Abstract
Bacteria often infect their hosts from environmental sources, but little is known about how environmental and host-infecting populations are related. Here, phylogenetic clustering and diversity were investigated in a natural community of rhizobial bacteria from the genus Bradyrhizobium. These bacteria live in the soil and also form beneficial root nodule symbioses with legumes, including those in the genus Lotus. Two hundred eighty pure cultures of Bradyrhizobium bacteria were isolated and genotyped from wild hosts, including Lotus angustissimus, Lotus heermannii, Lotus micranthus, and Lotus strigosus. Bacteria were cultured directly from symbiotic nodules and from two microenvironments on the soil-root interface: root tips and mature (old) root surfaces. Bayesian phylogenies of Bradyrhizobium isolates were reconstructed using the internal transcribed spacer (ITS), and the structure of phylogenetic relatedness among bacteria was examined by host species and microenvironment. Inoculation assays were performed to confirm the nodulation status of a subset of isolates. Most recovered rhizobial genotypes were unique and found only in root surface communities, where little bacterial population genetic structure was detected among hosts. Conversely, most nodule isolates could be classified into several related, hyper-abundant genotypes that were phylogenetically clustered within host species. This pattern suggests that host infection provides ample rewards to symbiotic bacteria but that host specificity can strongly structure only a small subset of the rhizobial community.
Symbiotic bacteria often encounter hosts from environmental sources (32, 48, 60), which leads to multipartite life histories including host-inhabiting and environmental stages. Research on host-associated bacteria, including pathogens and beneficial symbionts, has focused primarily on infection and proliferation in hosts, and key questions about the ecology and evolution of the free-living stages have remained unanswered. For instance, is host association ubiquitous within a bacterial lineage, or if not, do host-infecting genotypes represent a phylogenetically nonrandom subset? Assuming that host infection and free-living existence exert different selective pressures, do bacterial lineages diverge into specialists for these different lifestyles? Another set of questions addresses the degree to which bacteria associate with specific host partners. Do bacterial genotypes invariably associate with specific host lineages, and is such specificity controlled by one or both partners? Alternatively, is specificity simply a by-product of ecological cooccurrence among bacteria and hosts?
Rhizobial bacteria comprise several distantly related proteobacterial lineages, most notably the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium (52), that have acquired the ability to form nodules on legumes and symbiotically fix nitrogen. Acquisition of nodulation and nitrogen fixation loci has likely occurred through repeated lateral transfer of symbiotic loci (13, 74). Thus, the term “rhizobia” identifies a suite of symbiotic traits in multiple genomic backgrounds rather than a taxonomic classification. When rhizobia infect legume hosts, they differentiate into specialized endosymbiotic cells called bacteroids, which reduce atmospheric nitrogen in exchange for photosynthates from the plant (35, 60). Rhizobial transmission among legume hosts is infectious. Rhizobia can spread among hosts through the soil (60), and maternal inheritance (through seeds) is unknown (11, 43, 55). Nodule formation on hosts is guided by reciprocal molecular signaling between bacteria and plant (5, 46, 58), and successful infection requires a compatible pairing of legume and rhizobial genotypes. While both host and symbiont genotypes can alter the outcome of rhizobial competition for adsorption (34) and nodulation (33, 39, 65) of legume roots, little is known about how this competition plays out in nature.
Rhizobia can achieve reproductive success via multiple lifestyles (12), including living free in the soil (14, 44, 53, 62), on or near root surfaces (12, 18, 19, 51), or in legume nodules (60). Least is known about rhizobia in bulk soil (not penetrated by plant roots). While rhizobia can persist for years in soil without host legumes (12, 30, 61), it appears that growth is often negligible in bulk soil (4, 10, 14, 22, 25). Rhizobia can also proliferate in the rhizosphere (soil near the root zone) of legumes (4, 10, 18, 19, 22, 25, 51). Some rhizobia might specialize in rhizosphere growth and infect hosts only rarely (12, 14, 51), whereas other genotypes are clearly nonsymbiotic because they lack key genes (62) and must therefore persist in the soil. The best-understood rhizobial lifestyle is the root nodule symbiosis with legumes, which is thought to offer fitness rewards that are superior to life in the soil (12). After the initial infection, nodules grow and harbor increasing populations of bacteria until the nodules senesce and the rhizobia are released into the soil (11, 12, 38, 40, 55). However, rhizobial fitness in nodules is not guaranteed. Host species differ in the type of nodules they form, and this can determine the degree to which differentiated bacteroids can repopulate the soil (11, 12, 38, 59). Furthermore, some legumes can hinder the growth of nodules with ineffective rhizobia, thus punishing uncooperative symbionts (11, 27, 28, 56, 71).
Here, we investigated the relationships between environmental and host-infecting populations of rhizobia. A main objective was to test the hypothesis that rhizobia exhibit specificity among host species as well as among host microenvironments, specifically symbiotic nodules, root surfaces, and root tips. We predicted that host infection and environmental existence exert different selective pressures on rhizobia, leading to divergent patterns of clustering, diversity, and abundance of rhizobial genotypes.
MATERIALS AND METHODS
Collection of Lotus plants for sampling of rhizobial bacteria.
Rhizobial bacteria were isolated from symbiotic nodules and root surface communities of wild-growing Lotus spp., including Lotus angustissimus (an annual plant not native to California), Lotus corniculatus (a nonnative perennial), Lotus micranthus, Lotus strigosus, and Lotus wrangelianus (native annuals), and L. heermannii (a native perennial). Sampling occurred at two adjacent sites on the Northern California coast, the Bodega Marine Reserve and the Sonoma Coast State Park during January to April 2005. Whole plants were excavated from the soil, GPS coordinates were recorded for each collected plant, and excavated plants were stored in sealed plastic bags in coolers for transport to the lab. Once in the laboratory, plants were stored at 4°C for no more than 48 h before they were photographed and dissected and rhizobia were cultured from them.
Isolation of rhizobia from nodules and root surface communities of Lotus hosts.
Plant roots were washed with tap water to until no soil particles were apparent. Plants were dried lightly with new paper towels and spread on 1-mm-grid graph paper so that roots, nodules, and locations for root surface bacterial sampling could be labeled and digitally photographed for archiving (see the supplemental material). Subsequent to this step, plants were handled and dissected using sterilized materials. Nodules that exhibited no signs of herbivory or damage were dissected from roots using scalpels and forceps, and dissected nodules were immediately placed in presterilized centrifuge tubes (sterilization of laboratory supplies and liquids was performed by autoclaving unless otherwise noted). Dissected nodules were surface sterilized in bleach (5% sodium hypochlorite) for 2 to 3 minutes (depending on the size) and then rinsed three times in sterile deionized-distilled water (ddH2O). Nodules were then individually crushed with a flame-sterilized glass rod on petri plates, and each resultant slurry was streaked onto two replica plates containing 25 ml of solid modified arabinose-gluconate (MAG) medium (modified by P. van Berkum from method described in reference 7; see the supplemental material). A total of 301 nodules from 34 plants were cultured. Single rhizobial colonies were sampled from each nodule because naturally occurring legume nodules usually contain single rhizobial genotypes (33, 56).
