. oligandrum ITS | up_F1b | TGCTTCGTCGCAAGACT | 124 |
| lo_146d | CGTATTCGGAGTATAGTTCAGT | ||
| 142_LNAc | FAM-AGTCTGCGTCTATTTTGGA-BHQ1 | ||
| P. dissotocum ITS | up_504d | GTTTGGATCGCTTTGCT | 215 |
| lo_702d | CCGAAGCTAGAGCGCTT | ||
| 183_LNAc | FAM-TGACTGGAGTTGTTTTCTGTT-BHQ1 |
Standard curves were generated by plotting the cycle threshold values for a 10-fold dilution series of known amounts of genomic DNA of the targeted Pythium species versus the logarithm of the concentration. A regression line was drawn to determine the concentration of the target DNA in the test sample from its cycle threshold value. Opticon Monitor software (version 3.1.32; Bio-Rad Laboratories, Inc.) was used for data analysis.
The amounts of Pythium spp. detected in control and inoculated plants during the growing season were compared by a signed rank test (11) in which the means of the two repetitions per date and per treatment were used as paired data.
ISSR amplification of Pythium oligandrum DNA.
At the end of the cropping season, 90 isolates of P. oligandrum were isolated from the inoculated tomato roots as described above. After plate counting and identification based on the presence of echinulated oospores, P. oligandrum strains were isolated, purified, and transferred into the same liquid medium as that used for the preparation of the inoculum. After 15 days in the dark and at 25°C, the biomass was separated from the medium, rinsed with distilled sterile water, dried, and stored at −20°C until DNA extraction.
The primer used for ISSR analysis was [GACA]4. The reaction mixture (20 μl) was composed of 50 ng of DNA template, 2 μM of primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 2 mM MgCl2, 4 μl of 5× Green buffer (Promega), and 0.15 μl of GoTaq DNA polymerase (5 U/μl; Promega). A PTC-100 thermocycler (MJ Research, Inc.) was used for PCR, with the following conditions: initial denaturation at 95°C for 4 min and 30 cycles of 30 s at 95°C, 45 s at 58°C, and 2.5 min at 72°C.
For each mixture, the amplification products were analyzed by electrophoresis of 12-μl aliquots in a 2% agarose gel (Invitrogen) in 0.5× Tris-borate-EDTA (Promega) buffer, staining with ethidium bromide, and visualization under UV light. A molecular size ladder (100-bp BenchTop ladder; Promega) was used as the size standard.
Analysis of the rhizosphere fungal community by SSCP analysis.
Pairs of primers recognizing the 28S ribosomal DNA (rDNA) gene (U1/U2), the ITS region (ITS1/ITS2), and the mitochondrial large-subunit rDNA gene (ML1/ML2) were used for SSCP (Table 2). DNA was amplified by PCR in a PTC-100 thermocycler (MJ Research, Inc.) in a reaction mixture (25-μl final volume) consisting of 1 μl of DNA template, a 0.2 mM concentration of each deoxynucleoside triphosphate, 2 ng/μl of each primer, 2.5 μl of 10× Pfu Turbo buffer, and 0.05 Unit of Pfu Turbo DNA polymerase (Stratagene, The Netherlands). The cycling conditions were as follows: enzyme activation at 95°C for 2 min; 35 cycles of denaturation at 95°C for 30 s, hybridization for 30 s (Table 2), and extension at 72°C for 30 s; and a final extension at 72°C for 10 min. The PCR products were visualized by 1% Tris-borate-EDTA agarose gel electrophoresis prior to SSCP analysis. The lengths of the fragments yielded by amplification of DNAs from all fungi tested were in the range of 200 to 300 bp.
TABLE 2.
Pairs of universal primers used in SSCP analyses of native rhizosphere fungal populations
| Gene | Primer | Primer sequence (5′-3′)d | Amplicon size (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| Mitochondrial large-subunit rDNA | ML1a | FAM-GTACTTTTGCATAATGGGTCAGC | 253 | 58 |
| ML2a | FAM-TATGTTTCGTAGAAAACCAGC | |||
| ITS | ITS1a | TCCGTAGGTGAACCTGCGG | 300 | 62 |
| ITS2a | FAM-GCTGCGTTCTTCATCGATGC | |||
| 28S rDNA | U1b,c | FAM-GTGAAATTGTTGAAAGGGAA | 260 | 56 |
| U2b,c | GACTCCTTGGTCCGTGTT |
SSCP analyses were performed on an ABI Prism 310 genetic analyzer (Applied Biosystems) with a 47-cm-long capillary. One microliter of a PCR product was mixed with 18.8 μl Hi-Di formamide (Applied Biosystems) and 0.2 μl of the Genescan 400 HD ROX standard internal DNA molecular size marker (Applied Biosystems). The sample mixture was denatured at 95°C for 5 min, immediately cooled on ice, and then loaded onto the instrument. The nondenaturing polymer consisted of 5% POP conformational analysis polymer (Applied Biosystems), 10% glycerol, EDTA buffer (10×), and water (Applied Biosystems). The migration time was set to 45 min, the voltage was 12 kV, and the temperature was 32°C.
Samples were allowed to comigrate with the fluorescent size standard (GeneScan 400 ROX) to allow comparison of migration profiles between samples. Patterns were aligned with the SAFUM program (66) and studied by principal component analysis (PCA) with Statbox Pro (version 6.6; Grimmersoft, Paris, France).
RESULTS
Molecular characterization of P. oligandrum strains.
