Abstract
Cilia and flagella are cell organelles that are highly conserved throughout evolution. For many years, the green biflagellate alga Chlamydomonas reinhardtii has served as a model for examination of the structure and function of its flagella, which are similar to certain mammalian cilia. Proteome analysis revealed the presence of several kinases and protein phosphatases in these organelles. Reversible protein phosphorylation can control ciliary beating, motility, signaling, length, and assembly. Despite the importance of this posttranslational modification, the identities of many ciliary phosphoproteins and knowledge about their in vivo phosphorylation sites are still missing. Here we used immobilized metal affinity chromatography to enrich phosphopeptides from purified flagella and analyzed them by mass spectrometry. One hundred forty-one phosphorylated peptides were identified, belonging to 32 flagellar proteins. Thereby, 126 in vivo phosphorylation sites were determined. The flagellar phosphoproteome includes different structural and motor proteins, kinases, proteins with protein interaction domains, and many proteins whose functions are still unknown. In several cases, a dynamic phosphorylation pattern and clustering of phosphorylation sites were found, indicating a complex physiological status and specific control by reversible protein phosphorylation in the flagellum.
Cilia and flagella, which are essentially identical, are among the most ancient cellular organelles, providing motility for primitive eukaryotic cells living in aqueous environments. The assembly and motility of flagella have been studied extensively with the unicellular biflagellate green alga Chlamydomonas reinhardtii. This alga uses flagella for motility and for cell-cell recognition during mating. In basal land plants, such as bryophytes and pteridophytes, the only flagellated cells are motile sperm cells, which require water to swim to the egg. With the evolution of pollen tubes in higher gymnosperms and angiosperms, these plant species lost the ability to assemble flagella (24, 42). Flagella of animals have acquired new functions in multicellular organizations during evolution (6). In mammals, cilia and flagella can be motile or immotile. Motile cilia can be found, for example, in airways (respiratory cilia), in the brain (ependymal cilia), or in the male reproductive system (sperm flagella). Defects in cilia in humans can cause severe diseases, such as polycystic kidney disease, retinal degeneration, hydrocephalus, or changes in the left-right symmetry of organs, collectively known as ciliopathies (20, 32).
Although C. reinhardtii and mammals are separated by more than 109 years of evolution, C. reinhardtii flagella are amazingly similar in structure and function to the 9+2-type axonemes of most motile mammalian flagella and cilia (42). They are composed of nine microtubular doublets surrounding two central microtubular singlets. The axoneme of motile flagella includes substructures such as dynein arms and radial spokes that generate and control axoneme bending (31). The flagellum also contains matrix proteins that are not tightly associated with the flagellar membrane or the axoneme. They serve diverse functions and can be involved in intraflagellar transport (IFT) (37).
Proteome analyses of cilia, including, for example, a human cilium, a mouse photoreceptor sensory cilium, and the flagella of the green alga Chlamydomonas reinhardtii, have unraveled hundreds of so far unknown proteins of this organelle (18, 29, 33) and have paved the way to further study the functions of these proteins. Several kinases and phosphatases were found in these proteomes, suggesting that reversible protein phosphorylation plays an important role in signaling in this organelle. This is underlined by earlier studies showing that phosphorylation and dephosphorylation control flagellar motility (35), signaling (30), length, and assembly (37, 53) in C. reinhardtii. Some phosphoproteins known or assumed to be involved in these processes, such as outer dynein arm heavy chain alpha (13), inner dynein arm intermediate chain protein IC138 (7), and central pair kinesin KLP1 (61), were characterized, but the exact in vivo phosphorylation sites were not determined. From earlier studies, it is known that >80 protein spots, representing axonemal components, are labeled by 32P by two-dimensional electrophoretic techniques (34), but many of them have not been identified so far. In the past years, the relevance of some of the flagellar kinases has been shown. For example, silencing of casein kinase 1 (CK1) disturbs flagellum formation, among several other effects (41). One of its targets is IC138 (54). Glycogen synthase kinase 3 was suggested to regulate the assembly and length of flagella (53). Also, in mammalian cilia, reversible protein phosphorylation plays an important role in ciliary beating. Second messengers such as cyclic AMP (cAMP) and cGMP, which activate special kinases, are known to be relevant there (39).
An understanding of how reversible protein phosphorylation influences the function of cilia and their role in diseases will require increased information not only about the nature of the phosphoproteins but also on their in vivo phosphorylation sites. In order to gain insight into the phosphoproteome of a eukaryotic cilium, we used the green alga C. reinhardtii, whose entire genome has been sequenced, as a model (23). This organism has many advantages for biochemical and molecular genetic studies of the flagellum. Importantly, as mentioned before, its flagellar proteome is known (33), and in addition, the proteome of the centriole that anchors the flagella is also known (11, 12).