From a subset of 20 plants, root sections were dissected (8 to 20 sections, ∼1 cm in length, including root tips and old roots), placed individually in presterilized centrifuge tubes containing a sterile solution of 0.01% Tween 20 (Fisher Scientific, Fair Lawn, NJ), incubated for ∼20 min, and vortexed at full power for 1 min to remove the remaining bacteria. The wash solution was then serially diluted and plated on a glucose-based rhizobium-defined medium (GRDM, with 100 mg/liter cycloheximide as an antifungal agent) (62), which favors the growth of Bradyrhizobium, Mesorhizobium, Methylobacterium, and Rhizobium bacteria (63; J. L. Sachs, unpublished data). These bacteria are referred to as “root surface isolates,” since they were closely associated with either the root surface or the root surface biofilm (18, 19). Approximately 20,000 colonies were scored and selectively isolated by growth rate (emerging on plates in 5 to 9 days), morphology (glossy in texture), and color (off-white to yellow) to match the characteristics of colonies cultured from host nodules. The 647 colonies that passed this selection were replica plated on GRDM agar medium, yeast-mannitol agar medium (57), and Luria-Bertani (LB) medium (57), since rhizobia from nodules actively grew on GRDM and yeast-mannitol agar media but not on LB medium (J. L. Sachs, unpublished data), yielding 395 clones. Root surface isolates and clonal cultures from nodules were grown individually in 15 ml of liquid MAG at 29°C and 180 rpm until logarithmic growth was achieved, and the cultures were centrifuged, washed, and archived in 25% glycerol-MAG at −80°C.
Molecular methods.
Genomic DNA was purified from 25 μl of each archived isolate using DNA isolation kits (no. K0512; Fermentas, Hanover, MD). DNA was amplified at the ITS locus (between 16S and 23S ribosomal subunits) using the bacterium-specific primers 450 and 1440 (66). PCR analysis was carried out in 25-μl reaction volumes containing 2.5 μl 10× PCR buffer, 2.5 μl deoxynucleoside triphosphates (2.5 mM), and 0.25 μl Taq polymerase (Invitrogen, Carlsbad, CA). The PCR protocol was as follows: initial denaturation of 94°C for 2 minutes; 16 cycles of denaturation (30 s at 94°C), annealing (70°C for 40 s, reduced by 0.5°C per cycle to 60°C), and extension (90 s at 72°C); followed by 30 more cycles with an annealing temperature of 60°C and a final extension (4 min at 72°C). Amplification products were visualized on 1.5% agarose gels stained with ethidium bromide. PCR fragments were sequenced using an Applied Biosystems 96-capillary 3730xl DNA analyzer (Foster City, CA) at the UC Berkeley DNA Sequencing Facility and assembled using DNAStar software (Madison, WI) with an average of 2× coverage. Only sequences that were visually unambiguous were included in the study (e.g., with a clear, single nucleotide [nt] peak at each site).
All cultured colonies were successfully sequenced, and the species level identities of >300 sequences were verified by the GenBank database (NCBI BLAST) (2). L. wrangelianus and L. corniculatus were discovered to harbor only Mesorhizobium spp. in their nodules and were not evaluated further in this study. All nodules from the remaining hosts harbored only Bradyrhizobium japonicum (i.e., the top five GenBank matches were B. japonicum), which also comprised the majority of root surface isolates. Approximately 10% of root surface isolates did not belong to Bradyrhizobium spp. (or any known lineage of rhizobial bacteria) (52) and were also not considered further for this study. The analysis focused on plants that harbored Bradyrhizobium spp. and for which a substantial portion of the root surface was sampled that resulted in 280 isolates from 24 plants (see Table S3 in the supplemental material).
ITS sequences were aligned with ClustalW using the collected cultures plus 17 Bradyrhizobium ITS sequences from GenBank (63), using default parameters. GenBank sequences included 12 B. japonicum isolates (accession numbers AF208504, AF208505, AF208508, AF208511, AF208515, AF208517, AF293377, AF293378.1, AF293381, AF293382.1, U69638.3, and Z35330.1), 1 Bradyrhizobium liaoningense sequence (AF208513), and 4 Bradyrhizobium elkanii isolates (AF208512, AF208518, AF293376, and AF293380.1) and comprised all the Bradyrhizobium sequences on GenBank that overlapped completely with the amplified locus. The alignment produced multiple small gaps that were clustered in two variable regions. Gaps were treated as missing data and only unambiguously aligned positions were used to construct the phylogenetic hypothesis. Duplicate sequences were not used when generating the phylogenetic trees, and the final sample size of 86 represents the number of unique ITS genotypes rather than the total number of analyzed sequences.
Phylogenetic reconstruction.
Model fitting was performed with likelihood ratio tests (Mr Modeltest 2.2) (41) using the Akaike information criterion (1). The ITS locus was divided into seven regions based on functional differences (16S rRNA, intergenic region 1 [IG1], isoleucine tRNA, IG2, alanine tRNA, IG3, and 23S rRNA), and nucleotide substitution models were fitted separately to each region, resulting in four different evolutionary models. The nucleotide locations listed for each chosen data partition (i to iv) are based on the ITS sequence of B. japonicum accession no. AF208508 (66). The DNA substitution models for each DNA partition are as follows: (i) K80 + G (IG1, nt 1473 to 1658), (ii) JC (IG2, nt 1736 to 1767), (iii) HKY + I + G (IG3, nt 1845 to 2255), and (iv) GTR + I + G (23S, nt 2256 to 2644) (26, 29, 47, 72). The 16S, tRNA-Ile, and tRNA-Ala regions were found to be invariable (nt 1428 to 1472, 1659 to 1735, and 1768 to 1844, respectively). Phylogenetic trees were reconstructed with Bayesian methods using MrBayes 3.1.2 (24) and by treating the four B. elkanii isolates as a monophyletic outgroup (67). Two parallel MrBayes runs with 106 generations were employed, each starting with a random tree, with eight simultaneous chains, a heating temperature of 0.02, and a “burnin” of the first 9,001 sampled trees (sample frequency = 100). A plot of log likelihood scores of sampling points against the generation number was observed to ensure that stationarity had been reached during the burnin period. Using a sample of the 100,000 post-burnin trees, a majority rule consensus tree was reconstructed using the “sumt” command on MrBayes.
Phylogenetic structure analysis.