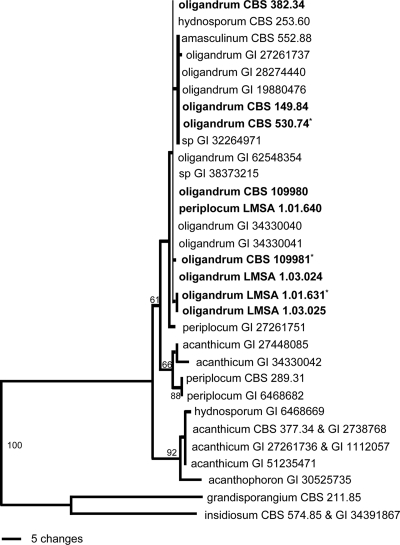
The sequences of the strains of P. oligandrum studied here shared substantial similarity with that of the CBS 382.34 strain used for species description by van der Plaats-Niterink (60). The P. oligandrum sequences fall within the range of variations previously observed within this species (Fig. 1); thus, these strains belong to the P. oligandrum complex. The ITS sequence of P. periplocum LMSA 1.01.640 suggests a misidentification based on morphological features.
FIG. 1.
Phylogenetic tree of the Pythium strains used in this study (in bold), based on ITS-1 and -2 and the 5.8S gene of nuclear rDNA. The sequences of representative strains (CBS numbers) and strains from the LUBEM laboratory (LMSA numbers) were compared against all available entries in GenBank (GI numbers), as described by Lévesque and De Cock (26). Recently reported P. oligandrum sequences are included. The strains used in this study are shown in bold with an asterisk. This is one of 18 equally parsimonious trees (length, 147; consistency index, 0.925; retention index, 0.926; rescaled consistency index, 0.856).
Morphological and biochemical characterization of P. oligandrum strains.
The production of oospores, tryptamine, and oligandrin by the strains of P. oligandrum under study was tested (Table 3). All P. oligandrum strains tested, except for strain CBS 382.34, produced oospores. CBS 109981 produced the most (2 × 105 to 2.5 × 105 oospores ml−1), followed by LMSA 1.01.631 (1.5 × 105 to 2.0 × 105 oospores ml−1), and CBS 530.74 produced the least (0.5 × 105 to 1.1 × 105 oospores ml−1).
TABLE 3.
Evaluation of oospores, tryptamine, and oligandrin produced by the strains of Pythium species used in this study
| Pythium species | Strain code | Collection reference no.b | No. of oospores (103 ml−1)c | Tryptamine concn (nmol mg−1)c,d | Oligandrin productione |
|---|---|---|---|---|---|
| P. oligandrum | 126 | LMSA 1.03.024 | 193EF | 82.5AB | + |
| 981a | CBS 109981 | 235F | 0A | ++ | |
| 530a | CBS 530.74 | 84B | 72.5AB | +++ | |
| 147 | LMSA 1.03.025 | 171DE | 15.7A | ++ | |
| 1133a | LMSA 1.01.631 | 139CD | 200C | +++ | |
| 149 | CBS 149.84 | 107BC | 64.6AB | +++ | |
| 382 | CBS 382.34 | 0A | 0A | + | |
| 980 | CBS 109980 | 217F | 48.7AB | + | |
| P. periplocum | Pp | LMSA 1.01.640 | 0.3A | 0A | − |
| P. dissotocum | Pd | LMSA 1.01.536 | ND | ND | − |
| P. ultimum | Pu | LMSA 1.01.653 | ND | ND | − |
| P. aphanidermatum | Pa | LMSA 1.01.476 | ND | 0A | − |
| P. violae | Pv | LMSA 1.01.639 | ND | 0A | − |
These strains were used for biocontrol assays with the rhizosphere of hydroponically grown tomato plants.
CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; LMSA, Souchotèque de Bretagne, ESMISAB, Brest, France.
Values are means for three replicates per strain. The absence of a significant difference (threshold set at P = 0.05), as assessed by the Duncan multiple test, is indicated by the use of the same letter in a column. ND, not determined.
Concentrations of tryptamine are given per mg of mycelium (dry weight).
+, positive and proportional immunoreaction; −, no reaction with anti-oligandrin antibodies.
All P. oligandrum strains except for CBS 109981 and CBS 382.34 produced tryptamine in the presence of tryptophan, a precursor of indole acetic acid metabolism. The highest tryptamine concentrations were obtained with strains LMSA 1.01.631 (200 nmol mg−1) and CBS 530.74 (72.5 nmol mg−1). Note that tryptamine production correlates with the metabolization of tryptophan. Large amounts of tryptophan were detected in the filtrates of cultures of CBS 109981 and CBS 382.34, consistent with their inability to synthesize tryptamine (data not shown).
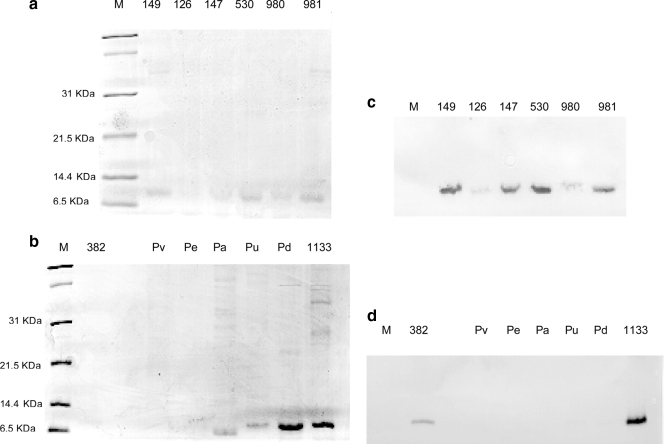
An electrophoresis-based method was used to assess oligandrin production by these strains (Fig. 2). A newly synthesized protein of about 10 kDa was detected in the culture filtrates of P. oligandrum strains and in those of the pathogenic species P. violae, P. ultimum, P. aphanidermatum, and P. dissotocum (Fig. 2a and b). Rabbit anti-oligandrin serum (Fig. 2c and d) recognized the protein produced by strains of P. oligandrum but not proteins of similar molecular masses in culture filtrates of the other Pythium species. Thus, only P. oligandrum strains produced oligandrin.