For the identification of the targets of the kinases and phosphatases in the flagella, phosphoproteomics can be applied. However, phosphoproteome analysis has been and still is a challenging task (19, 36, 47). This is due to a few facts, as follows. (i) Phosphoproteins can have more than one phosphorylation site, and the phosphorylation status of these sites can fluctuate depending on the physiological conditions of the cell. (ii) Only a small portion of a given protein in the cell can be phosphorylated. (iii) Furthermore, phosphoproteins, especially those of signaling pathways, are often proteins found in low abundance. Therefore, it is necessary to enrich the phosphopeptides. Among different methods, immobilized metal affinity chromatography (IMAC) is frequently used for phosphopeptide enrichment. In C. reinhardtii, phosphopeptides from proteins of the cellular, thylakoid, and eyespot phosphoproteomes were identified by this way (49, 50, 51, 52). Thereby, it became obvious that biochemical enrichment of subcellular fractions as it was done with the eyespot apparatus results in an increase of phosphopeptide identification (52). In this study, we used IMAC and tandem mass spectrometry (MS/MS) along with the acquisition of data-dependent neutral loss (MS/MS/MS spectra) to identify phosphopeptides from isolated flagella of C. reinhardtii. In this way, we identified 32 flagellar phosphoproteins, including different functional categories, along with 126 in vivo phosphorylation sites. In many cases, a dynamic phosphorylation pattern within one peptide was observed.
MATERIALS AND METHODS
Cell culture.
C. reinhardtii strain 137c was grown in Tris-acetate-phosphate medium (8) with a growth cycle of 12 h of light and 12 h of dark, with a light intensity of 71 microeinsteins/m2/s at 24°C. Cells were pelleted by centrifugation (1,100 × g, 5 min, 4°C) at a cell density of 2 × 106 to 3 × 106 cells/ml and resuspended in a one-half volume of minimal medium (15). The culture was then put under constant conditions of dim light (15 microeinsteins/m2/s). Cells were harvested (700 × g, 15 min, 4°C) after they had been kept under these conditions for 29 h (subjective day).
Isolation of flagella.
Isolation of the flagella was done as previously described (55), with some modifications. The isolated flagella were purified by at least two sucrose cushion steps. For these steps, Complete proteinase inhibitor cocktail (Roche Applied Science) was added to HMDS buffer (55) according to the directions in the user's manual. After each sucrose cushion step, the purity of the isolated flagella was checked by differential interference contrast microscopy, and if no contaminations by intact cells or cell debris were visible, the fraction was further used. The purified flagella were resuspended and solubilized in HMD buffer (55) containing a set of phosphatase inhibitors [microcystin LR, cantharidin, (−)-p-bromotetramisole, vanadate, molybdate, tartrate, and imidazole; Sigma] to prevent potential dephosphorylation of the proteins prior to solubilization. Flagella were demembranated by treatment with 1% Nonidet NP-40 for 10 min at 4°C.
Protein digestion and reverse-phase chromatography.
Proteins from the membrane, matrix, and axoneme (MMA) fraction were reduced and carboxyamidomethylated according to the method of Zhang et al. (62) prior to tryptic digestion. Briefly, 1 mg protein was dissolved in HMD buffer containing phosphatase inhibitors and Nonidet NP-40 to reach a final volume of 100 μl. After the addition of 100 μl 6 M guanidinium hydrochloride and 2 μl of 1 M dithiothreitol, the solution was incubated for 1 h at 56°C. Subsequently, 20 μl of 0.5 M iodoacetamide was added to reach a final concentration of 50 mM. After 1 h of incubation at room temperature, the sample was diluted 1 to 5 in 100 mM NH4HCO3, followed by an overnight incubation at 37°C with 10 μg trypsin (Promega).
The resulting peptides were fractionated on a reverse-phase column (Source 15 RPC; GE Healthcare) on a fast protein liquid chromatography system. After the addition of acetonitrile (2% final concentration) and formic acid (0.1% final concentration) to the peptide mix, the peptides were separated from insoluble material by centrifugation, loaded onto the column, and eluted with 0.5 ml 80% acetonitrile and 0.1% formic acid. The tryptic peptides were subjected to the IMAC procedure.
Enrichment of proteins by IMAC and peptide identification by nLC-ESI-MS.
IMAC was done similar to that described by Wagner et al. (52). Briefly, 50 μl of Poros 20 MC (Applied Biosystems) metal chelating resin (66% [wt/wt] slurry) was transferred into Eppendorf gel loading tips. After charging of the column with 150 μl of 100 mM GaCl3 followed by a washing step, the peptides were loaded onto the activated IMAC column. The column was washed several times, and the phosphopeptides were eluted with 200 mM Na2HPO4 and desalted using a ZipTip microtip (Perseptive Biosystems). The ZipTip contained C18 reversed-phase chromatographic medium and was additionally loaded with 50 μl of a Poros R2 (Applied Biosystems) mixture (10 μl Poros R2 plus 50 μl 60% methanol). To improve binding of the phosphopeptides to the reversed-phase column, formic acid (final concentration, 5%) was added to the sample. After two washing steps, the acidified sample was applied to the ZipTip, followed by two washes. Peptides were eluted twice with 50 μl 60% methanol-5% formic acid. The eluate was dried in a Speed-Vac overnight and stored at −80°C. The dried phosphopeptide pellet was dissolved in 5 μl 5% dimethyl sulfoxide-5% formic acid. Phosphopeptides were subjected to nano-high-performance liquid chromatography-electrospray ionization-MS (nLC-ESI-MS), using an UltiMate 3000 nano-high-performance liquid chromatograph (Dionex Corporation) with a flow rate of 300 nl/min coupled online with a linear ion-trap ESI-MS (Finnigian LTQ; Thermo Electron Corp.) as previously described (52). The instrument was run by the data-dependent neutral loss method, cycling between one full MS and MS/MS scans of the four most abundant ions. After each cycle, these peptide masses were excluded from analysis for 10 s. The detection of a neutral loss fragment (98, 49, or 32.66 Da) in the MS/MS scans triggered an MS/MS/MS scan of the neutral loss ion representing the dephosphorylated peptide.