Trait values of each unique genotype were mapped onto the Bayesian phylogenetic tree, including abundances (multiple isolates sharing a genotype), host species, and sampling microenvironment (nodule, root tip surface, or old root surface), and these values were used to calculate indices of community phylogenetic structure (68, 69) with the program Phylocom (70). The term “community” is used to refer to isolates co-occurring at some scale, such as host species or sampling microenvironment (23).
Phylogenetic clustering was investigated within the sampled rhizobial communities (i.e., not including the GenBank genotypes) using the abundance-weighted and presence/absence versions of the net relatedness index (NRI) and the nearest taxon index (NTI). These indices measure the degree to which a group of selected samples exhibits significant phylogenetic clustering relative to a null model of community assembly (69). Specifically, NRI = −(MPD − rndMPD)/(sd_rndMPD), where MPD is the mean pairwise phylogenetic distance among all n taxa (presence/absence version) or n isolates (abundance weighted version) present in a sample, and rndMPD and sd_rndMPD are the mean and standard deviation of the distance MPD for n taxa randomly distributed on the phylogeny. Phylogenetic distances among sampled communities were measured as the MPD between isolates from different communities, weighted by the isolate abundance obtained from each community (70). Similarly, NTI = −(MNTD − rndMNTD)/(sd_rndMNTD) and is a comparison of the observed MNTD (mean nearest taxon phylogenetic distance; the average distance to each taxon's or isolate's closest relative in a sample) to the mean (rndMNTD) and standard deviation (sd_rndMNTD) of the MNTD for n taxa or isolates randomly distributed on the phylogeny. For both indices, positive values indicate taxon samples that are more closely related on average than expected from random samples of the population, and negative values indicate samples that are less related on average (phylogenetic evenness or overdispersion). NRI detects tree-wide patterns of clustering and evenness, while NTI is more sensitive to clustering near the tips of the phylogeny (69). The degree of clustering among genotypes in each sample was tested for significant deviation from the clustering observed for 1,000 randomly drawn samples of the same number of isolates from the sampled populations. A method analogous to an analysis of molecular variance (AMOVA) (15) or nonparametric multivariate analysis of variance (37) implemented in the vegan package (version 1.8-8; J. Oksanen, R. Kindt, P. Legendre, B. O'Hara, M. Henry, and H. Stevens, vegan: community ecology package, [http://r-forge.r-project.org/projects/vegan/]) was used to partition variation in phylogenetic relatedness among samples into portions explained by host plant species, the microenvironment, and their interaction (70).
Strain richness analysis.
Genotype richness was compared between nodule and root surface isolates using sample-based genotype rarefaction curves and 95% confidence intervals calculated for each microenvironment. The rarefaction curves describe the expected number of taxa in a subset collection of n individuals drawn at random from a large pool of N individuals (21), and all isolates collected from a particular microenvironment on an individual plant comprise a single sample. The program EstimateS (8) was used to calculate these values and their standard deviations. Insufficient sample size prevented comparisons of genotype richness between host species.
Inoculation assays.
Inoculation assays were used to test the nodulation status of 60 isolates from the root surfaces and nodules of multiple hosts. L. strigosus was used for inoculations because of its small size and rapid growth and because the majority of the Bradyrhizobium cultures were isolated from this host species (>55%). Seeds obtained from ripe fruits of L. strigosus at the Bodega Marine Reserve were surface sterilized in bleach for 2 min, rinsed in sterile water, nick scarified, and germinated in sterile ddH2O (15°C for 5 to 7 days). Seedlings were planted into bleach-sterilized Conetainers (Stuewe & Sons, Corvallis, OR) filled with prewashed, autoclaved play sand (natural alluvial silica sand; Basalite, Tracy, CA). Axenic seedlings were incubated (20°C with 80% relative humidity, a 12:12 day/night cycle, and twice daily misting) in a growth chamber for 14 days (Conviron, Winnipeg, Canada) and then transferred to a greenhouse under ∼50% shade for hardening (14 days with twice daily misting) before inoculation. Once in the greenhouse, plants were fertilized weekly with Jensen's nitrogen-free solution (56), beginning with 2 ml per seedling, increasing by 1 ml each week until a maximum of 5 ml per plant was reached and used thereafter. Bradyrhizobium cultures of the selected isolates were initiated from ∼2 μl of original frozen stock inoculated into 50 ml of liquid MAG medium and incubated until cultures reached logarithmic-phase growth (29°C at 180 rpm for 72 h). Bacterial concentrations were estimated via light absorbance at 600 nm on a spectrophotometer. Grown cultures were centrifuged (4,000 rpm for 20 min) and resuspended in prewarmed sterile ddH2O to concentrations of 108 cells ml−1. For each rhizobial isolate, five plants were inoculated with 5 ml of resuspended cultures and five control plants were inoculated with 5 ml of sterile ddH2O. Plants were grown for 8 weeks in a greenhouse before they were uprooted to check for nodulation under a dissecting microscope.
ITS sequence accession numbers.
The GenBank ITS sequence accession numbers of the cultures collected here are FJ766023 to FJ766091.
RESULTS
Phylogenetic reconstruction and clustering within sampled communities.
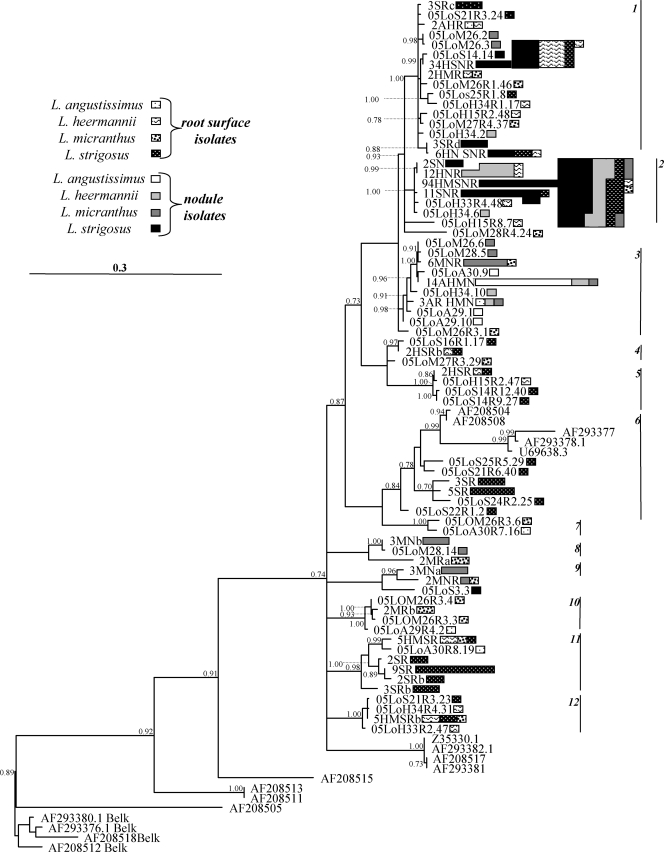
More than half of the nodes in the phylogeny were supported with high posterior probabilities (Fig. 1). For the sake of discussion, 12 independent, well-supported clades (minimum posterior support = 0.84) were identified and numbered. The majority of isolates were concentrated into clades 1 to 3 (58, 123, and 30 isolates, respectively) (Fig. 1). Many of the analyzed rhizobial communities exhibited positive and significant phylogenetic clustering. The results were similar regardless of whether genotype abundances or presence/absence data were considered, so only the former are reported (Table 1). Overall, a slightly smaller subset of communities exhibited significant NRI values than significant NTI values, indicating that phylogenetic clustering was stronger closer to the tips of the phylogenetic tree. None of the rhizobial communities exhibited significant phylogenetic evenness.
