Abstract
Cyanobacteria are photosynthetic prokaryotes capable of protecting themselves from UV radiation through the biosynthesis of UV-absorbing secondary metabolites, such as the mycosporines and scytonemin. Scytonemin, a novel indolic-phenolic pigment, is found sequestered in the sheath, where it provides protection to the subtending cells during exposure to UV radiation. The biosynthesis of scytonemin is encoded by a previously identified gene cluster that is present in six cyanobacterial species whose genomes are available. A comparison of these clusters reveals that two major cluster architectures exist which appear to have evolved through rearrangements of large sections, such as those genes responsible for aromatic amino acid biosynthesis and through the insertion of genes that potentially confer additional biosynthetic capabilities. Differential transcriptional expression analysis demonstrated that the entire gene cluster is transcribed in higher abundance after exposure to UV radiation. This analysis helps delineate the cluster boundaries and indicates that regulation of this cluster is controlled by the presence or absence of UV radiation. The findings from an evolutionary phylogenetic analysis combined with the fact that the scytonemin gene cluster is distributed across several cyanobacterial lineages led to our proposal that the distribution of this gene cluster is best explained through an ancient evolutionary origin.
Cyanobacteria are photosynthetic prokaryotes thought to be among the most ancient organisms on the planet (5). Their photosynthetic ability has long been speculated to have played a role in the oxygenation of the atmosphere, allowing the development of many other life forms. However, before the presence of oxygen, cyanobacteria lived in an environment where the absence of a planetary ozone layer allowed exposure to high levels of harmful UV radiation (18). The presence of high UV exposure levels early in the evolutionary history of cyanobacteria certainly presented these organisms with a major environmental pressure and resulted in the development of multiple UV defense adaptations which allow them to thrive in areas exposed to extremely high light and UV levels. These adaptations include avoidance, active repair mechanisms such as the SOS repair response, removal of reactive oxygen species by carotenoids, and biosynthesis of UV-absorbing secondary metabolites, such as mycosporine amino acids and scytonemin. These adaptations are used in combination to avoid both acute cell damage (e.g., carotenoids) and the harmful effects of long-term UV radiation exposure (e.g., mycosporines and scytonemin) (6).
Scytonemin is an extracellular pigment first observed in 1849, when Nägeli described a yellow-green pigmentation in the sheaths of cyanobacteria (7). In 1993, its chemical structure was elucidated and found to consist of an unprecedented dimeric indole-phenolic structure (9). In pharmacological screens, this unique molecule was found to have both anti-inflammatory and antiproliferative activity (13, 14). Scytonemin is considered to be a true sunscreen agent due to its passive UV absorption properties (4) in the UV-A region (in vivo λmax = 370 nm) (9). Thus, 85 to 90% of the incident UV-A is absorbed by scytonemin in the sheaths of cyanobacteria, providing an effective protection to the subtending cells. Remarkably, this pigment has been found in more than 300 species of cyanobacteria from various geographic locations and environments, leading to intriguing questions concerning its evolutionary history (7).
In 2007, a cluster of genes involved in the biosynthesis of scytonemin was identified through the analysis of a non-scytonemin-producing mutant of Nostoc punctiforme ATCC 29133 obtained through transposon mutagenesis (12). This mutation was embedded within a cluster of 18 open reading frames (ORFs) (NpR1276 to NpR1259) that were all transcribed in the same direction, thus suggesting this to be the functional genetic unit involved in scytonemin biosynthesis. This cluster contains genetic functions predicted to be involved in the biosynthesis of aromatic amino acids such as tryptophan, as well as other putative functions involved in the assembly of scytonemin. However, information concerning the number of these genes that are transcribed during biosynthesis is lacking.
In this study, we provide evidence on the boundaries of the scytonemin biosynthetic gene cluster through a transcriptional expression analysis after exposure to UV radiation and through a comparison of the gene cluster as found in six cyanobacterial species. The conservation of this pathway across these cyanobacterial lineages also enabled an analysis of the evolution of these genetic elements and provides support for the ancient origin of the scytonemin biosynthetic gene cluster.
MATERIALS AND METHODS
Cyanobacterial strains and culture techniques.
The cyanobacterium Nostoc punctiforme ATCC 29133 was obtained from the American Type Culture Collection (ATCC). A culture was maintained in unialgal condition in liquid BG-11 freshwater medium at 29°C under a light intensity of approximately 19 μmol m−2 s−1 and a light/dark cycle of 16 h/8 h.
Transcriptional expression analyses.