Data analysis.
Data analysis was done using Proteome Discoverer software (version 1.0) from Thermo Electron Corp., including the SEQUEST algorithm (16). The software parameters were set to detect a modification of 79.96 Da in Ser, Thr, or Tyr in MS/MS and MS/MS/MS spectra. For the database searches with MS/MS/MS data, modifications of −18 Da on Ser and Thr residues, representing the neutral loss, were additionally used. Furthermore, detection of a modification of 16 Da on Met, representing its oxidized form, was enabled, and carboxyamidomethylation of Cys residues was enabled as a static modification. Peptide mass tolerance was set to 1.5 Da in MS mode. In MS/MS and MS/MS/MS modes, fragment ion tolerance was set up to 1 Da. The parameters for all database searches were set to achieve a false discovery rate (FDR) of no more than 1% for each individual analysis. Thereby, the Proteome Discoverer software (version 1.0) generates a reversed “decoy” database from the chosen database, and any peptide passing the initial filtering parameter that was derived from this decoy database is defined as a false-positive result. It then automatically adjusts the minimum cross-correlation factor (Xcorr) filter for each individual charge state (+1, +2, or +3) separately in order to optimally meet the predetermined target FDR of 1%, based on the number of random false-positive matches from the reversed “decoy” database. Additionally, scores for the Xcorr (5) were set to the following limits: Xcorr of ≥2.0 if the charge of the peptide was 1, Xcorr of ≥2.5 if the charge of the peptide was 2, and Xcorr of ≥3.0 if the charge of the peptide was 3.
Data were searched against the flagellar proteome database (33; http://labs.umassmed.edu/chlamyfp/index.php). Additionally, the JGI database (version 2 and version 3) was used for data evaluation, as well as previous C. reinhardtii proteome data deposited on a protein network site of C. reinhardtii that is available at http://www2.uni-jena.de/biologie/chlamy/index.php. The protein sequences of the gene models were compared to the NCBI protein database by use of BLAST (1). An internal cutoff E value of 1e−05 for positive identification of proteins and functional domain prediction was used.
Preparation of crude extracts for immunoblots.
Protein extracts were prepared according to the method of Zhao et al. (63). The concentration of proteins was measured according to the method of Neuhoff et al. (26). Immunoblots were done with antibodies against phosphothreonine or against phosphotyrosine (Cell Signaling Technology) according to the method of Wagner et al. (52). Immunoblots with anti-C1 antibodies (63), anti-VIPP1 antibodies (17), or anti-CK1 peptide antibodies (41) were done as described before, along with chemiluminescence detection.
RESULTS
Isolation of flagella from Chlamydomonas reinhardtii and characterization of their phosphoproteins.
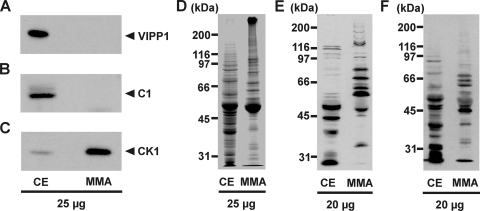
For identification of phosphorylated flagellar proteins of C. reinhardtii, we first isolated flagella from vegetative cells, with deflagellation by the dibucaine method (55). Flagella were then purified by sucrose cushions and demembranized by Nonidet NP-40 treatment in the presence of phosphatase inhibitors (see Materials and Methods). The resulting MMA fraction was first analyzed by immunodetection to find out if contaminating proteins were present. For this purpose, antibodies directed against the chloroplastic vesicle-inducing protein in plastids 1 (VIPP1) (17) and against the cytosolic C1 subunit of the RNA-binding protein CHLAMY1 (63) were used. While these antibodies detected VIPP1 and C1 in a protein crude extract, they did not show any reaction with proteins from the MMA fraction (Fig. 1A and B). Thus, contaminations of the MMA fraction with proteins of these major subcellular compartments, and probably also of others, should be minor. As a positive control, an antibody against CK1, which is enriched in flagella in comparison to a crude extract (41), was used (Fig. 1C). It showed a strong signal in the MMA fraction. A comparison of proteins from a crude extract and proteins from the MMA fraction by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) corroborated the enrichment of specific proteins (Fig. 1D).
FIG. 1.
Enrichment and purification of flagellar proteins and analysis of phosphoproteins in the flagellar MMA fraction. Totals of 20 μg and 25 μg of proteins from a crude extract (CE) and the flagellar MMA fraction, respectively, were separated by 9% SDS-PAGE along with a molecular mass standard and immunoblotted with specific antibodies against the C1 subunit of CHLAMY1 (A), VIPP1 (B), CK1 (C), phospho-Thr (E), and phospho-Tyr (F) or stained with Coomassie brilliant blue (D).