FIG. 1.
Phylogram of 86 Bradyrhizobium genotypes inferred from 1,310 nt of the ITS. This is a 50% majority consensus phylogeny based on 1,000 sampled trees from a Bayesian analysis. Clades with less than 0.50 posterior probabilities are collapsed and posterior probabilities greater than 0.70 are labeled on the nodes. The scale bar indicates the number of changes per site. Twelve well-supported clades are labeled with numbered vertical lines on the right. Genotype abundances are indicated by the sizes of the polygons next to each taxon label, with the sizes of the legend labels representing single isolates. Isolation source(s) of each genotype is indicated by the shading and pattern of each polygon. Taxon labels of the uniquely isolated genotypes begin with the year of isolation (e.g., 05 indicates 2005), host species (LoA, Lotus angustissimus; LoM, L. micranthus; LoH, L. heermannii; LoS, L. strigosus), plant number, and nodule or root surface number (the latter with R followed by root and isolate number). Taxon labels of the multiply isolated genotypes begin with the number of times the isolate was recovered followed by the host species (A, M, H, S) and isolate type (N, nodule; R, root surface). GenBank isolates are labeled with their accession numbers.
TABLE 1.
Phylogenetic clustering among communities using abundance-weighted genotypesc
| Sample group analyzeda | Sampleb | No. of genotypes | MPD | NRI | MPD quantile | MNTD | NTI | MNTD quantile |
|---|---|---|---|---|---|---|---|---|
| Host | A | 9 | 0.28 | 0.91 | 0.20 | 0.13 | 0.95 | 0.14 |
| H | 22 | 0.29 | 2.21 | 0.02 | 0.07 | 1.66 | 0.01 | |
| M | 24 | 0.57 | 0.16 | 0.41 | 0.14 | 0.78 | 0.23 | |
| S | 29 | 0.36 | 1.01 | 0.18 | 0.06 | 1.23 | 0.01 | |
| Nod/root | Nod | 25 | 0.20 | 2.27 | 0.01 | 0.04 | 1.52 | 0.00 |
| Root | 51 | 0.60 | −0.36 | 0.63 | 0.09 | 0.83 | 0.19 | |
| Nod/old/tip | Nod | 25 | 0.20 | 2.37 | 0.00 | 0.04 | 1.55 | 0.00 |
| Old | 23 | 0.54 | 0.69 | 0.24 | 0.10 | 2.12 | 0.01 | |
| Tip | 40 | 0.61 | −0.57 | 0.71 | 0.11 | 0.27 | 0.46 | |
| Host+nod/root | A nod | 4 | 0.04 | 2.16 | 0.00 | 0.05 | 1.45 | 0.00 |
| A root | 5 | 0.55 | −0.79 | 0.77 | 0.41 | −0.61 | 0.74 | |
| H nod | 9 | 0.13 | 2.50 | 0.00 | 0.06 | 1.87 | 0.00 | |
| H root | 16 | 0.53 | 0.47 | 0.30 | 0.13 | 1.89 | 0.03 | |
| M nod | 11 | 0.48 | 0.50 | 0.29 | 0.10 | 2.15 | 0.01 | |
| M root | 16 | 0.60 | −0.27 | 0.58 | 0.26 | −1.07 | 0.85 | |
| S nod | 8 | 0.09 | 2.36 | 0.00 | 0.05 | 1.61 | 0.00 | |
| S root | 25 | 0.58 | −0.42 | 0.64 | 0.08 | 1.86 | 0.00 | |
| Host+nod/old/tip | A nod | 4 | 0.04 | 2.16 | 0.00 | 0.05 | 1.45 | 0.00 |
| A old | 4 | 0.56 | −1.00 | 0.83 | 0.57 | −1.35 | 0.91 | |
| A tip | 1 | |||||||
| H nod | 9 | 0.13 | 2.51 | 0.00 | 0.06 | 1.88 | 0.00 | |
| H old | 10 | 0.50 | 0.54 | 0.28 | 0.18 | 1.33 | 0.09 | |
| H tip | 9 | 0.50 | 0.34 | 0.36 | 0.16 | 1.63 | 0.05 | |
| M nod | 11 | 0.48 | 0.50 | 0.29 | 0.10 | 2.19 | 0.01 | |
| M old | 2 | 0.32 | −0.33 | 0.52 | 0.72 | −0.33 | 0.52 | |
| M tip | 14 | 0.63 | −0.63 | 0.73 | 0.31 | −1.41 | 0.92 | |
| S nod | 8 | 0.09 | 2.36 | 0.00 | 0.05 | 1.61 | 0.00 | |
| S old | 10 | 0.47 | 0.65 | 0.24 | 0.10 | 2.22 | 0.01 | |
| S tip | 21 | 0.59 | −0.55 | 0.68 | 0.09 | 1.75 | 0.01 |
Host, the four host species; Nod/root, nodules versus root surface; Nod/old/tip, nodules versus root tips versus old (mature) roots; Host+nod/root, host species subdivided by nodules versus root surface; and Host+nod/old/tip, host species subdivided by nodules versus root tips versus old roots.
A, L. angustissimus; M, L. micranthus; H, L. heermannii; S, L. strigosus; nod, nodule samples; root, root surface samples; tip, root surface samples from the distal 1 cm of the root; old, root surface samples from proximal (non-tip) root sections.
“No. of genotypes” indicates number of genotypes in each community; MPD quantile, quantile of observed MPD versus 1,000 randomly generated communities (<0.05 indicates statistically significant clustering); MNTD, mean nearest taxon distance of the analyzed taxa; NTI, nearest taxon index; MNTD quantile, quantile of observed MPD versus 1,000 randomly generated communities (<0.05 indicates statistically significant clustering). Statistically significant values of MPD and MNTD (quantile < 0.05) are bold.