FIG. 2.
Electrophoretic patterns on polyacrylamide gels and corresponding immunoblots of concentrated culture filtrates of P. oligandrum strains. (a and b) Coomassie blue-stained 12.5% SDS-PAGE gels. The same culture filtrates were subjected to 12.5% SDS-PAGE, and the proteins were transferred to nitrocellulose membranes. (c and d) The membranes were probed with rabbit anti-oligandrin serum. Each lane was loaded with 25 μg of proteins. Lanes M, molecular mass standard (Bio-Rad); other lanes, culture filtrates of the Pythium strains listed in Table 3.
On the basis of these various analyses, P. oligandrum strains LMSA 1.01.631, CBS 109981, and CBS 530.74 were used as biocontrol agents for tomato plants grown by hydroponics.
Root colonization by P. dissotocum, assessed by plate counting and real-time PCR.
P. dissotocum was first detected by plate counting in May. The rates of root colonization of uninoculated control plants by P. dissotocum rose to a maximum of 60% in July and August before decreasing in September. At the end of the cropping season, approximately 40% of the root systems of the controls were still colonized. The pattern of colonization of roots of P. oligandrum-inoculated plants by P. dissotocum was generally similar to that for control plants, except that the colonization rates (maximum, 40%) were significantly lower (signed rank test; P = 0.029) (Fig. 3a1).
FIG. 3.
(a) Root colonization (%) by P. dissotocum (a1) and P. oligandrum (a2), assessed by plate counting. The values reported are means for eight samples collected each month. The signed rank test indicates preferential colonization of the control plants by P. dissotocum (P = 0.029) and a more extensive colonization of the inoculated plants by P. oligandrum (P = 0.0000123). (b) Root colonization by P. dissotocum (b1) and P. oligandrum (b2), assessed by real-time PCR. DNA quantities correspond to amounts of Pythium DNA (fg) per unit of tomato DNA (ng). Median values are represented on the graphs (medians are suitable markers for dissymmetrical distributions). P. oligandrum DNA quantities were higher on inoculated than on control plants (signed rank test; P = 0.0000123), and those of P. dissotocum on control and inoculated plants were not significantly different.
Real-time PCR analyses confirmed these findings. In both greenhouses and for all experimental conditions (inoculated and control plants), P. dissotocum was first detected in July, and the populations increased to the end of the growing season in September (Fig. 3b1). P. dissotocum populations on P. oligandrum-inoculated plants were smaller than those on controls, but the difference was not significant, as assessed by the signed rank test, possibly because the sample sizes were too small.
Assessment of the persistence of P. oligandrum by plate counting and real-time PCR.
P. oligandrum colonization of the rhizosphere of hydroponically grown tomato plants was assessed by plate counting (Fig. 3). Two weeks after the second inoculation (end of April), P. oligandrum was detected on 90% of roots and persisted (at ≥50%) throughout the cropping season (Fig. 3a2). At the end of the cropping season, P. oligandrum was very highly significantly more abundant on inoculated plants than on controls (signed rank test; P = 0.0000123).
These findings were confirmed by real-time PCR (Fig. 3b2). P. oligandrum was detected after its inoculation and was found in increasing quantities throughout the growing season. The amount of P. oligandrum on inoculated plants was significantly higher (P = 0.0000123) than that on control plants, which were virtually P. oligandrum-free.
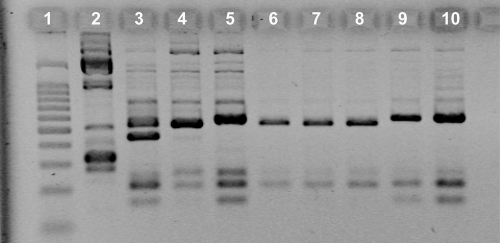
Discrimination of Pythium oligandrum strains by ISSR analysis.
The primer [GACA]4 and electrophoresis were used to differentiate between the three strains used to inoculate the tomato plants (Fig. 4). Among the 90 isolates collected at the end of the cropping season, 84 were identified as strain CBS 530.74, 4 as LMSA 1.01.631, and 2 as CBS 109981 strain (Table 3).
FIG. 4.
Pythium oligandrum ISSR electrophoretic patterns. Lane 1, BenchTop 100-bp ladder (Promega, France); lane 2, P. dissotocum LMSA 1.01.531; lane 3, P. oligandrum CBS 109981; lane 4, P. oligandrum CBS 530.74; lane 5, P. oligandrum LMSA 1.01.631; lanes 6 to 10, P. oligandrum strains collected from tomato roots at the end of the cropping season.
Effect of introduction of P. oligandrum on the genetic structure of the rhizosphere fungal community.
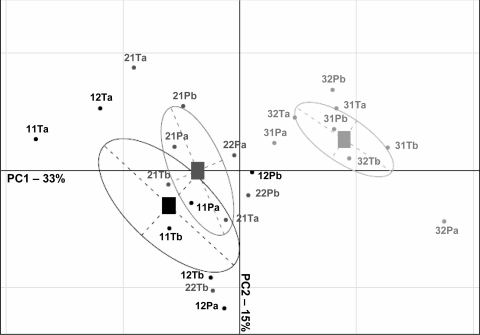
The 28S rDNA gene was studied to assess the genetic structure of the rhizosphere fungal communities. There were no differences in SSCP profiles between inoculated and control plants at any date of sampling (Fig. 5). Control and P. oligandrum-inoculated samples were not separated on either the first or the second PCA analysis axis (Fig. 6), which explains 33 and 15%, respectively, of the total fungal variability. The complexity of the fungal profiles was greater at the end of the cropping season than at the beginning for all experimental conditions (inoculated and control plants) (Fig. 5); thus, the genetic structure of the fungal community changed with time. The root samples collected at the beginning, the middle, and the end of the growing season were separated versus time on PC1, the first PCA axis (Fig. 6). Each condition was sampled twice at each time point, and these duplicate samples appeared distant from each other on the graph (points labeled “a” and “b,” respectively, in Fig. 6), indicating heterogeneity in the distribution of the fungal populations. Similar results were obtained when the fungal communities were analyzed according to the ITS region. Note that the apparent changes in the fungal community were less marked when the analysis was based on the mitochondrial large-subunit rDNA gene (data not shown).