For the analysis of phosphoproteins in the MMA fraction, we first performed immunoblotting with two different commercially available phospho-amino-acid-specific antibodies. Immunodetection of the proteins in the MMA fraction with a polyclonal anti-phosphothreonine antibody revealed about 18 major and some minor phosphorylated protein bands in the apparent molecular mass range of ≥30 kDa (Fig. 1E). Comparison with an immunoblot of a total crude extract of C. reinhardtii cells underlined the enrichment of flagellar phosphoproteins, which were not abundantly present in the crude extract, based on their molecular mass. Immunodetection with antibodies directed against phosphotyrosine revealed about 11 major and some minor phosphorylated protein bands (Fig. 1F). Again, a significant difference was visible in comparison to the crude extract. These results clearly demonstrate that considerable amounts of phosphoproteins were still present in the MMA fraction after the isolation procedure.
Identification of flagellar phosphopeptides by nLC-ESI-MS.
Proteins from the MMA fraction were reduced, and the cysteine residues were blocked to disrupt disulfide bonds and enhance the accessibility of the proteins for trypsin (see Materials and Methods). Enrichment of phosphopeptides, which are often low in abundance, is an essential initial step prior to their subsequent analysis (19, 36). Removal of the nonphosphorylated peptides increases the selectivity and detection sensitivity for phosphopeptides in MS analysis. For phosphopeptide enrichment, we applied IMAC based on Ga3+ as the metal ion, along with tryptic peptides (see Materials and Methods). Enriched phosphopeptides from four independent flagellum preparations were separated by nLC and analyzed by ESI-MS. Therefore, the mass spectrometer was run in the data-dependent neutral loss mode (see Materials and Methods).
To increase the reliability of the analysis for data assignment, we applied the following criteria for positive phosphopeptide identification. (i) The phosphopeptides and their in vivo phosphorylation sites were identified in at least two independent experiments. All spectra were manually validated and checked for the presence of b- and/or y-type ions representing the phosphorylation sites. In case a phosphorylation site was present on the first amino acid at the N terminus of the phosphopeptide and therefore not present in the b- and y-type ions, these phosphorylation sites were considered only when all other possible phosphorylation sites in the phosphopeptide were validated as phosphorylated or nonphosphorylated. (ii) The phosphopeptides could be identified within the protein sequence of a predicted gene model of the flagellar proteome of C. reinhardtii (33) (see Materials and Methods). The database searches were done with the newly released Proteome Discoverer program (version 1.0; Thermo Electron Corp.). The parameters were adapted to yield an FDR of no more than 1% (see Materials and Methods for details).
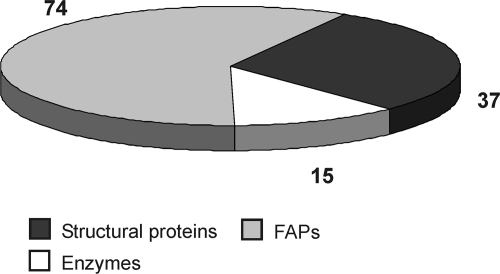
In this way, 141 phosphopeptides were detected, belonging to 32 flagellar phosphoproteins fulfilling the criteria mentioned above (Table 1; see Table S1 in the supplemental material). For the functional classification of identified phosphorylated flagellar proteins, we used information from the genome website of the Joint Genome Institute (JGI) (Vs2 and Vs3), along with the available annotation and information on conserved domains. Moreover, we did NCBI protein BLAST homology searches and BLASTp analysis that was offered by the flagellum database (33) (see Materials and Methods). The 32 identified proteins were functionally divided into three categories (Table 1), including (i) 8 structural proteins of the flagella, (ii) 4 enzymes, and (iii) 20 known flagellum-associated proteins (FAPs).
TABLE 1.
Functional categorization of flagellar phosphoproteinsa
| Category and protein | Abbreviation(s) |
|---|---|
| Structural proteins of the flagella | |
| Outer dynein arm docking complex subunit 1 | ODA3, ODA-DC1 |
| Outer dynein arm docking complex protein 2 | ODA1, ODA-DC2 |
| Inner dynein arm I1 intermediate chain IC138 | BOP5 |
| Tektin, associated with inner arm dynein | |
| Radial spoke protein 17 | |
| Flagellar central pair-associated protein PF6 | |
| Hydin-like proteinb | |
| Intraflagellar transport protein IFT43 | MOT41 |
| Enzymes | |
| cGMP-dependent protein kinase | |
| Mitogen-activated protein kinase 7 | |
| FAP262; Ser/Thr protein kinase domainc | |
| FAP39; plasma membrane calcium-transporting ATPasec | |
| FAPs | |
| FAP254; putative ankyrin-like protein | |
| FAP228; callose synthase-like protein; 1,3-beta-glucan synthase component | |
| FAP230; ankyrin repeats; ion transport protein domain | |
| FAP288; EF hand | |
| FAP190; sterile alpha motif | |
| FAP33d; ankyrin repeats | |
| FAP59d; RecF/RecN/SMC N-terminal domain | |
| FAP116d; microtubule-binding protein MIP-T3 domain | |
| FAP217 | |
| FAP83 | |
| FAP1b | |
| FAP152 | |
| FAP93 | |
| FAP55 | |
| FAP18 | |
| FAP147 | |
| FAP154 | |
| FAP184 | |
| FAP263 | |
| Hypothetical protein (partially overlaps with dynein heavy chain 9) |
The functions of depicted proteins are given as determined by NCBI BLASTp, along with their conserved domains.