Across four host species, communities of Bradyrhizobium bacteria in nodules exhibited significant phylogenetic clustering (NRI and NTI of nodule samples) (Table 1). In contrast, the pooled root surface communities showed no evidence of such clustering (root samples) (Table 1). For example, most nodule isolates were clustered in clades 1 and 2 whereas root surface isolates were spread throughout the tree. However, when the root surface communities were subdivided into old root and root tip microenvironments, the old root (NTI) but not the root tip communities exhibited significant phylogenetic clustering. The root surface communities of L. angustissimus and L. micranthus were poorly sampled across the microenvironments (Table 1), so this analysis was repeated without these two host species. Including only L. heermannii and L. strigosus, both old root and root tip microenvironments exhibited significant phylogenetic clustering (NTI). The clustering appeared to be stronger in the old root community than in the root tip community (NTI values of 2.24 and 1.89, respectively), but these differences were not statistically significant when tested with a post-hoc analysis of variance.
When analyzed by host species, communities of Bradyrhizobium bacteria (pooled across microenvironments) exhibited significant phylogenetic clustering in two of the four hosts, L. heermannii (NRI, NTI) and L. strigosus (NTI). Nodule communities exhibited significant clustering within all host species, but clustering was weaker in L. micranthus nodules (large NTI but small NRI). In particular, nodule isolates of L. strigosus, L. heermannii, and L. angustissimus were phylogenetically clustered and often represented largely by one or two related genotypes in each host, whereas L. micranthus nodule isolates were more evenly distributed across the tree (Fig. 1). In the three hosts for which there were adequate samples of root surface isolates (L. heermannii, L. strigosus, and L. micranthus), only root surface communities of L. heermannii and L. strigosus exhibited significant clustering. However, even in these two species, root surface isolates were more broadly distributed among clades than were nodule isolates (Table 1).
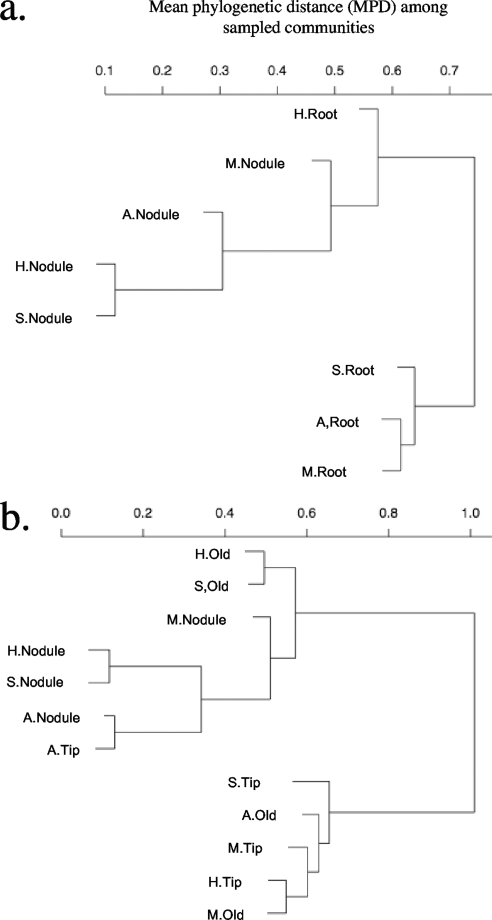
Phylogenetic clustering among sampled communities.
The nodule communities of all host species clustered together (Fig. 2a). The root surface community of L. heermannii also grouped with this cluster, whereas the other root surface communities occupied another cluster. When the samples from each host species were further subdivided into nodule, old root, and root tip communities, the old root samples of L. heermannii and L. strigosus and the single root tip sample from L. angustissimus clustered with the nodule communities of all of the host species (Fig. 2b). All other root isolates occupied the other branch of the dendrogram. Finally, an AMOVA indicated that the magnitude of variation in community phylogenetic relatedness among samples was approximately equal among hosts and among microenvironments (an R2 value of 0.12 for each), with no significant host/microenvironment interaction ( Table 2).
FIG. 2.
Cluster dendrograms depict the phylogenetic clustering among host plant species and microenvironments. The trees indicate the MPD among sampled communities taken from the nodules and root surfaces of each host (a) or with root surface isolates further subdivided into root tips and old (mature) roots (b). Also shown are the host species (A, L. angustissimus; M, L. micranthus; H, L. heermannii; L, L. strigosus). Root, root surface; Old, mature root; Tip, root tip.
TABLE 2.
Results of an AMOVA of community phylogenetic distancesa
| Sample location | Df | SS | MS | F | R2 | P |
|---|---|---|---|---|---|---|
| Microenvironment | 1 | 0.52 | 0.52 | 5.87 | 0.12 | <0.001 |
| Host | 3 | 0.51 | 0.17 | 1.90 | 0.12 | 0.028 |
| Microenvironment by host | 3 | 0.38 | 0.13 | 1.41 | 0.09 | 0.990 |
| Residuals | 33 | 2.95 | 0.09 | 0.68 | ||
| Total | 40 | 4.36 | 1.00 |
AMOVA of community phylogenetic distances among samples from different hosts and microenvironments. Df, degrees of freedom; SS, sums of squares; MS, mean squares; F, F statistic; R2, variation in community phylogenetic distances explained by the variable; P, significance of variable based on permutation test.
Relative richness of the sampled rhizobial communities.
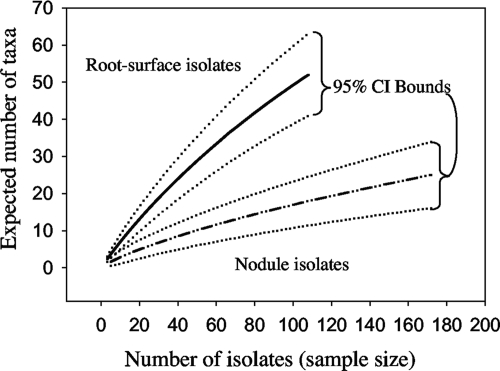
In all, 280 isolates comprised 69 genotypes. A few genotypes were abundant, but most were represented by only one or a few individuals (Fig. 1). Almost all the genotypes (88%) were recovered in the root surface communities, even though these isolates were in the minority (39%) compared to nodule isolates. Furthermore, 43 of the genotypes (62%) were recovered only in the root surface communities. In comparison, 18 genotypes (26%) were recovered only in nodules and only 8 genotypes were found in both nodule and root surface microenvironments. For these eight genotypes, there was a significant positive correlation between abundance in nodules and abundance in the root surfaces of hosts (R2 = 0.56; P = 0.03). Independent sample-based rarefaction curves for the nodule and root surface microenvironments exhibited significantly different rates of genotype accumulation; root surface isolates accumulated genotypes at about three times the rate of nodule isolates (Fig. 3). Neither genotype accumulation curve showed any sign of reaching an asymptote, although the rate of genotype accumulation in nodule isolates began to decline after about 50 isolates. Genotype richness among host species was not compared because poor power was expected.