N. punctiforme ATCC 29133 was initially grown for approximately 45 days. Following this growth period, a portion of this culture was transferred to a petri dish and allowed to acclimate for 30 days prior to initiating the experiment. A sample was then taken from this dish culture as the −UV sample, and then the cyanobacterial filaments were exposed to UV radiation for 48 h (0.64 mW/cm2; λmax = 365 nm). A second sample was taken from this dish after the 48-h exposure period, representing the +UV sample. The extended acclimation time in the petri dish cultures prior to taking samples for either the −UV or +UV samples controlled for pathway transcript levels between the two samples. Both −UV and +UV cultured biomass was extracted for RNA following a modified Trizol protocol (Invitrogen). The extracted RNA was treated with Turbo DNase (Ambion) and found to be free of genomic DNA contamination through control reverse transcription-PCRs (RT-PCRs) which lacked reverse transcriptase. RNA concentrations were determined using a Beckman Coulter DU800 spectrophotometer. Specific primers were designed to amplify regions of RNA ranging from 160 to 220 bp for 48 genes, including those reported to be part of the scytonemin biosynthetic cluster and surrounding neighbors (12). Primer sequences are available in Table S1 in the supplemental material. cDNA was synthesized following a standard protocol for Superscript III reverse transcriptase with an incubation at 50°C (Invitrogen) and using 350 ng of RNA as a template for every reaction. The PCR amplification was accomplished using a Taq polymerase master mix (Promega), two microliters of first-strand cDNA template, and an annealing temperature of either 55 or 56°C based on optimization for each primer set on genomic DNA. Genomic DNA was isolated and purified following a modified phenol protocol. For each of the 48 genes, four cDNA reactions were completed: −UV, +UV, negative control (no reverse transcriptase or RNA), and a positive control (created through PCR of genomic DNA). These four samples were visualized on a 1.5% agarose gel stained with ethidium bromide and documented using a Fisher Biotech transilluminator and a Kodak photographic system with a 5-s exposure optimized for ethidium bromide. The visualization of each cDNA sample was repeated three times to minimize variations from gel loading by pipette. Analysis of gel band intensity was completed using the Bio-Rad Quantity One software program (Bio-Rad). For each cDNA band, an average band intensity was calculated and normalized for the gel background by subtracting the average negative control intensity. In order to minimize the effects of primer bias, the DNA amplicons were normalized to one another by calculating a percentage of the positive control for each of the −UV and +UV bands. The percent band intensity of the −UV gel band was then subtracted from the percent band intensity of the +UV gel band and graphically represented using the Excel software program (Microsoft). The standard error was calculated based on technical replicates by comparing the three gels run for each of the 48 primer sets, and any negative values are represented as zero values. An additional experimental replicate was also completed using the same methods described above and confirmed the results of the trend in increased expression discussed below (see Fig. S1 in the supplemental material).
Bioinformatic analysis.
Using the Joint Genome Institute and National Center for Biotechnology Information (NCBI) web databases, 32 complete and 17 incomplete cyanobacterial genomes were examined for the scytonemin biosynthetic gene cluster using sequence similarity BLAST searches (NCBI) with the ORFs Np1264 (S5) and Np1276 (S17) from the N. punctiforme ATCC 29133 scytonemin biosynthetic cluster (12). Five additional scytonemin biosynthetic gene clusters were located, and the ORFs found in each of these were identified through available genome annotations and through manual identification using the NCBI ORF Finder. The six scytonemin gene clusters were each assembled using the Vector NTI software program (Invitrogen) and the amino acid sequences for individual ORFs compared to one another using bl2seq BLAST searches (NCBI) and through alignments created in Vector NTI.
Phylogenetic analyses.
Phylogenetic analyses were completed using the MEGA 4.0 software program (15). All sequence alignments were performed using ClustalW algorithms with the Gonnet protein weight matrix for amino acids and the IUB DNA weight matrix for nucleotide-based alignments. Sequence alignments were manually edited to exclude ambiguous regions. Neighbor-joining, minimum-evolution, and maximum-parsimony trees were created and evaluated with 10,000 bootstrap replicates. Outgroups (not shown) used in the creation of the phylogenetic trees were obtained by identifying homologues by BLAST searches. The basis of evolutionary selection for ORFs S6, S7, S16, and S17 was calculated using the number of nonsynonymous substitutions per nonsynonymous site (KA) and the number of synonymous substitutions per synonymous site (KS) in MEGA 4.0 (15).
RESULTS
Transcriptional expression of the scytonemin biosynthetic pathway.
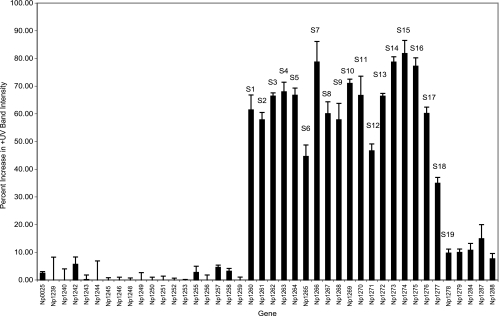
The previously identified ORFs of the scytonemin biosynthetic gene cluster (12), as well as those ORFs from the surrounding region in the N. punctiforme ATCC 29133 genome, were analyzed for relative changes in their transcriptional levels after exposure to UV radiation. A total of 42 ORFs spanning 63.7 kb of the annotated genome from Np1239 to Np1288 were evaluated in this manner. Transcripts from the putative biosynthetic cluster (Np1260 to Np1276 and annotated in Table S1 in the supplemental material) were substantially increased in cultures irradiated with UV light, as visualized by semiquantitative reverse transcription-PCR on agarose gels (Fig. 1). Conversely, transcripts from outside of the reported cluster, Np1239 to Np1259, showed a less than 10% increase in the UV-stimulated cultures over the UV-deficient cultures. This low level of intensity difference between +UV and −UV cDNA is similar to that for the gyrase control ORF Np0025. Because gyrase represents a housekeeping gene, it is not expected to show a transcriptional variation in response to UV radiation. The difference in the intensity of the +UV cDNA and that of the −UV cDNA for the ORFs Np1260 to Np1276 ranged from 44% for Np1265 to 82% for Np1274. ORF Np1277, a putative histidine kinase, showed an increase in intensity upon UV radiation stimulation of approximately 35%. ORFs Np1278 to Np1288 showed very modest increases in cDNA in the UV-stimulated cultures, generally less than 20%. Thus, the major ORFs increased in expression by growth under UV light are those previously identified as the scytonemin biosynthetic gene cluster, with the addition of one downstream gene (Np1277), which is likely involved in regulation of the cluster.