FIG. 5.
SSCP fingerprints of fungal microfloras from control and P. oligandrum-inoculated plants at the beginning (April) and end (September) of the cropping season in the two greenhouses.
FIG. 6.
Comparison of genetic structures of fungal communities of the rhizosphere associated with inoculated and control plants by PCA, using fungal 28S rRNA profiles obtained by SSCP. For each sample, the first number represents the date of sampling (1, start; 2, middle; 3, end of the cropping season), the second number refers to the compartment (1, greenhouse 1; 2, greenhouse 2), the letters T and P are for control plants and P. oligandrum-inoculated plants, respectively, and the last letter indicates the sample.
Tomato yields of controls and P. oligandrum-inoculated plants.
The numbers of bunches per tomato plant and fruits per bunch were similar for controls and P. oligandrum-inoculated plants (Table 4). However, the tomato yield and the mean weight of tomatoes were both slightly (but not significantly) lower for inoculated plants than for controls.
TABLE 4.
Agronomic resultsa
| Greenhouse no. | Treatment | Tomato yield (kg/m2) | No. of bunches/plant | Mean wt (kg) | No. of fruits/bunch |
|---|---|---|---|---|---|
| 1 | Control | 48.39 | 38.1 | 111 | 4.9 |
| Inoculation | 45.45 | 37.7 | 106 | 4.9 | |
| 2 | Control | 48.76 | 38.3 | 112 | 4.9 |
| Inoculation | 47.23 | 38.5 | 108 | 4.9 |
There were no significant differences at a P value cutoff of 0.05 by the Newman-Keuls test.
DISCUSSION
The aim of this study was to optimize P. oligandrum establishment on roots of hydroponically grown tomato plants. Three strains of P. oligandrum were selected (CBS 530.74, LMSA 1.01.631, and CBS 109981), partly because they produce oospores expected to favor both colonization and persistence of this antagonistic oomycete in the rhizosphere. However, ISSR analysis of P. oligandrum strains isolated from root samples collected at the end of the cropping season showed that the strain that produced the fewest oospores was by far the most abundant (90% of all P. oligandrum organisms present). This raises serious doubts about the relevance of selecting P. oligandrum strains according to their production of oospores in vitro. Oospores may exhibit extended dormancy and then germinate when the environmental conditions become more favorable (61). Reported percentages of germination are divergent: McQuilken et al. (29) described up to 30% germination under optimal laboratory conditions, but Cliquet and Tirilly (9) reported a value below 5%. Our findings suggest that there is no direct relationship between oospore production in vitro and colonization of the rhizosphere. Oospore germination in the field can be affected by diverse environmental factors, including root exudates, plant age, and the cultivar used. These effects are not easily reproducible in laboratory experiments. The reason that only one of the three P. oligandrum strains tested appeared to be well adapted to the environmental conditions in soilless tomato culture remains to be determined.
Plate counting indicated that the colonization of the root system by these P. oligandrum strains was 50% or higher. Real-time PCR demonstrated high rates of persistence in the rhizosphere throughout the cropping season (April to September). Both colonization and persistence rates were thus higher than the results reported by other investigations (23, 25). The development of P. oligandrum can be inhibited by fast growth of P. dissotocum on the selective medium used for plate counting (25), and this may have affected the results. Indeed, the colonization of the rhizosphere by P. oligandrum may be underestimated. The three P. oligandrum strains used were found in very different quantities in the rhizosphere: it is possible that the growth of two strains was inhibited by P. dissotocum, whereas that of the most abundant, strain CBS 530.74, was not. Some P. dissotocum complex strains grow quickly, whereas others do not (42). The level of root colonization, as assessed in a selective medium, may be the result of the association of P. oligandrum mycoparasitism with moderate development of P. dissotocum strains. It may therefore be possible to enhance the persistence of P. oligandrum on tomato roots by using mixtures of strains.
There is a very diverse set of interactions between P. oligandrum and other fungi or oomycetes (49). Interactions differ between genera, between species of the same genus, and sometimes between strains of the same species. For instance, within the genus Phytophthora, P. oligandrum attacks the species Phytophthora parasitica through mycoparasitism and Phytophthora megasperma through antibiosis. The microbial responses of these organisms to prevent such attacks are also divergent: P. parasitica reinforces its wall, whereas P. megasperma is unable to trigger host-like defenses. P. aphanidermatum strains show diverse behaviors compared to P. oligandrum: the P. oligandrum strain CBS 1.01.631 mycoparasitizes the P. aphanidermatum strain used by Benhamou and coworkers (3), but P. aphanidermatum also develops a mycoparasite-like behavior against another P. oligandrum strain (20). It is clear that the relationships between microorganisms or closely related fungi are highly complex. Therefore, a promising approach to biocontrol is the use of antagonistic mixtures including various strains, each displaying its own antagonistic properties against a range of potentially pathogenic microorganisms. We found that inoculation with P. oligandrum and its persistence in the rhizosphere both tend to reduce P. dissotocum populations. This antagonistic effect was the strongest when these pathogenic populations emerged; on the other hand, it was less marked at the end of the cropping season (in September). The following two explanations can be proposed: (i) the huge genetic diversity in P. dissotocum strains (62) resulted in very diverse interactions between P. dissotocum and P. oligandrum strains, making control of this minor pathogenic group difficult; or (ii) the concentrations of P. oligandrum were too low to affect P. dissotocum populations significantly throughout the season.