FIG. 3.
Rarefaction curves for the nodule and root surface microenvironments, with 95% confidence intervals indicated with dotted lines. Each curve describes the expected number of taxa (species richness) in a subset of n individuals drawn at random from the total pool of isolates (21). The confidence intervals of the curves do not overlap, indicating that root surfaces were significantly more diverse than nodules.
Nodulation assays.
Thirty-six of the 60 tested strains successfully nodulated L. strigosus. All 16 nodule isolates formed nodules, including two isolates each from L. heermannii and L. micranthus. In contrast, only 20 of 44 isolates cultured from the root surface successfully nodulated. The root surface rhizobia could be roughly divided into two groups by genetic affinity. Twenty of the root surface isolates exhibited genotypes that were also found in nodule isolates in the field, whereas 24 were represented by genotypes found only in root surface samples (i.e., never in nodules). Of the 20 isolates with genotypes that were found in both nodules and root surfaces, 17 successfully nodulated L. strigosus. In contrast, among genotypes found only in root surfaces in the field, 3 of 24 isolates successfully nodulated L. strigosus (see Table S3 in the supplemental material).
DISCUSSION
The phylogenetic data in this study support a key conclusion of inoculation studies, which is that certain rhizobial genotypes are more likely than others to adsorb to (34) and ultimately infect (33, 39, 65) specific host species. The pattern of phylogenetic clustering in the sample of nodule rhizobia indicates that each of the Lotus hosts studied usually forms nodules with a small, more genetically related subset of the population of rhizobia that are available to it in its zone near the root. Unlike most previous studies of rhizobial specificity, which are based upon inoculation experiments (with a few strains isolated from nodules), this study examined in situ specificity by comparing naturally occurring nodule inhabitants with rhizobial strains isolated from the root surfaces of the same host individuals. The data presented here support theoretical predictions (45) that rhizobial populations on host root surfaces are phylogenetically richer than those found in nodules. All 12 well-supported rhizobial clades reconstructed from the cultured samples included root surface isolates, and 7 contained no nodule isolates.
Mutualism theory predicts that host specificity is promoted by selection to minimize conflict among symbionts, which favors host mechanisms that restrict symbiont diversity to one or a few related genotypes (64). Here, Lotus host species exhibited rhizobial specificity, and the data suggest that phylogenetically conserved traits determine which of the small minority of root surface-dwelling rhizobial genotypes can form nodulating symbioses with Lotus hosts. Furthermore, hosts varied in their degree of specificity, as has been found by other researchers (39, 65). For instance, almost 80% of the 93 L. strigosus nodule isolates are concentrated into only three genotypes, two from clade 2 (genotype 94hmsnr, with 47 isolates, and genotype 11snr, with 10 isolates) and one from clade 1 (genotype 34hsnr, with 16 isolates), and isolates from L. heermannii nodules were restricted to clades 1, 2, and 3 (Fig. 1). In contrast, L. micranthus nodules were occupied by isolates from small clusters of closely related strains that were scattered across five divergent clades. This pattern suggests that the ability to nodulate L. micranthus is more evolutionarily labile (relative to the ITS region used to estimate the phylogeny) than the ability to nodulate L. strigosus or L. heermannii. However, the data do not allow the determination of whether these traits are (i) ancestral but frequently lost, (ii) novel and repeatedly evolved, or (iii) mobile and acquired via recombination (horizontal transfer). Its broader symbiont range also suggests that L. micranthus might be more dependent on postnodulation partner choice to constrain rhizobial cheating (11, 49, 55).
Lotus strigosus and L. heermannii, the two host species with the largest numbers of sampled root isolates, exhibited root surface communities with significant phylogenetic clustering, as indicated by the significant NTI. Unlike nodule isolates, however, the MPD between root surface strains was no different than expected by chance (values of NRI were not significant). This pattern suggests that although nodule specialization to these hosts is phylogenetically conserved, the traits needed to persist on their root surfaces are evolutionarily labile relative to the ITS region used to estimate the phylogeny. The evolutionary process by which these traits evolved cannot be discerned.
Spatial or ecological co-occurrence of partners could bias in situ measures of host specificity. For example, edaphic conditions often influence soil bacterial communities (16), and L. strigosus and L. heermannii were collected in xeric mobile sand dunes whereas L. micranthus and L. angustissimus were found in wet dunal swales. These two habitats differ in organic matter (54), which strongly influences microbial community composition in coastal dunes (73). However, although L. strigosus and L. heermannii were both collected from mobile dunes, their root surface rhizobia did not cluster together (Fig. 2). Instead, L. strigosus root surface rhizobia clustered with those found on the root surfaces of species collected in wet dunal swales (L. micranthus and L. angustissimus). This pattern refutes the hypothesis that host specificity detected here is a by-product of ecological co-occurrence among bacteria and hosts.
The trade-off theory predicts that traits that promote fitness in one environment may cause a poor performance in other environments (3, 9, 17, 20, 36) and lead to specialization for either habitat. Yet, it has been argued that symbiotic rhizobia spend so much time in the soil that they should be as well adapted to the soil environment as nonnodulating strains (12). More than 70% of the 109 isolates obtained from root surfaces exhibited a genotype that was never recovered from a nodule. This pattern supports the trade-off theory because it suggests that traits needed to survive in the soil and on the root surface differ from those needed to succeed as a nodule symbiont. On the other hand, genotypes associated with the ability to nodulate were hyper-abundant in the sample, with some being recovered many times both in nodules and in the surrounding root surfaces. Among the eight genotypes found in both microenvironments, a positive correlation between abundance on the root surface and in nodules is consistent with the idea that competitiveness in the rhizosphere is correlated with successful nodulation (31, 34). Together, these datasets suggest that host-derived rewards might drive a few symbiotic strains to flourish in the population near the root and that the fitness rewards of nodulating are greater than those of the competing strategy, which involves staying in the soil and avoiding host infection entirely (12).
One possible mechanism that could account for these apparently conflicting results is the frequent and rapid loss of nodulation ability via degradation or wholesale loss of symbiotic loci (50, 51). However, the extent to which this occurs in nature is unknown. Genotypes found only on root surfaces might be (i) symbiotically competent strains that are out-competed for nodulation sites, (ii) successful root surface competitors that are specialized symbionts of non-Lotus hosts, or (iii) nonsymbiotic strains. The inoculation assays suggest that these genotypes are most likely either nonsymbiotic or non-Lotus symbionts, since 88% of them (e.g., 21/24 of genotypes isolated only in root surface samples) failed to form nodules on L. strigosus. If so, then ecological conditions (such as an influx of soil nutrients or local extinction of hosts) that decrease the availability of nodule microhabitats would favor mutants that abandon symbiotic interaction (50).