FIG. 1.
Graphical comparison of the variation in transcriptional levels of the scytonemin biosynthetic pathway, as well as surrounding ORFs in the N. punctiforme ATCC 29133 genome. This analysis shows the percent difference between the band intensity for the +UV cDNA over that of the −UV cDNA compared to that of the genomic DNA control. Each bar represents an individual gene in the pathway, and Np0025 is the control representing gyrase from N. punctiforme ATCC 29133.
Organization of the scytonemin biosynthetic gene cluster.
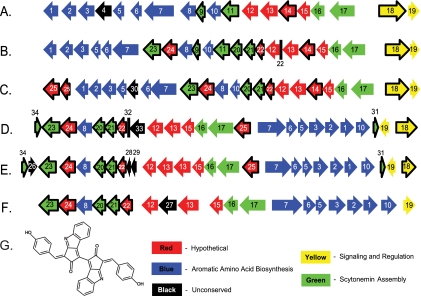
Forty-nine partial or complete cyanobacterial genomes were analyzed for the scytonemin biosynthetic cluster using Np1276 as a query for BLAST searches. Np1276 is a thiamine diphosphate-containing enzyme found within this cluster from N. punctiforme (accession no. NC_010628). As a result, five additional cyanobacterial species were found to contain a closely related gene cluster: Nodularia spumigena CCY9414 (accession no. NZ_AAVW01000004), Cyanothece sp. strain PCC7822 (accession no. NZ_ABVE01000001), Cyanothece sp. strain PCC7424 (accession no. ABOY01000036), Lyngbya sp. strain PCC8106 (accession no. AAVU01000005), and the previously identified gene cluster from Nostoc sp. strain PCC7120 (accession no. NC_003272) (12). Many of the ORFs in the clusters from these other species are from the same conserved protein family and share a high percentage of amino acid sequence similarity (Table 1). However, the ORFs found both upstream and downstream of the scytonemin cluster from N. punctiforme have no significant sequence similarity to the regions surrounding the putative scytonemin cluster from these other species, thus further helping to delineate the boundaries of the cluster. The scytonemin biosynthetic gene clusters for all six cyanobacterial species are displayed in Fig. 2, where each ORF is annotated with respect to its involvement in similar biosynthetic functions across these six species (S1 to S34).
TABLE 1.
Best BLAST results and amino acid comparison of conserved ORFs in the scytonemin biosynthetic gene clustera
| ORF designation | Locus tag for N. punctiforme | Avg size of product (aa) | Conserved protein family | Best BLAST hit | % Identity (no. of aa) | Amino acid % identity for conserved proteins |
|---|---|---|---|---|---|---|
| S1 | Np1260 | 355 | AroA, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase | Carboxysome formation protein, Cyanothece sp. strain ATCC 51142 | 80 (345) | 55 |
| S2 | Np1261 | 368 | TrpD, anthranilate phosphoribosyltransferase | Anthranilate phosphoribosyltransferase, Anabaena variabilis ATCC 29413 | 67 (361) | 46 |
| S3 | Np1262 | 416 | TrpB, tryptophan synthase beta chain | Tryptophan synthase subunit beta, Anabaena variabilis ATCC 29413 | 90 (394) | 70 |
| S4 | Np1263 | 409 | Tyrosinase | Hypothetical protein DDBDRAFT_0185605, Dictyostelium discoideum AX4 | 31 (404) | NAb |
| S5 | Np1264 | 274 | TrpA, tryptophan synthase alpha chain | Tryptophan synthase, alpha subunit, Microcoleus chthonoplastes PCC7420 | 73 (263) | 54 |
| S6 | Np1265 | 287 | TrpC, indole-3-glycerol phosphate synthase | Indole-3-glycerol-phosphate synthase, Anabaena variabilis ATCC 29413 | 70 (262) | 44 |
| S7 | Np1266 | 738 | TrpE, anthranilate/para-aminobenzoate synthase component I | Anthranilate synthase, Anabaena variabilis ATCC 29413 | 77 (726) | 55 |
| S8 | Np1267 | 400 | AroB, 3-dehydroquinate synthetase | 3-Dehydroquinate synthase, Cyanothece sp. strain CCY0110 | 66 (389) | 45 |
| S9 | Np1268 | 216 | DsbA_FrnE, DsbA family, FrnE subfamily | Hypothetical protein OA307_1438, Octadecabacter antarcticus 307 | 36 (203) | 70 |
| S10 | Np1269 | 345 | TyrA, bifunctional chorismate mutase/prephenate dehydrogenase | COG0287, prephenate dehydrogenase, Yersinia mollaretii ATCC 43969 | 38 (354) | 27 |