Protein was present in the cellular phosphoproteome of C. reinhardtii.
Two FAPs with a conserved kinase and ATPase domain were included in the enzyme category.
Predicted functional domains are present only in the Vs3 model.
Table S1 in the supplemental material summarizes all identified peptides along with their corresponding phosphorylation sites, including their positions within the open reading frame, their protein ID numbers, charges, and Xcorr values, and the biological function (if known) of the corresponding protein models.
Twenty-three phosphoproteins were covered by more than one phosphorylated peptide or by different phosphorylation sites within a given peptide. In nine cases, phosphoproteins were identified by only one peptide with one or more phosphorylation sites. In total, 141 phosphopeptides along with 126 in vivo phosphorylation sites were identified that belonged to 32 flagellar proteins of different functional categories (Fig. 2). Identical peptides with different phosphorylation sites were also counted. Phosphorylation events occurred 89 times on Ser, 29 times on Thr, and 8 times on Tyr residues.
FIG. 2.
Distribution of in vivo phosphorylation sites of flagellar proteins belonging to different functional categories.
Variability of phosphorylation sites.
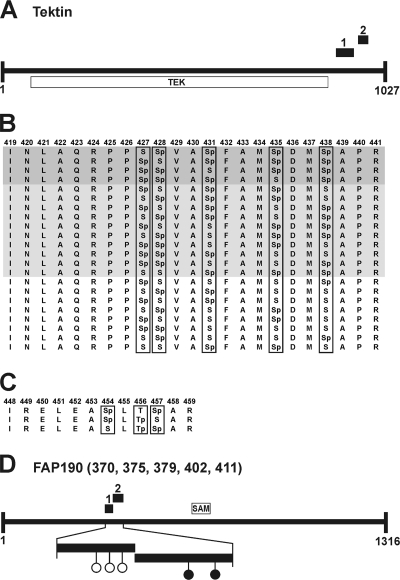
Twenty-six peptides were identified more than once, but with variable phosphorylation sites. Notably, variable phosphorylation sites were sometimes identified in different phosphopeptides that were situated closely together within one protein. A typical example is tektin, which belongs to a family of fibrous proteins that form specialized protofilaments in flagellum microtubules (28). Two phosphopeptides from tektin were identified that cluster at the C terminus outside the tektin domain (Fig. 3A). Phosphopeptide 1, which is 23 amino acids long, was identified with 22 different patterns of phosphorylation sites (Fig. 3B). In four cases, four phosphorylation sites were found within this peptide and varied among Ser-427, Ser-428, Ser-431, Ser-435, and Ser-438. In 10 cases, three phosphorylation sites were found, and in 8 cases, only two phosphorylation sites were present in this peptide, varying again at Ser residues 427, 428, 431, 435, and 438. Phosphopeptide 2 of tektin is 12 amino acids long and located close to phosphopeptide 1 (Fig. 3A and C). It was identified three times, but again with different phosphorylation sites, with positions at Ser-454, Thr-456, and Ser-457. This displays a complex physiological status of reversible phosphorylation within the flagellum and could play an important role in the regulation of the affected protein. The distribution of phosphorylation sites outside conserved domains in combination with the clustering of the sites in small areas of the protein was observed several times in the identified flagellar phosphoproteins. Another typical example is FAP190, with a sterile alpha motif (SAM) domain (Fig. 3D). It was identified with two different phosphopeptides that are located close together. Thereby, a total of five in vivo phosphorylation sites were identified in the N-terminal region of the protein (see Table S1 in the supplemental material).
FIG. 3.
Examples of variances and clustering of phosphorylation sites. Black boxes mark identified phosphopeptides. (A) Predicted domain structure of tektin along with its tektin domain (TEK). (B) Variances of phosphorylation sites in phosphopeptide 1 of tektin. The amino acid positions are mentioned at the top. Dark and light gray backgrounds mark peptides with four and three phosphorylation sites, respectively. No background is used for the peptides with two phosphorylation sites. “p” indicates in vivo phosphorylation sites. (C) Variances of phosphorylation sites in phosphopeptide 2 of tektin. (D) Predicted structure of FAP190 along with its SAM domain. Black circles indicate nonvariable in vivo phosphorylation sites, and open circles indicate variable in vivo phosphorylation sites on Ser residues at different positions (numbers in parentheses) within the phosphopeptides.
Structural flagellar phosphoproteins.