SSCP fingerprinting can reveal rapid changes in microbial communities even if their composition is unknown. It is therefore a useful technique for following the changes in genetic structure of the fungal populations in the rhizosphere. This method also avoids the biases associated with culture methods. SSCP analyses of three different DNA regions indicated increases in the number and areas of peaks as the cropping season progressed. This implies increases in the complexity and size of the microflora with time. Biological processes in the rhizosphere are strongly affected by plant root exudates, which consist of easily degradable organic carbon compounds that attract specific microbial populations and stimulate their growth. Maloney et al. (27) found correlations between the changes within the culturable rhizobacterial populations of lettuce and tomato and the plant growth stage, carbon availability, and nitrogen concentration. The most marked aspect of this “rhizosphere effect” was quantitative. In this experiment, it seems likely that the changes in fungal populations over the course of the cropping season were induced by plant root exudates.
There is no consensus about whether shifts in the rhizosphere microflora can result from pathogenic attacks. Indeed, Naseby et al. (33) and Hagn et al. (16) suggested that changes in the microbial communities of the rhizosphere are a consequence of both root damage caused by Pythium ultimum and secondary colonization due to the resulting nutrient leakage. Calvo-Bado et al. (6) reported that the microbial communities established early on the roots of tomatoes grown in soilless systems are robust and resistant to the effect(s) of the introduction of oomycete pathogens or of switching from a recirculating to a run-to-waste nutrient supply. However, this idea, arising from experiments conducted over only 1.5 months, is contradicted by the observation of changes in the microbial community over the 6 months of our experiments. However, both studies suggest that there are no substantial changes in the genetic structure of the native fungal populations after root inoculation with the nonpathogenic oomycete P. oligandrum or the pathogenic oomycetes Pythium group F, P. aphanidermatum, and P. cryptogea (6).
The P. oligandrum strains used were expected to promote plant resistance to pathogens via the stimulation of plant defense systems and to promote plant growth by boosting the auxinic pathway (22). Therefore, P. oligandrum strains were selected as producers of oligandrin, a glycoprotein of the elicitin family, and of the auxinic compound tryptamine.
All P. oligandrum strains produce oligandrin. Strain LMSA 1.01.640, identified by morphological characters as P. periplocum, another mycoparasite, and the pathogens P. dissotocum, P. ultimum, P. intermedium, and P. violae do not. Proteins of similar molecular mass to oligandrin (about 10 kDa) were detected in culture media of some plant-pathogenic Pythium species, but they were not recognized by oligandrin antibodies. Thus, only P. oligandrum strains produce oligandrin, an important part of their metabolism (37). The production of oligandrin may serve as a marker for P. oligandrum identification. Our results support the findings of Colas et al. (10), who reported a high specificity of elicitins produced by Phytophthora species. Elicitin-like expressed sequence tag assemblies found in P. ultimum had a much greater sequence similarity to elicitin-like proteins from P. sylvaticum than to those from P. oligandrum. This is consistent with the closer phylogenetic relationship between P. ultimum and P. sylvaticum than that between these species and P. oligandrum (8).
Le Floch et al. (22) demonstrated a correlation between tryptamine production by P. oligandrum and an enhancement of root development of tomato plantlets. We observed large differences in tryptamine production between P. oligandrum strains. The tryptamine pathway is common in the Pythium genus and has been found in P. oligandrum strains and in plant-pathogenic Pythium species. For instance, the pathogens P. dissotocum and P. ultimum produce indole-3-acetic acid and tryptophol from tryptophan (47, 52). This study shows that under the same experimental conditions, P. oligandrum synthesized only tryptamine from a tryptophan precursor, with no other auxin metabolites detected. Further investigations are required to determine whether the observed differences in tryptamine production are associated with complete or incomplete tryptamine pathways in P. oligandrum strains.
Although root colonization by P. oligandrum was substantial and constant throughout the cropping season, this had no beneficial effect on tomato yield. It is difficult to assess the protection conferred by P. oligandrum against P. dissotocum from this experiment. The rhizosphere of both control and inoculated plants was colonized by P. dissotocum. The plants were not severely attacked, and as usual with this minor pathogen, infections were asymptomatic and probably limited to the outer part of roots (44). Such asymptomatic attacks on roots can cause yield losses when they last for a period of several months (43), but this was not the case in this experiment.
The strain CBS 530.74 was the most abundant of the inoculated strains in the rhizosphere. It produced tryptamine in quantities (72.5 nmol mg−1) three times lower than those for the other two strains in vitro. However, it is difficult to extrapolate from in vitro experiments to the rhizosphere of plants grown hydroponically. In vitro experiments (22) demonstrated that tryptamine production by P. oligandrum has a positive effect on root growth of tomato plantlets. However, no such auxinic effect in the rhizosphere of mature tomato plants was observed. The protection and growth promotion of plants by P. oligandrum may well depend on a combination of factors, including plant species, environmental conditions, and cropping practices, with all of them having an influence on yield.
In conclusion, this work describes the successful establishment of the three selected P. oligandrum strains, but only one of them seemed particularly adapted for colonizing the rhizosphere of plants grown hydroponically. P. oligandrum was able to colonize and persist in the complex microbial ecosystem without greatly modifying the indigenous fungal populations, other than a reduction of the population of pathogenic P. dissotocum. The beneficial effects of these strains on plant development remain to be determined, particularly for plants grown under normal environmental conditions that are attacked by minor pathogens.
Acknowledgments
This research work was financially supported by the Brittany and Pays de la Loire Regional Councils (Pôle Agronomique Ouest, Pathorac program).