| S11 | Np1270 | 430 | Glycos_transf_1, glycosyltransferases group 1 | Glycosyltransferase, group 1, Anabaena variabilis ATCC 29413 | 74 (426) | 77 |
| S12 | Np1271 | 395 | Uncharacterized conserved protein | NHL repeat-containing protein, “Candidatus Methanoregula boonei” 6A8 | 38 (246) | 30 |
| S13 | Np1272 | 453 | None | Conserved hypothetical protein, Streptomyces ambofaciens | 28 (423) | 21 |
| S14 | Np1273 | 422 | None | Conserved hypothetical protein, Streptomyces ambofaciens | 31 (353) | 55 |
| S15 | Np1274 | 325 | None | Phosphoenolpyruvate carboxykinase, Heliobacterium modesticaldum Ice1 | 29 (120) | 52 |
| S16 | Np1275 | 351 | ELFV_dehydrog, glutamate/leucine/phenylalanine/valine dehydrogenase | Leucine dehydrogenase, Geobacillus thermodenitrificans NG80-2 | 53 (333) | 52 |
| S17 | Np1276 | 625 | TPP_enzymes, thiamine diphosphate enzyme family | Acetolactate synthase large subunit, Plesiocystis pacifica SIR-1 | 51 (586) | 41 |
| S18 | Np1277 | 553 | HATPase_c, histidine kinase-like ATPases | PAS fold family, Microcoleus chthonoplastes PCC 7420 | 40 (246) | 24 |
| S19 | Np1278 | 295 | REC, signal receiver domain | Two-component AraC family transcriptional regulator, Anabaena variabilis ATCC 29413 | 67 (276) | 7 |
| S20 | 306 | UbiA, 4-hydroxybenzoate polyprenyltransferase and related prenyltransferases | Prenyltransferase, UbiA family, Synechococcus sp. strain PCC 7335 | 60 (299) | 35 | |
| S21 | 304 | TatD_DNAse, TatD-like proteins | Hypothetical protein CY0110_32085, Cyanothece sp. strain CCY0110 | 81 (292) | 67 | |
| S22 | 258 | None | Hypothetical protein S7335_3653, Synechococcus sp. strain PCC 7335 | 52 (234) | 39 | |
| S23 | 460 | Phosphodiest, type I phosphodiesterase/nucleotide pyrophosphatase | Hypothetical protein CY0110_09261, Cyanothece sp. strain CCY0110 | 71 (455) | 53 | |
| S24 | 419 | None | Hypothetical protein CY0110_09256, Cyanothece sp. strain CCY0110 | 59 (396) | 40 | |
| S25 | 423 | None | Predicted: hypothetical protein, Rattus norvegicus | 30 (130) | 17 | |
| S26 | 323 | None | Hypothetical protein MC7420_5057, Microcoleus chthonoplastes PCC 7420 | 39 (322) | NA | |
| S27 | 451 | Arginosuccinate synthase | Conserved hypothetical protein, Streptomyces ambofaciens ATCC 23877 | 28 (392) | NA | |
| S28 | 130 | None | Pep-cten putative exosortase interaction domain protein, Microcoleus chthonoplastes PCC 7420 | 54 (35) | NA | |
| S29 | 162 | None | Hypothetical protein RRC374, uncultured methanogenic archaeon | 36 (87) | NA | |
| S30 | 319 | Qur, NADPH:quinone reductase and related Zn-dependent oxidoreductases | Zinc-containing alcohol dehydrogenase superfamily protein, Anabaena variabilis ATCC 29413 | 50 (318) | NA | |
| S31 | 117 | None | Hypothetical protein EHI_148590, Entamoeba histolytica HM-1:IMSS | 45 (37) | 74 | |
| S32 | 183 | None | Putative transposase, Cyanothece sp. strain PCC7425 | 59 (44) | NA | |
| S33 | 400 | None | Hypothetical protein RRC373, uncultured methanogenic archaeon | 21 (208) | NA | |
| S34 | 127 | None | Unnamed protein product, Microcystis aeruginosa PCC7806 | 73 (123) | 83 |
Gene designations are assigned based on the original designation from the N. punctiforme ATCC 29133 gene cluster and are used uniformly throughout the five other identified gene clusters. The architecture of these genes across the six gene clusters is diagrammed in Fig. 2. The identity column represents the percent identity of the amino acid sequence to that of its nearest nonscytonemin gene cluster-containing homolog by BLAST analysis and is followed by the number of amino acids being compared in the ORF. The values in the column labeled “Amino acid % identity for conserved proteins” were determined through the alignment of the ORFs from all six gene clusters. aa, amino acids.
NA, not applicable (protein found in only one gene cluster).
FIG. 2.