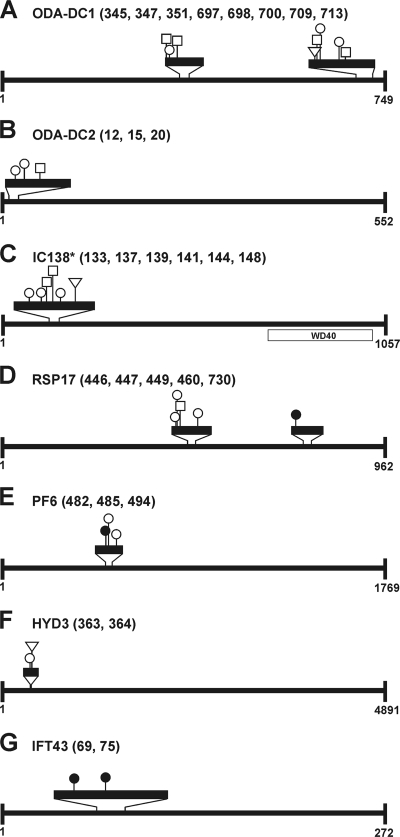
Eight proteins were structurally associated with the flagella, including the already mentioned tektin protein. The outer dynein arm docking complex (ODA-DC), which is microtubule associated and targets the outer dynein arm to its binding site on the flagellar axoneme (48), is composed of three proteins, designated DC1, DC2, and DC3. We could identify ODA-DC1 with two different phosphopeptides and eight in vivo phosphorylation sites and ODA-DC2 with one peptide and three sites of phosphorylation (Fig. 4A and B; Table 1). In both proteins, the phosphopeptides showed a dynamic phosphorylation pattern, with 10 (ODA-DC1) and 3 (ODA-DC2) variants overall, respectively (see Table S1 in the supplemental material). A functionally different group of dyneins that are also associated with the nine doublet microtubules include the inner dynein arm I1. Here we identified the 138-kDa intermediate chain (IC138) of this arm as a phosphoprotein. Six variable phosphorylation sites were found in one peptide of IC138 that is situated at its N terminus (Fig. 4C).
FIG. 4.
In vivo phosphorylation sites in structural flagellar phosphoproteins. Identified phosphopeptides are marked by black boxes in 4-fold (A, B, D, E, and G), 8-fold (C), and 16-fold (F) amplitudes. Circles indicate Ser residues, squares Thr residues, and triangles Tyr residues, which are phosphorylated at positions (numbers in parentheses) within the amino acid sequence of Vs2 protein models. Black circles, squares, or triangles mark nonvariable in vivo phosphorylation sites, and white circles, squares, or triangles mark variable in vivo phosphorylation sites (see Table S1 in the supplemental material). The domain structure was analyzed by NCBI protein BLAST homology searches. *, the depicted protein represents the Vs3 model for IC138.
The regulation of dynein arms involves other structures in the axoneme, such as radial spokes and central apparatuses (35). In our analysis, the radial spoke protein RSP17 was detected with two different phosphopeptides along with four variable and one fixed phosphorylation site, respectively (Fig. 4D). The central pair has an important function in implementation of dynein-induced microtubule sliding into complex ciliary and flagellar wave forms (46). We also identified one phosphopeptide with three phosphorylation sites, two of which are variable, in the PF6 protein of the central pair projection (Fig. 4E).
A large number of human diseases are a result of defects in ciliary assembly (20). Hydin is a component of the C2 projection of the central pair of microtubules. It is required for normal flagellar motility in C. reinhardtii, and defects in hydin cause hydrocephalus in humans (14). We detected hydin with one peptide with two dynamic phosphorylation sites (Fig. 4F).
An important mechanism to maintain the assembly and function of cilia and flagella is the already mentioned IFT (37). IFT43 of the IFT complexes was identified with one peptide and two in vivo phosphorylation sites (Fig. 4G).
Enzymes, including three kinases and an ATPase, are subject to phosphorylation.
Kinases play a very important role in various regulatory processes, including cell motility, gene expression, cell differentiation, cell division, and metabolism. Protein kinases are regulated themselves by different mechanisms, including their own phosphorylation (10). We identified one cGMP-dependent protein kinase with one peptide and three in vivo phosphorylation sites located at its N terminus outside the kinase domain (Fig. 5A). In contrast, a Ser/Thr protein kinase, which was annotated in the JGI database as mitogen-activated kinase 7 (MAK7), was found with one peptide and four variable phosphorylation sites located directly in the catalytic kinase domain (Fig. 5B). FAP262 also has a Ser/Thr protein kinase catalytic domain. In that protein, one phosphorylated peptide along with six variable phosphorylation sites was detected, situated in the Ser/Thr protein kinase catalytic domain (Fig. 5C). Moreover, a plasma membrane calcium-transporting ATPase (FAP39) was found with one phosphopeptide and two distinct in vivo phosphorylation sites (Fig. 5D; Table 1; see Table S1 in the supplemental material).
FIG. 5.
Positions of in vivo phosphorylation sites of flagellar kinases and one ATPase. Identified phosphopeptides are marked by black boxes in fourfold (A and D) and eightfold (B and C) amplitudes. Circles indicate Ser residues, squares Thr residues, and triangles Tyr residues, which are phosphorylated at positions (numbers in parentheses) within the amino acid sequence of Vs2 protein models. Black circles, squares, or triangles mark nonvariable in vivo phosphorylation sites, and open circles, squares, or triangles mark variable in vivo phosphorylation sites. The domain structure was adapted from NCBI protein BLAST homology searches. CAP, cAMP receptor protein effector domain; Ser/Thr Kin, Ser/Thr protein kinase catalytic domain; E1-E2 ATPase, E1-E2 ATPase domain; P-ATP, soluble P-type ATPase domain; A-C, cation-transporting ATPase C-terminal domain.
FAPs.