We are grateful to Padraig Nally (Centre for Epidemiology and Risk Analysis, Veterinary Laboratories Agency, United Kingdom) and Marie-Paule Friocourt (Université de Brest, France) for critical reviews of the manuscript.
Footnotes
Published ahead of print on 15 May 2009.
REFERENCES
- 1.Allain-Boulé, N., R. Tweddell, M. Mazzola, R. Bélanger, and C. A. Lévesque. 2004. Pythium attrantheridium sp. nov.: taxonomy and comparison with related species. Mycol. Res. 108:795-805. [DOI] [PubMed] [Google Scholar]
- 2.Benhamou, N., P. Rey, M. Chérif, J. Hockenhull, and Y. Tirilly. 1997. Treatment with the mycoparasite Pythium oligandrum triggers induction of defense-related reactions in tomato roots when challenged with Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathology 87:108-122. [DOI] [PubMed] [Google Scholar]
- 3.Benhamou, N., P. Rey, K. Picard, and Y. Tirilly. 1999. Ultrastructural and cytochemical aspects of the interaction between the mycoparasite Pythium oligandrum and soilborne plant pathogens. Phytopathology 89:506-517. [DOI] [PubMed] [Google Scholar]
- 4.Benhamou, N., R. R. Bélanger, P. Rey, and Y. Tirilly. 2001. Oligandrin, the elicitin-like protein produced by the mycoparasite Pythium oligandrum, induces systemic resistance to Fusarium crown and root rot in tomato plants. Plant Physiol. Biochem. 39:681-698. [Google Scholar]
- 5.Bonnet, P., E. Bourdon, M. Ponchet, J. P. Blein, and P. Ricci. 1996. Acquired resistance triggered by elicitins in tobacco and other plants. Eur. J. Plant Pathol. 102:181-192. [Google Scholar]
- 6.Calvo-Bado, L. A., G. Petch, N. R. Parsons, J. A. W. Morgan, T. R. Pettitt, and J. M. Whipps. 2006. Microbial community responses associated with the development of oomycete plant pathogens on tomato roots in soilless growing systems. J. Appl. Microbiol. 100:1194-1207. [DOI] [PubMed] [Google Scholar]
- 7.Chatterton, S., J. C. Sutton, and G. J. Boland. 2004. Timing Pseudomonas chlororaphis applications to control Pythium aphanidermatum, Pythium dissotocum, and root rot in hydroponic peppers. Biol. Control 30:360-373. [Google Scholar]
- 8.Cheung, F., J. Win, J. M. Lang, J. Hamilton, H. Vuong, J. F. Leach, S. Kamoun, and C. A. Lévesque. 2008. Analysis of the Pythium ultimum transcriptome using Sanger and pyrosequencing approaches. BMC Genomics 9:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliquet, S., and Y. Tirilly. 2002. Development of a defined medium for Pythium oligandrum oospore production. Biocontrol Sci. Technol. 12:455-467. [Google Scholar]
- 10.Colas, V., I. Lacourt, P. Ricci, F. Vanlerberghe-Masutti, P. Venard, A. Poupet, and F. Panabières. 1997. Diversity of virulence in Phytophthora parasitica on tobacco, as reflected by nuclear RFLPs. Phytopathology 88:205-212. [DOI] [PubMed] [Google Scholar]
- 11.Dagnelie, P. 1975. Théorie et méthodes statistiques, vol. 2. Presses Agronomiques, Gembloux, Belgium.
- 12.Déniel, F., P. Rey, M. Chérif, A. Guillou, and Y. Tirilly. 2004. Inoculations of filter unit with antagonistic- and PGPR-bacteria improve slow filtration efficiency in soilless culture. Can. J. Microbiol. 50:499-508. [DOI] [PubMed] [Google Scholar]
- 13.Ehret, D. L., B. Alsanius, W. Wohanka, J. G. Menzies, and R. Utkhede. 2001. Disinfestations of recirculating nutrient solutions in greenhouse horticulture. Agronomy 21:323-339. [Google Scholar]
- 14.Foley, M. F., and J. W. Deacon. 1986. Susceptibility of Pythium spp and other fungi to antagonism by the mycoparasite Pythium oligandrum. Soil Biol. Biochem. 18:91-95. [Google Scholar]
- 15.Godfrey, S. A. C., R. D. Monds, D. T. Lash, and J. W. Marshall. 2003. Identification of Pythium oligandrum using species-specific ITS rDNA PCR oligonucleotides. Mycol. Res. 107:790-796. [DOI] [PubMed] [Google Scholar]
- 16.Hagn, A., M. Engel, B. Kleikamp, J. C. Munch, M. Schloter, and C. Bruns. 2008. Microbial community shifts in Pythium ultimum-inoculated suppressive substrates. Biol. Fertil. Soils 44:481-490. [Google Scholar]
- 17.Hase, S., S. Takahashi, S. Takenaka, K. Nakaho, T. Arie, S. Seo, Y. Ohashi, and H. Takahashi. 2008. Involvement of jasmonic acid signalling in bacterial wilt disease resistance induced by biocontrol agent Pythium oligandrum in tomato. Plant Pathol. 57:870-876. [Google Scholar]
- 18.Herrero, M. L., A. Hermansen, and O. N. Elen. 2003. Occurrence of Pythium spp. and Phytophthora spp. in Norwegian greenhouses and their pathogenicity on cucumber seedlings. J. Phytopathol. 151:36-41. [Google Scholar]
- 19.Jeffers, S. N., and S. B. Martin. 1986. Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 70:1038-1043. [Google Scholar]
- 20.Jones, E. E., and W. Deacon. 1995. Mycoparasite-like behaviour of the plant pathogen Pythium aphanidermatum in vitro. Plant Pathol. 44:396-405. [Google Scholar]
- 21.Kucharek, T., and D. Mitchell. 2000. Diseases of agronomic and vegetable crops caused by Pythium. Plant pathology fact sheet PP-53. University of Florida, Gainesville, FL.