Architectural comparison of ORFs involved in the biosynthesis of scytonemin across six cyanobacterial species. (A) Nostoc punctiforme ATCC 29133; (B) Nodularia spumigena CCY9414; (C) Nostoc sp. strain PCC7120; (D) Cyanothece sp. strain PCC7822; (E) Cyanothece sp. strain PCC7424; (F) Lyngbya sp. strain PCC8106; (G) chemical structure of scytonemin. Arrows and lines represent individual ORFs, with corresponding numbers annotated in Table 1. Colors represent predicted gene functions as outlined in the key. Arrows with black outlines represent open reading frames not conserved between the scytonemin pathways in all six of the cyanobacterial species.
These six putative scytonemin clusters range in size from 33.2 kbp in Cyanothece sp. strain PCC7424 to 27.7 kbp in Lyngbya sp. strain PCC8106 (Table 2). There are 14 ORFs found conserved in all 6 cyanobacterial species, including the genes involved in aromatic amino acid biosynthesis (S1 to S3, S5 to S8, and S10), hypothetical proteins (S12, S13, and S15), a dehydrogenase (S16), a thiamine diphosphate-containing enzyme (S17), and a response regulator (S19). Five of these clusters contain an additional set of conserved ORFs (S20, S21, and S26) which encode a prenyltransferase, a putative hydrolase, and a hypothetical protein.
TABLE 2.
Comparison of six cyanobacterial genomes found to contain the scytonemin biosynthetic gene cluster
| Cyanobacteriuma | Genome
|
Scytonemin pathway
|
|||||
|---|---|---|---|---|---|---|---|
| Size (Mb) | % G+C Content | Status | Sequence location | Size (kb) | No. of ORFs | Location in genome | |
| Nostoc punctiforme ATCC 29133* | 8.2 | 41 | Complete | JGI | 28 | 19 | 1516115-1544157 |
| Nostoc sp. strain PCC7120 (= ATCC 27893) | 6.4 | 41 | Complete | Kazusa DNA Research Institute | 31.7 | 24 | 475640-507374 |
| Nodularia spumigena CCY9414 | 5.3 | 42 | Incomplete | J. Craig Venter Institute | 31.6 | 23 | Unknown |
| Cyanothece sp. strain PCC7424 | 6.4 | 39 | Incomplete | JGI | 33.2 | 25 | Unknown |
| Cyanothece sp. strain PCC7822 | 5.7 | 40 | Incomplete | JGI | 29.5 | 23 | Unknown |
| Lyngbya sp. strain PCC 8106 (= L. aestuarii CCY9616)* | 10 | 41 | Incomplete | J. Craig Venter Institute | 27.7 | 21 | Unknown |
Strains marked with an asterisk are reported to produce scytonemin.
Phylogenetic analyses of the biosynthesis of scytonemin.
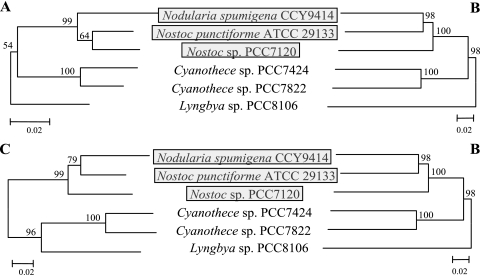
Two fused subsets were created in silico from the scytonemin gene cluster for phylogenetic analyses. The first contained 6,887 bp for the genes involved in aromatic amino biosynthesis (S1 to S3, S5 to S7, and S10), and the second contained 2,565 bp for the genes likely involved in scytonemin assembly (S16 and S17). These were used to analyze the percent difference in mutations per site compared to the total number of amino acids found within each section of the scytonemin biosynthetic pathway (Fig. 3). Based on a minimum evolution phylogenetic analysis, the genes encoding aromatic amino acid biosynthesis showed approximately a twofold increase in mutations per site compared to the scytonemin assembly genes. In a second analysis, phylogenetic markers were created using 1,476 bp from the 16S rRNA and 1,859 bp from the rpoC1 gene, both of which are in common use for phylogenetic classification of cyanobacteria (11), and compared to the sequence produced by fusing ORFs S6 and S7 (approximately 3,129 bp) and S16 and S17 (approximately 3,135 bp). These gene sets were used to evaluate phylogenetic congruence between the phylogenetic marker genes and those encoding scytonemin biosynthesis (Fig. 4). Compared to the phylogenetic marker genes, all six scytonemin gene clusters appear to be fairly congruent, with a similar branching pattern based on sequence divergence. The only differences appear to be the timing of the divergence and the variation in clustering of the two Nostoc species, which is also apparent when the phylogenetic marker genes are compared. In order to determine if S6, S7, S16, and S17 are under positive or negative selection pressure, we compared the KA number to the KS number (10). The KA/KS ratios for all genes were well below a value of 1.0, indicating that this pathway is under purifying selection.
FIG. 3.