Another group represents FAPs (20 proteins) that were not functionally characterized in C. reinhardtii or any other species. FAP262, which has a kinase domain, and FAP39, an ATPase (see above), were included in the enzyme category and therefore were not counted for this statistical analysis. FAPs with predicted functional domains (E value cutoff of 1e−05) include, for example, proteins with ankyrin repeats and SAM domains, which can mediate protein-protein interactions (FAP33, FAP190, FAP254, and FAP230), and a protein with a Ca2+-binding domain that belongs to the EF hand superfamily (FAP288). All predicted domains are described in Table 1. Dynamic patterns of phosphorylation sites and clustering of the sites were also observed in this group. Examples of proteins with clustering of at least two phosphopeptides include the already mentioned FAP190 with a SAM domain (Fig. 3D), as well as FAP230. Several examples of proteins with dynamic patterns of phosphorylation sites were found in this category, including the EF hand protein FAP288. It has seven variants of one phosphopeptide showing four differential in vivo phosphorylation sites. Variances in phosphorylation sites were also found in FAP1, FAP18, FAP116, FAP154, FAP147, FAP184, FAP228, FAP230, FAP263, and a hypothetical protein.
DISCUSSION
Reversible protein phosphorylation controls flagellar motility (35), signaling (30), length, and assembly (37, 53) in C. reinhardtii. Several kinases and phosphatases have been found in flagella/cilia, and some appear to be anchored in the axoneme close to the motor docking complex (33, 35, 60). One challenge to understand the control of motility has been to identify the relevant target sites among the known phosphoproteins and to identify unknown phosphoproteins discovered by labeling procedures (34). For this purpose, we analyzed the flagellar phosphoproteome of C. reinhardtii. We identified 141 phosphopeptides which belong to 32 proteins that were already identified in the flagellar proteome of C. reinhardtii (33). In total, 126 in vivo phosphorylation sites were determined. Three additional predicted/hypothetical phosphoproteins with unknown functions were found in the genome of C. reinhardtii and are not listed in the flagellar proteome (data not shown), indicating that the isolated fraction was relatively pure.
With regard to the dynamics of phosphorylation and the low abundance of many phosphoproteins, the enrichment of phosphorylated peptides via IMAC prior to MS analysis was a prerequisite. The activities of specific phosphatases, which could change the phosphorylation pattern during the preparation procedure, were prevented by adding a phosphatase inhibitor cocktail prior to demembranization of isolated flagella (see Materials and Methods). Immunodetection with anti-Thr and anti-Tyr antibodies revealed the presence of ca. 18 phosphoprotein bands with the anti-Thr antibodies and ca. 11 phosphoprotein bands with the anti-Tyr antibodies. In the current analysis, we identified 17 phosphoproteins with at least one phosphorylated Thr and 6 phosphoproteins with at least one phosphorylated Tyr. Each phosphoprotein band may include more than one protein. On the other hand, differentially phosphorylated proteins may run as different bands in SDS-PAGE. Thus, the phosphorylated proteins determined in this study seem in rough agreement with those in immunological assays.
Nevertheless, it is clear that we have not identified a complete set of phosphorylation sites within the flagella. More than 80 axonemal phosphoprotein spots were found by radiolabeling (34). They may include some that belong to the same protein but migrate differently in a two-dimensional gel due to changes in their isoelectric point upon differential phosphorylation. The dynamic phosphorylation pattern found in this study also supports such a possibility. Some phosphoproteins which are known from biochemical analysis, such as outer dynein arm heavy chain alpha (13) and the central pair kinesin KLP1 (61), did not show up in our analysis when we applied an FDR rate of 1% (see Materials and Methods). The currently available software tool by Thermo, named Proteome Discoverer (version 1.0), includes this FDR. With less stringent parameters, we could also detect KLP1 with one phosphopeptide and the outer dynein arm heavy chain alpha with three different phosphopeptides (data not shown). Two of them are located exactly in the predicted areas of phosphorylation (13). However, in these cases, we would have to apply an FDR of >1%, and thus the percentage of false-positive results would also be increased. Thus, we limited the FDR to 1%, taking into account that we may miss some positive results.
Another limitation affecting the yield of phosphopeptides is based on the methodology for phosphopeptide enrichment. Therefore, IMAC, used frequently for this purpose, was applied. It has been reported to enrich more multiple phosphorylated peptides than monophosphopeptides (19). Indeed, we found only 10 monophosphopeptides in our study, in agreement with such a bias, which was strengthened by one of the applied criteria for reliable results, that a phosphopeptide had to be identified in at least two independent experiments. The latter should be compensated to some extent by using four independent flagellum preparations for our analysis.
There has been some discussion about the misidentification of phospho- and sulfopeptides because sulfopeptides also display an 80-Da mass increase (22). There are several reasons that render it unlikely that sulfated sites instead of phosphorylation sites were determined in our study. In contrast to phosphorylated peptides, sulfated peptides show no affinity for IMAC (25) and thus should be present only in minor traces in the analyzed samples. ESI analyses in most cases lead to losses of the sulfo moiety of tyrosine in the interface/skimmer region before acquisition of the full MS (25). Sulfation on serine and threonine is more stable, leading to a precursor ion with a mass difference of 80 Da from the unmodified one. But during collision-induced dissociation, O-sulfopeptides undergo a gas-phase rearrangement reaction that completely eliminates the sulfate, making them indistinguishable from nonmodified molecules (21). Thus, they would not be identified by MS/MS and manual validation of the spectra as phosphopeptides, and they would also not be neutral loss triggered for MS/MS/MS in the setup used in our study, because our parameters are based on a difference of −98 Da, not −80 Da (see Materials and Methods).