Epidemic distributions of symbionts—with a few very abundant genotypes (Fig. 1)—have been uncovered in Bradyrhizobium species previously (67). Past studies have suggested that the microgeographic distribution of epidemic bacterial clones is very restricted (6, 42, 67), perhaps suggesting that clones are limited by ecological barriers. Yet, the most abundant genotype (genotype 94HMSNR) (Fig. 1; see Table S3 in the supplemental material) was found broadly over the entire sampling range (>3 km) and was recovered at least once in the nodules of three of the four host species.
While more isolates were obtained from nodules than from root surfaces, the overall diversity of nodule genotypes was low. The greater diversity of rhizobia isolated directly from root surfaces suggests that the competition and conflict occurring on legume roots are more complex and potentially more intense than previously predicted. While much research that has examined competition for nodulation has logically focused on nodulating rhizobial strains, these results suggest that nonsymbiotic rhizobia could play an important role in competition to colonize host root surfaces. Nonsymbiotic strains, which appeared to dominate the root surface in this study, represent an important aspect of rhizobial biology that has largely been neglected.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for helpful comments.
This research was supported by funding from NIH (NRSA GM77892-01 to J.L.S.) and from NSF (DEB no. 0108708 to E.L.S. and DEB no. 0816663 to J.L.S.). S.W.K. was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 29 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akaike, H. 1973. Information theory and an extension of the maximum likelihood principle, p. 267-281. In B. N. Petrov and F. Csaki (ed.), Second international symposium on information theory. Akademiai Kiado, Budapest, Hungary.
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, A. F., and R. E. Lenski. 1993. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution 47:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Brockwell, J., R. J. Roughley, and D. F. Herridge. 1987. Population dynamics of Rhizobium japonicum strains used to inoculate three successive crops of soybeans. Aust. J. Agric. Res. 38:61-74. [Google Scholar]
- 5.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, J. C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, M. A., and G. H. Elkan. 1973. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob. Agents Chemother. 4:248-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. University of Connecticut, Storrs, CT.
- 9.Cooper, V. S., and R. E. Lenski. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736-739. [DOI] [PubMed] [Google Scholar]
- 10.Del Papa, M. F., M. Pistorio, L. J. Balague, W. O. Draghi, C. Wegener, A. Perticari, K. Niehaus, and A. Lagares. 2003. A microcosm study on the influence of pH and the host-plant on the soil persistence of two alfalfa-nodulating rhizobia with different saprophytic and symbiotic characteristics. Biol. Fertil. Soils 39:112-116. [Google Scholar]
- 11.Denison, R. F. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 156:567-576. [DOI] [PubMed] [Google Scholar]
- 12.Denison, R. F., and E. T. Kiers. 2004. Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol. Lett. 237:187-193. [DOI] [PubMed] [Google Scholar]
- 13.Dobert, R. C., B. T. Brei, and E. W. Triplett. 1994. DNA sequence of the common nodulation genes of Bradyrhizobium elkanii and their phylogenetic relationship to those of other nodulating bacteria. Mol. Plant-Microbe Interact. 7:564-572. [DOI] [PubMed] [Google Scholar]
- 14.Duodu, S., T. V. Bhuvaneswari, J. Gudmundsson, and M. M. Svenning. 2005. Symbiotic and saprophytic survival of three unmarked Rhizobium leguminosarum biovar trifolii strains introduced into the field. Environ. Microbiol. 7:1049-1058. [DOI] [PubMed] [Google Scholar]
- 15.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA genotypes: application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry, J. D. 1990. Trade-offs in fitness on different hosts: evidence from a selection experiment with a phytophagous mite. Am. Nat. 136:569-580. [Google Scholar]
- 18.Fujishige, N. A., N. N. Kapadia, and A. M. Hirsch. 2006. A feeling for the micro-organism: structure on a small scale. Biofilms on plant roots. Bot. J. Linnean Soc. 150:79-88. [Google Scholar]
- 19.Fujishige, N. A., N. N. Kapadia, P. L. De Hoff, and A. M. Hirsch. 2006. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 56:195-206. [DOI] [PubMed] [Google Scholar]
- 20.Futuyma, D. J., and G. Moreno. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19:207-233. [Google Scholar]
- 21.Gotelli, N. J., and R. K. Colwell. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4:379-391. [Google Scholar]
- 22.Hagen, M., A. Puhler, and W. Selbitschka. 1997. The persistence of bioluminescent Rhizobium meliloti L1 (RecA−) and L33 (RecA+) in non-sterile microcosms depends on the soil type, on the co-cultivation of the host legume alfalfa and on the presence of an indigenous R. meliloti population. Plant Soil 188:257-266. [Google Scholar]
- 23.Horner-Devine, M. C., and B. J. M. Bohannan. 2006. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87:S100-S108. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 25.Hynes, R. K., D. C. Jans, E. Bremer, N. Z. Lupwayi, W. A. Rice, G. W. Clayton, and M. M. Collins. 2001. Rhizobium population dynamics in the pea rhizosphere of rhizobial inoculant strain applied in different formulations. Can. J. Microbiol. 47:595-600. [DOI] [PubMed] [Google Scholar]
- 26.Jukes, T., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 27.Kiers, E. T., R. A. Rousseau, S. A. West, and R. F. Denison. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425:78-81. [DOI] [PubMed] [Google Scholar]
- 28.Kiers, E. T., R. A. Rousseau, and R. F. Denison. 2006. Measured sanctions: legume hosts detect quantitative variation in rhizobium cooperation and punish accordingly. Evol. Ecol. Res. 8:1077-1086. [Google Scholar]
- 29.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 30.Kuykendall, L. D. 1989. Influence of Glycine max nodulation on the persistence in soil of a genetically marked Bradyrhizobium japonicum strain. Plant Soil 116:275-277. [Google Scholar]
- 31.Kuykendall, L. D., F. M. Hashem, and W. J. Hunter. 1996. Enhanced competitiveness of Bradyrhizobium japonicum mutant strain improved for nodulation and nitrogen fixation. Plant Soil 186:121-125. [Google Scholar]
- 32.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 33.Lieven-Antoniou, C. A., and T. S. Whittam. 1997. Specificity in the symbiotic association of Lotus corniculatus and Rhizobium loti from natural populations. Mol. Ecol. 6:629-639. [Google Scholar]