Comparison of the sequence divergence between gene set S1 to S3, S5 to S7, and S10, involved in scytonemin-associated aromatic amino acid biosynthesis (A), and gene set S16 and S17, conserved in the proposed scytonemin assembly portion of the cluster (B). Percentages indicate the number of substitutions for each branch compared to the total number of amino acids used in the analysis. Phylogenetic trees were created using minimum evolution criteria and supported by neighbor-joining and maximum parsimony. Outgroups used in the analysis are not shown (for S16-S17, Anabaena variabilis ATCC 29413; and for 16S-rpoC1, Thermosynechococcus elongatus BP-1).
FIG. 4.
Comparison of the evolution of phylogenetic markers, 16S rRNA (A) and rpoC1 (C), with that of four genes found in the scytonemin biosynthetic pathway (B), S6, S7, S16, and S17. Phylogenetics are shown using minimum evolution criteria with bootstrap values from 10,000 replicates. These tree topologies are supported by both neighbor-joining and maximum-parsimony criteria, with bootstrap values greater than 50 for all branches (results not shown). Gray boxes represent those cyanobacteria having the scytonemin gene cluster architecture where all ORFs are transcribed in one direction.
DISCUSSION
The increase in the level of transcription for Np1260 through Np1276 in N. punctiforme ATCC 29133 after exposure to UV radiation is consistent with these genes forming the scytonemin biosynthetic gene cluster (Fig. 1). Previous efforts utilized a mutagenesis approach of a single gene within this pathway to propose that these genes may form a biosynthetic functional unit (12); however, this report provides the first evidence that all 17 of these genes are expressed after exposure to UV radiation. This differential transcriptional analysis shows that there is an average 65% increase in the level of transcription for Np1260 through Np1276, which suggests that this gene cluster is under tight regulation and ultimately controlled by the presence of UV radiation. The regulation of the scytonemin gene cluster by an environmental parameter also suggests the presence of a unique signaling mechanism which responds to UV radiation. The presence of a conserved histidine kinase and response regulator just downstream of this cluster in five of the six identified pathways may indicate that this is the two-component signaling mechanism responsible for its regulation. Although further studies, including quantitative PCR, are needed to better understand how variation in the wavelength of light affects this proposed regulatory histidine kinase, these semiquantitative results indicate that the expression of this gene increases after exposure to UV radiation, although at a lower level than that for the other genes in the cluster. The apparent lower level of transcription may be a result of the phosphorylation kinetics of the two-component system. Because the phosphorylated state can have a half-life in the range of hours, this decreases the turnover time for proteins involved in the signaling response (17). In this study, the general increase in the level of transcription of the kinase and the presence of a conserved motif known as a Per Arnt Sim (PAS) fold, which has been shown to play a role in light induced regulation (16), provide support for this hypothesis. A better understanding of UV-induced signaling elements, such as the potential histidine kinase involved in scytonemin biosynthesis, is important to appreciating how organisms adapt to harsh light regimes. These signaling elements may also provide mechanistic insights into the transcriptional regulation of various classes of UV protectant molecules in other organisms and could have broad biotechnological applications as an inducible regulatory system.
Based on our differential transcriptional analysis, it appears that the enzymatic functions for the biosynthesis of scytonemin are found between Np1260 and Np1276. Previously the biosynthesis of scytonemin was suggested to derive from a tryptophan-derived subunit and a phenylpropanoid-derived subunit (9). The presence of eight genes involved in the biosynthesis of aromatic amino acids (Np1260 to Np1262, Np1264 to Np1267, and Np1269) is consistent with this hypothesis. Interestingly, these eight aromatic amino acid biosynthetic genes are all clearly upregulated in the presence of UV radiation, indicating that their functionality is primarily for the biosynthesis of scytonemin. Consequently, the second copies of these aromatic amino acid biosynthetic genes, which are found scattered throughout these six cyanobacterial genomes, must be responsible for the primary metabolic needs of the cell. Previous work suggests that this biosynthetic gene cluster could be considered a supraoperon due to the nesting of the tryptophan operon within this cluster (19). This supraoperon thus encodes for a specialized metabolic capability that provides a selective advantage to cyanobacteria (19).
Clarification of the boundaries for transcriptional regulation of the scytonemin biosynthetic gene cluster provides insights into some of the mechanistic steps that may be involved in its biosynthesis in N. punctiforme ATCC 29133. For example, the conserved thiamine diphosphate-containing enzyme (Np1276) is likely involved in the decarboxylative coupling of two aromatic amino acid-derived precursors to form an acyloin intermediate. This reactivity has recently been confirmed through an in vitro study of the product of Np1276, which catalyzes the condensation of the two alpha-keto acids derived from tryptophan and tyrosine (1). Based on this in vitro data, the precursors are expected to couple in a similar manner in vivo, with the resulting acyloin intermediate subsequently cyclizing to the tetracyclic system present in each half of the scytonemin molecule (Fig. 2G). Dimerization to the completed scytonemin molecule may either occur spontaneously or result from the tyrosinase activity found present in the N. punctiforme ATCC 29133 gene cluster.