- 22.Le Floch, G., P. Rey, E. Benizri, N. Benhamou, and Y. Tirilly. 2003. The impact of auxin-compounds produced by the antagonistic oomycete, Pythium oligandrum or the minor pathogen, Pythium group F on plant growth. Plant Soil 257:459-470. [Google Scholar]
- 23.Le Floch, G., P. Rey, F. Déniel, N. Benhamou, K. Picard, and Y. Tirilly. 2003. Enhancement of development and induction of resistance in tomato plants by the antagonist, Pythium oligandrum. Agronomie 23:455-460. [Google Scholar]
- 24.Le Floch, G., N. Benhamou, E. Mamaca, M. I. Salerno, Y. Tirilly, and P. Rey. 2005. Characterisation of the early events in atypical tomato root colonisation by a biocontrol agent, Pythium oligandrum. Plant Physiol. Biochem. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Le Floch, G., J. Tambong, J. Vallance, Y. Tirilly, C. A. Lévesque, and P. Rey. 2007. Rhizosphere persistence of three Pythium oligandrum strains in tomato soilless culture assessed by DNA macroarray and real-time PCR. FEMS Microbiol. Ecol. 61:317-326. [DOI] [PubMed] [Google Scholar]
- 26.Lévesque, C. A., and A. W. A. M. De Cock. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108:1363-1383. [DOI] [PubMed] [Google Scholar]
- 27.Maloney, P. E., A. H. C. van Bruggen, and S. Hu. 1997. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb. Ecol. 34:109-117. [DOI] [PubMed] [Google Scholar]
- 28.McPherson, G. M., M. R. Harriman, and D. Pattison. 1995. The potential for spread of root diseases in recirculating hydroponic systems and their control with disinfection. Med. Fac. Landbouww Univ. Gent 60/2b:371-379. [Google Scholar]
- 29.McQuilken, M. P., J. M. Whipps, and R. C. Cooke. 1990. Oospores of the biocontrol agent Pythium oligandrum bulk-produced in liquid culture. Mycol. Res. 94:613-616. [Google Scholar]
- 30.McQuilken, M. P., J. M. Whipps, and R. C. Cooke. 1992. Nutritional and environmental factors affecting biomass and oospore production of the biocontrol agent Pythium oligandrum. Enzyme Microb. Technol. 14:106-111. [Google Scholar]
- 31.Mohamed, N., J. Lherminier, M. J. Farmer, J. Fromentin, N. Béno, V. Houot, M. L. Milat, and J. P. Blein. 2007. Defense responses in grapevine leaves against Botrytis cinerea induced by application of a Pythium oligandrum strain or its elicitin, oligandrin, to roots. Biochem. Cell Biol. 97:611-620. [DOI] [PubMed] [Google Scholar]
- 32.Möhlenhoff, P., L. Müller, A. A. Gorbushina, and K. Petersen. 2001. Molecular approach to the characterisation of fungal communities: methods for DNA extraction, PCR amplification and DGGE analysis of painted art objects. FEMS Microbiol. Lett. 195:169-173. [DOI] [PubMed] [Google Scholar]
- 33.Naseby, D. C., J. A. Pascual, and J. M. Lynch. 2000. Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. J. Appl. Microbiol. 88:161-169. [DOI] [PubMed] [Google Scholar]
- 34.Paulitz, T. C. 1997. Biological control of root pathogens in soilless and hydroponic systems. HortScience 32:193-196. [Google Scholar]
- 35.Paulitz, T. C., and R. R. Bélanger. 2001. Biological control in greenhouse systems. Annu. Rev. Phytopathol. 39:103-133. [DOI] [PubMed] [Google Scholar]
- 36.Pettitt, T. R., A. J. Wakeham, M. F. Wainwright, and J. G. White. 2002. Comparison of serological, culture, and bait methods for detection of Pythium and Phytophthora zoospores in water. Plant Pathol. 51:720-727. [Google Scholar]
- 37.Picard, K., M. Ponchet, J. P. Blein, P. Rey, Y. Tirilly, and N. Benhamou. 2000. Oligandrin. A proteinaceous molecule produced by the mycoparasite Pythium oligandrum induces resistance to Phytophthora parasitica infection in tomato plants. Plant Physiol. 124:379-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poncet, C., R. Brun, M. Offroy, and G. Bonnet. 2001. Disinfection of recycling water in rose cultures. Acta Horticult. 547:121-126. [Google Scholar]
- 39.Postma, J., E. A. Van Os, and G. Kritzman. 1999. Prevention of root diseases in closed soilless growing systems by microbial optimization. Med. Fac. Landbouww Univ. Gent 64/3b:431-440. [Google Scholar]
- 40.Postma, J., M. J. E. I. M. Willemsen de Klein, and J. D. van Elsas. 2000. Effect of the indigenous microflora on the development of root and crown rot caused by Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 90:125-133. [DOI] [PubMed] [Google Scholar]
- 41.Postma, J., B. P. J. Geraats, R. Pastoor, and J. D. van Elsas. 2005. Characterization of the microbial community involved in the suppression of Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 95:808-818. [DOI] [PubMed] [Google Scholar]
- 42.Rafin, C., and Y. Tirilly. 1995. Characteristics and pathogenicity of Pythium spp. associated with root rot of tomatoes in soilless culture in Brittany, France. Plant Pathol. 44:779-785. [Google Scholar]
- 43.Rey, P., P. Nodet, and Y. Tirilly. 1997. Pythium F induce a minor but ubiquitous disease in tomato soilless cultures. J. Plant Pathol. 79:173-180. [Google Scholar]
- 44.Rey, P., N. Benhamou, and Y. Tirilly. 1998. Ultrastructural and cytochemical investigation of asymptomatic infection by Pythium sp. Phytopathology 88:234-244. [DOI] [PubMed] [Google Scholar]
- 45.Rey, P., N. Benhamou, E. Wulff, and Y. Tirilly. 1998. Interactions between tomato (Lycopersicon esculentum) root tissues and the mycoparasite Pythium oligandrum. Physiol. Mol. Plant Pathol. 53:105-122. [Google Scholar]
- 46.Rey, P., K. Picard, F. Déniel, N. Benhamou, and Y. Tirilly. 1999. Development of an IPM system in soilless culture by using slow sand filtration and a biocontrol agent, Pythium oligandrum. IOBC/WPRS Bull. 22:205-208. [Google Scholar]
- 47.Rey, P., S. Leucart, H. Désilets, R. Bélanger, J. P. Larue, and Y. Tirilly. 2001. Production of auxin and tryptophol by Pythium ultimum and minor pathogen, Pythium group F: possible role in pathogenesis. Eur. J. Plant Pathol. 107:895-904. [Google Scholar]
- 48.Rey, P., N. Benhamou, G. Le Floch, M. I. Salerno, E. Thuillier, and Y. Tirilly. 2005. Different interactions between the mycoparasite Pythium oligandrum and two sclerotia-forming plant pathogenic fungi: Botrytis cinerea and Sclerotinia minor. Mycol. Res. 109:779-788. [DOI] [PubMed] [Google Scholar]
- 49.Rey, P., G. Le Floch, N. Benhamou, and Y. Tirilly. 2008. Pythium oligandrum biocontrol: its relationships with fungi and plants, p. 43-67. In E. Ait Barka and C. Clément (ed.), Plant-microbe interactions. Research Signpost, Kerala, India.