- 34.Lodeiro, A. R., and G. Favelukes. 1999. Early interactions of Bradyrhizobium japonicum and soybean roots: specificity in the process of adsorption. Soil Biol. Biochem. 31:1405-1411. [Google Scholar]
- 35.Lodwig, E. M., A. H. F. Hosie, A. Bordes, K. Findlay, D. Allaway, R. Karunakaran, J. A. Downie, and P. S. Poole. 2003. Amino-acid cycling drives nitrogen fixation in the legume: Rhizobium symbiosis. Nature 422:722-726. [DOI] [PubMed] [Google Scholar]
- 36.MacLean, R. C., G. Bell, and P. B. Rainey. 2004. The evolution of a pleiotropic fitness tradeoff in Pseudomonas fluorescens. Proc. Natl. Acad. Sci. USA 101:8072-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McArdle, B. H., and M. J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297. [Google Scholar]
- 38.Mergaert, P., T. Uchiumi, B. Alunni, G. Evanno, A. Cheron, O. Catrice, A. E. Mausset, F. Barloy-Hubler, F. Galibert, A. Kondorosi, and E. Kondorosi. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA 103:5230-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mpepereki, S., A. G. Wollum, and F. Makonese. 1996. Diversity in symbiotic specificity of cowpea rhizobia indigenous to Zimbabwean soils. Plant Soil 186:167-171. [Google Scholar]
- 40.Müller, J., A. Wiemken, and T. Boller. 2001. Redifferentiation of bacteria isolated from Lotus japonicus root nodules colonized by Rhizobium sp. NGR234. J. Exp. Bot. 52:2181-2186. [DOI] [PubMed] [Google Scholar]
- 41.Nylander, J. A. A., F. Ronquist, J. P. Huelsenbeck, and J. L. Nieves-Aldrey. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53:47-67. [DOI] [PubMed] [Google Scholar]
- 42.Oda, Y., B. Star, L. A. Huisman, J. C. Gottschal, and L. J. Forney. 2003. Biogeography of the purple nonsulfur bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 69:5186-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker, M. A. 1999. Mutualism in metapopulations of legumes and rhizobia. Am. Nat. 153:S48-S60. [DOI] [PubMed] [Google Scholar]
- 44.Pongsilp, N., N. Teaumroong, A. Nuntagij, N. Boonkerd, and M. J. Sadowski. 2002. Genetic structure of indigenous non-nodulating and nodulating populations of Bradyrhizobium in soils from Thailand. Symbiosis 33:39-58. [Google Scholar]
- 45.Provorov, N. A., and N. I. Vorobyov. 2006. Interplay of Darwinian and frequency-dependent selection in the host-associated microbial populations. Theor. Popul. Biol. 70:262-272. [DOI] [PubMed] [Google Scholar]
- 46.Radutoiu, S., L. H. Madsen, E. B. Madsen, H. H. Felle, Y. Umehara, M. Gronlund, S. Sato, Y. Nakamura, S. Tabata, N. Sandal, and J. Stougaard. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585-592. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 48.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 49.Sachs, J. L., U. G. Mueller, T. P. Wilcox, and J. J. Bull. 2004. The evolution of cooperation. Q. Rev. Biol. 79:135-160. [DOI] [PubMed] [Google Scholar]
- 50.Sachs, J. L., and E. L. Simms. 2006. Pathways to mutualism breakdown. Trends Ecol. Evol. 21:585-592. [DOI] [PubMed] [Google Scholar]
- 51.Sachs, J. L., and E. L. Simms. 2008. The origins of uncooperative rhizobia. Oikos 117:961-966. [Google Scholar]
- 52.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing symbionts. J. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 53.Segovia, L., D. Pinero, R. Palacios, and E. Martinez-Romero. 1991. Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl. Environ. Microbiol. 57:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shumway, S. W., and C. R. Banks. 2001. Species distributions in interdunal swale communities: the effects of soil waterlogging. Am. Midland Naturalist 145:137-146. [Google Scholar]
- 55.Simms, E. L., and D. L. Taylor. 2002. Partner choice in nitrogen-fixing mutualisms of legumes and rhizobia. Integr. Comp. Biol. 42:369-380. [DOI] [PubMed] [Google Scholar]
- 56.Simms, E. L., D. L. Taylor, J. Povich, R. P. Shefferson, J. L. Sachs, M. Urbina, and Y. Tausczik. 2006. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc. R. Soc. B 273:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somasegaran, P., and J. Hoben. 1994. Handbook for Rhizobia. Springer-Verlag, New York, NY.
- 58.Spaink, H. P. 1995. The molecular basis of infection and nodulation by rhizobia—the ins and outs of sympathogenesis. Ann. Rev. Phytopathol. 33:345-368. [DOI] [PubMed] [Google Scholar]
- 59.Sprent, J. I. 2001. Nodulation in legumes. Royal Botanic Gardens, Kew, United Kingdom.
- 60.Sprent, J. I., J. M. Sutherland, and S. M. Faria. 1987. Some aspects of the biology of nitrogen-fixing organisms. Philos. Trans. R. Soc. Lond. B 317:111-129. [Google Scholar]
- 61.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene-transfer in the environment. Proc. Natl. Acad. Sci. USA 92:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan, J. T., B. D. Eardly, and P. van Berkum. 1996. Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl. Environ. Microbiol. 62:2818-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, J. N. 2005. The geographic mosaic of coevolution. University of Chicago Press, Chicago, IL.
- 65.Turk, D., and H. H. Keyser. 1992. Rhizobia that nodulate tree legumes: specificity of the host for nodulation and effectiveness. Can. J. Microbiol. 38:451-460. [Google Scholar]
- 66.van Berkum, P., and J. J. Fuhrmann. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165-2172. [DOI] [PubMed] [Google Scholar]
- 67.Vinuesa, P., C. Silva, D. Werner, and E. Martinez-Romero. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34:29-54. [DOI] [PubMed] [Google Scholar]
- 68.Webb, C. O. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156:145-155. [DOI] [PubMed] [Google Scholar]
- 69.Webb, C. O., D. D. Ackerly, M. A. McPeek, and M. J. Donoghue. 2002. Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 33:475-505. [Google Scholar]
- 70.Webb, C. O., D. D. Ackerly, and S. W. Kembel. 2008. Phylocom: software for the analysis of community phylogenetic structure and trait evolution. Bioinformatics 18:2098-2100. [DOI] [PubMed] [Google Scholar]
- 71.West, S. A., E. T. Kiers, E. L. Simms, and R. F. Denison. 2002. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. Lond. B 269:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, Z. H. 1993. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10:1396-1401. [DOI] [PubMed] [Google Scholar]
- 73.Yoshitake, S., and T. Nakatsubo. 2008. Changes in soil microbial biomass and community composition along vegetation zonation in a coastal sand dune. Aust. J. Soil Res. 46:390-396. [Google Scholar]
- 74.Young, J. P. W., and K. E. Haukka. 1996. Diversity and phylogeny of rhizobia. New Phytol. 133:87-94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.