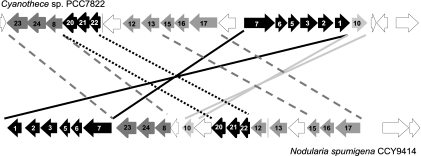
The putative scytonemin biosynthetic gene cluster is present in six species of cyanobacteria whose genomes are available (Fig. 2). Based on the overall structure of these six gene clusters, they appear to have evolved by two major mechanisms: (i) the rearrangement of large sections of the cluster and (ii) the insertions of individual genes or small clusters of genes which confer additional biosynthetic capabilities. The two variations produced by rearrangements separate Lyngbya sp. strain PCC8106, Cyanothece sp. strain PCC7424, and Cyanothece sp. strain PCC7822 from the other three cyanobacterial species containing the scytonemin biosynthetic gene cluster. This rearrangement switches the transcriptional direction of S1 to S3, S5 to S7, and S10, which are involved in the biosynthesis of the aromatic amino acids, and moves them from upstream of S17 to downstream of this position (Fig. 5). This rearrangement is similar to what has been found in the microcystin biosynthetic gene cluster as identified in Microcystis aeruginosa and Planktothrix agardhii. The microcystin biosynthetic genes in M. aeruginosa are organized into two units which are transcribed in opposite directions, whereas most of the microcystin genes in P. agardhii are transcribed in the same direction and as a single operon (3). The presence of these two types of rearranged scytonemin gene clusters suggests that this pathway must also involve two types of pathway promotion in order to regulate the production of this molecule. Based on the precedent formed by the microcystin biosynthetic gene cluster wherein there are two different promoters (3), one of these promoter regions may be a bidirectional promoter involved in initiating transcription of both the upstream and downstream parts of the scytonemin biosynthetic pathway.
FIG. 5.
Examples of the two major types of scytonemin biosynthetic gene clusters (top, Cyanothece sp. strain PCC7822; bottom, Nodularia spumigena CCY9414). For purposes of clarity only, ORFs forming subclusters are shown in various shades of white, gray, and black. Lines between the clusters identify the rearrangement and inversion of ORFs involved in the biosynthesis of aromatic amino acids as well as other major differences between the two types of pathways.
The two architectures of the scytonemin biosynthetic gene cluster result in a splitting of the pathway into two subclusters, one containing the genes involved in the biosynthesis of aromatic amino acids and the other containing the putative scytonemin assembly genes. Based on a minimum evolution phylogenetic analysis of these two portions of the scytonemin gene cluster (Fig. 3), the number of mutational changes between them is slightly different following the last evolutionary divergence. In this analysis, the genes involved in the biosynthesis of aromatic amino acids contain about 10% more substitutions per amino acid site than the scytonemin assembly genes. These results suggest that the S16 and S17 genes may be under a tighter selection pressure and less able to accommodate mutational changes than the genes involved in aromatic amino acid biosynthesis. The weaker selection pressure imposed on the subcluster involving aromatic amino acid biosynthesis may be a result of the more dynamic properties reported for the trp operon (20). It has been speculated that the trp operon has undergone a constant process of fine-tuning over evolutionary time, because the operon encodes for biochemically expensive enzymatic reactions (20).
Comparison of the six scytonemin gene clusters suggests that the insertion of individual genes or small clusters of genes has contributed to the evolution of the pathway. These inserted elements include an isomerase (S9) and glycosyltransferase (S11) in N. punctiforme ATCC 29133 and N. spumigena CCY9414 and a tyrosinase (S4) which is found only in N. punctiforme ATCC 29133. Because a set of genes (S20, S21, and S24) encoding a prenyltransferase, a putative hydrolase, and a hypothetical protein is found in all of the scytonemin gene clusters except that of N. punctiforme ATCC 29133, we speculate that gene deletions are also involved in modifications to the pathway. However, there is a core set of genes involving aromatic amino acid biosynthesis (S1 to S3, S5 to S8, and S10) and scytonemin assembly (S12, S13, and S15 to S17) which is highly conserved within all six gene clusters, and this likely signifies that these are key biosynthetic reactions for the production of UV sunscreen pigments. We speculate that the variable genes in these pathways represent enzymatic functions that are creating as yet uncharacterized structural variants of the scytonemin molecule with altered physiological or UV-screening properties.
A comparison of the 16S rRNA and rpoC1 data sets with those of four genes found in the scytonemin biosynthetic gene cluster, S6 and S7 from the aromatic amino acid biosynthesis section and S16 and S17 from the scytonemin assembly portion, indicates that this pathway may be of ancient origin due to the congruence of the branching pattern between all of these species of cyanobacteria (Fig. 4). These six gene clusters have a branching pattern similar to that of the phylogenetic marker genes except in the comparison of N. punctiforme ATCC 29133, Nostoc sp. strain PCC7120, and N. spumigena CCY9414, where variation is found compared to the 16S rRNA evolutionary marker gene. However, this apparent evolutionary divergence may result from the fact that the 16S rRNA gene sometimes poorly defines fine-scale phylogenetic relationships of closely related species due to intragenomic variations between its multiple gene copies. In such cases, alternative phylogenetic marker genes or use of gene combinations such as multilocus sequence typing can clarify these relationships (2). These results are similar to the findings for the microcystin gene cluster, where the biosynthetic sequences and the phylogenetic sequences (16S rRNA and rpoC1) have a high degree of congruence and similar levels of divergence; this is consistent with an ancient origin of the microcystins (10). In the case of scytonemin, the congruence between the scytonemin data set and that of the evolutionary marker genes is also indicative of the ancient origin of this gene cluster.