- 50.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 11:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schurko, A. M., L. Mendoza, C. A. Lévesque, N. L. Desaulniers, W. A. M. De Cock, and G. R. Klassen. 2003. A molecular phylogeny of Pythium insidiosum. Mycol. Res. 107:537-544. [DOI] [PubMed] [Google Scholar]
- 52.Shimada, A., S. Takeuchi, A. Nakajima, S. Tanaka, T. Kawano, and Y. Kimura. 1999. Phytotoxicity of indole-3-acetic acid produced by the fungus Pythium aphanidermatum. Biosci. Biotechnol. Biochem. 63:187-189. [DOI] [PubMed] [Google Scholar]
- 53.Stanghellini, M. E., and S. L. Rasmussen. 1994. Hydroponics, a solution for zoosporic pathogens. Plant Dis. 78:1129-1138. [Google Scholar]
- 54.Sutton, J. C., C. R. Sopher, T. N. Owen-Going, W. Liu, B. Grodzinski, J. C. Hall, and R. L. Benchimol. 2006. Etiology and epidemiology of Pythium root rot in hydroponic crops: current knowledge and perspectives. Summa Phytopathol. 32:307-321. [Google Scholar]
- 55.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 56.Takenaka, S., Z. Nishio, and Y. Nakamura. 2003. Induction of defence reactions in sugar beet and wheat by treatment with cell wall protein fractions from Pythium oligandrum. Phytopathology 93:1228-1232. [DOI] [PubMed] [Google Scholar]
- 57.Takenaka, S., H. Sekiguchi, K. Nakaho, M. Tojo, A. Masunaka, and H. Takahashi. 2008. Colonization of Pythium oligandrum in the tomato rhizosphere for biological control of bacterial wilt disease analyzed by real-time PCR and confocal laser-scanning microscopy. Phytopathology 98:187-195. [DOI] [PubMed] [Google Scholar]
- 58.Tambong, J. T., A. W. A. M. De Cock, N. A. Tinker, and C. A. Lévesque. 2006. An oligonucleotide array for identification and detection of Pythium species. Appl. Environ. Microbiol. 72:2691-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu, J. C., A. P. Papadopoulos, X. Hao, and J. Zheng. 1999. The relationship of Pythium root rot and rhizosphere microorganisms in a closed circulating and an open system in rockwool culture of tomato. Acta Horticult. 481:577-583. [Google Scholar]
- 60.van der Plaats-Niterink, J. A. 1981. Monograph of the genus Pythium. Studies Mycol. 21:1-242. [Google Scholar]
- 61.Van West, P., A. A. Appiah, and N. A. R. Gow. 2003. Advances in research on oomycete root pathogens. Physiol. Mol. Plant Pathol. 62:99-113. [Google Scholar]
- 62.Vasseur, V., P. Rey, E. Bellanger, Y. Brygoo, and Y. Tirilly. 2005. Molecular characterization of Pythium group F isolates by ribosomal- and intermicrosatellite-DNA regions analysis. Eur. J. Plant Pathol. 112:301-310. [Google Scholar]
- 63.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-332. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, NY.
- 64.Wulff, E. G., A. T. H. Pham, M. Chérif, P. Rey, Y. Tirilly, and J. Hockenhull. 1998. Inoculation of cucumber roots with zoospores of mycoparasitic and plant pathogenic Pythium species: differential zoospore accumulation, colonization ability and plant growth response. Eur. J. Plant Pathol. 104:69-76. [Google Scholar]
- 65.Yang, L., A. Pan, J. Jia, J. Ding, J. Chen, H. Cheng, C. Zhang, and D. Zhang. 2005. Validation of a tomato-specific gene, LAT52, used as an endogenous reference gene in qualitative and real-time quantitative PCR detection of transgenic tomatoes. J. Agric. Food Chem. 53:183-190. [DOI] [PubMed] [Google Scholar]
- 66.Zemb, O., B. Haegeman, J. P. Delgenes, P. Lebaron, and J. J. Godon. 2007. SAFUM: statistical analysis of SSCP fingerprints using PCA projections, dendrograms and diversity estimators. Mol. Ecol. Notes 7:767-770. [Google Scholar]