The evolution of the scytonemin biosynthetic gene cluster from an ancient origin is further supported by its being under purifying selection pressure. A high KA/KS ratio would indicate positive selection pressure due to the helpful influence of these mutations on an organism's fitness; however, the observed low KA/KS ratio indicates a purifying selection pressure, which signifies that mutations affecting the protein sequence are deleterious and negatively affect an organism's fitness (10). This purifying selection pressure is consistent with an ancient origin of the scytonemin biosynthetic gene cluster (8).
The tight transcriptional regulation of the scytonemin biosynthetic gene cluster by UV radiation and its apparent ancient evolution are evidence of this molecule's ecological significance as a sunscreen pigment. The idea that cyanobacteria would support a biosynthetic pathway of high energetic requirements, due to the use of aromatic amino acids, throughout their evolution even after the formation of the protective ozone layer is further support for the importance of scytonemin in expanding the habitat of these diverse organisms. The presence of the scytonemin biosynthetic gene cluster across cyanobacterial lineages and the variations between individual pathways indicate that scytonemin is an adaptive and functional sunscreen that supports the expansion of cyanobacteria into many different ecological niches exposed to high levels of UV radiation. Indeed, in the current era, wherein greater levels of UV irradiation are reaching the earth's surface, this adaptation is of great significance to cyanobacterial survival and growth in new habitats. Finally, a better understanding of the signaling and biosynthetic mechanisms involved in this widespread and naturally occurring sunscreen molecule may allow the development of important biotechnological advances in both drug discovery and agricultural biology.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health predoctoral fellowship through Training Grant in Marine Biotechnology no. T32GM067550 and the California Sea Grant program (SG-100-TECH-N) and previous support from the Oregon Sea Grant program, no. R/BT-40.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
We acknowledge E. Esquenazi and P. Dorrestein at the University of California, San Diego, for help identifying scytonemin production by matrix-assisted laser desorption mass spectrometry and J. Wells and family for general support to the laboratory. We also acknowledge L. Gerwick for helpful discussions on phylogenetics and molecular biological techniques.
Footnotes
Published ahead of print on 29 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Balskus, E. P., and C. T. Walsh. 2008. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 130:15260-15261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Case, R. J., Y. Boucher, I. Dahllöf, C. Holmström, W. F. Doolittle, and S. Kjelleberg. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christiansen, G., J. Fastner, M. Erhard, T. Börner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockell, C. S., and J. Knowland. 1999. Ultraviolet radiation screening compounds. Biol. Rev. Camb. Philos. Soc. 74:311-345. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, Y., and M. Gurevitz. 2006. The cyanobacteria—ecology, physiology and molecular genetics. Prokaryotes 4:1074-1098. [Google Scholar]
- 6.Ehling-Schulz, M., and S. Scherer. 1999. UV protection in cyanobacteria. Eur. J. Phycol. 34:329-338. [Google Scholar]
- 7.Garcia-Pichel, F., and R. W. Castenholz. 1991. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. J. Phycol. 27:395-409. [Google Scholar]
- 8.Massingham, T., and N. Goldman. 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169:1753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proteau, P. J., W. H. Gerwick, F. Garcia-Pichel, and R. Castenholz. 1993. The structure of scytonemin, an ultraviolet sunscreen pigment from the sheaths of cyanobacteria. Experientia 49:825-829. [DOI] [PubMed] [Google Scholar]
- 10.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo, P. S., and A. Yokota. 2003. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, rpoC1 and rpoD1 gene sequences. J. Gen. Appl. Microbiol. 49:191-203. [DOI] [PubMed] [Google Scholar]
- 12.Soule, T., V. Stout, W. D. Swingley, J. C. Meeks, and F. Garcia-Pichel. 2007. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 189:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson, C. S., E. A. Capper, A. K. Roshak, B. Marquez, C. Eichman, J. R. Jackson, M. Mattern, W. H. Gerwick, R. S. Jacobs, and L. A. Marshall. 2002. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharmacol. Exp. Ther. 303:858-866. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson, C. S., E. A. Capper, A. K. Roshak, B. Marquez, K. Grace, W. H. Gerwick, R. S. Jacobs, and L. A. Marshall. 2002. Scytonemin—a marine natural product inhibitor of kinases key in hyperproliferative inflammatory diseases. Inflamm. Res. 51:112-114. [DOI] [PubMed] [Google Scholar]
- 15.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 16.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 18.Wynn-Williams, D. D., H. G. M. Edwards, E. M. Newton, and J. M. Holder. 2002. Pigmentation as a survival strategy for ancient and modern photosynthetic microbes under high ultraviolet stress on planetary surfaces. Int. J. Astrobiol. 1:39-49. [Google Scholar]
- 19.Xie, G., C. A. Bonner, T. Brettin, R. Gottardo, N. O. Keyhani, and R. A. Jensen. 2003. Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella species and in heterocystous cyanobacteria. Genome Biol. 4:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.