In the cellular phosphoproteome analysis (51), several phosphoproteins from the flagella were found that are missing here. Only hydin and FAP1 had also been found before, but with different phosphopeptides. However, for the cellular phosphoproteome analysis, we had grown the cells for 29 h in the presence of a phosphatase inhibitor. In this study, we focused on the physiological state of the cell and its flagella with regard to the time of cell harvesting and the applied growth conditions (see Materials and Methods). This will allow us to compare different physiological conditions and their influence on in vivo phosphorylation sites in future. Only during the purification procedure did we add different phosphatase inhibitors to prevent phosphatases that are active in the flagellar lysate. Nevertheless, we identified 30 novel flagellar phosphoproteins along with their in vivo phosphorylation sites in this study. The increase in positive phosphoprotein yield after enrichment of organelles was also found with the eyespot phosphoproteome (52). It is also in agreement with the immunological assays with anti-phospho-amino-acid antibodies (Fig. 1E and F), where different phosphoprotein patterns were visible for flagellar and crude extract fractions.
Our data indicate that regulation of ciliary motility may include rapid dynamic phosphorylation and dephosphorylation events. The large number of in vivo phosphorylation sites that showed a dynamic pattern presumably reflects such short-term modifications. Thus, the same phosphopeptide was identified in independent scans but had the phosphorylation sites on different residues within a given peptide. Also, in several cases where different phosphopeptides per protein were found, a clustering of these peptides in certain regions of the protein occurred. Altogether, these observations implicate a complex physiology and importance of specific phosphorylation sites for function within the flagella. It also suggests that certain areas of some flagellar proteins represent exposed regions for kinases and protein phosphatases, which may be connected with the exact localization of these proteins within this cell organelle.
Structural elements of the flagella and motor proteins.
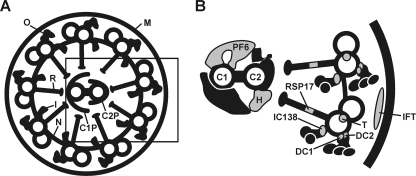
Eukaryotic flagella and cilia are composed of a 9+2 array of microtubules plus more than 250 accessory proteins that form the axoneme. A cross section of a flagellum is shown in Fig. 6A, and the positions of known structural phosphoproteins identified in the present study are depicted in gray (Fig. 6B). One structural part of the axoneme consists of dyneins, which are members of molecular motors. Flagella and cilia contain the following three classes of dyneins: (i) cytoplasmic dynein, (ii) axonemal outer dynein, and (iii) axonemal inner dynein (48). The outer dynein arms are attached to specific sites on the A tubules of the flagellar doublet microtubules and repeat at 24-nm intervals along the length of the doublet (48). This mechanism is realized by a microtubule-associated structure, the ODA-DC, that targets the outer dynein arm to its binding site on the flagellar axoneme. The ODA-DC of C. reinhardtii is composed of three proteins (DC1, DC2, and DC3), and DC1 and DC2 were identified as phosphorylated proteins in our analysis. Phosphorylation of ODA-DC1 and -DC2 was hypothesized before, based on the gel mobility behavior of these proteins (48), but in vivo phosphorylation sites were not known.
FIG. 6.
Components of the flagella and the positions of its phosphoproteins (in gray). A schematic diagram of a cross section of a flagellum from C. reinhardtii (A) and a more detailed view (B), represented by the black box in panel A, are shown. The locations of the components were adapted from several studies (27, 28, 32, 33, 45, 48, 54, 58, 61). FM, flagellar membrane; O, outer dynein arm; I, inner dynein arm; R, radial spoke; N, nexin bridge; C1P, C1 central pair projection; C2P, C2 central pair projection. (B) Identified phosphorylated proteins have been depicted in gray. C1, C1 central pair of microtubules; C2, C2 central pair of microtubules; PF6, PF6 protein; H, hydin; RSP17, radial spoke protein 17; DC1, outer dynein arm docking complex 1; DC2, outer dynein arm docking complex 2; T, tektin; IC138, inner dynein arm intermediate chain protein 138; IFT, intraflagellar transport particle including intraflagellar transport protein 43.
The inner arm dyneins are structurally and functionally more heterogeneous than the outer arm dyneins and are precisely organized in a 96-nm repeat (27, 35). The heterogeneous composition of inner arm dyneins reveals itself by the presence of at least eight different inner arm dynein heavy chains that are organized with various intermediate and light chains into seven distinct complexes (27). One complex is built by a two-headed dynein, called I1, that is composed of two motor units (1α- and 1β-HC), three intermediate chains (IC140, IC138, and IC97), and five light chain subunits (54). In our study, we identified six in vivo phosphorylation sites of IC138 at its N terminus. Phosphorylation of IC138 was shown before by radiolabeling (7). Changes in the phosphorylation pattern of IC138 are involved in the regulation of motility of flagella and adjust the dynein-driven microtubule sliding. Notably, IC138 is hyperphosphorylated in paralyzed flagellar mutants lacking radial spoke and central pair components, suggesting a role of these elements in the regulation of phosphorylation of IC138 (9).
It was suggested that CK1, protein kinase A (PKA), and protein phosphatases 1 and 2A contribute to the phosphorylation pattern of IC138 and have functional roles in regulating the activity of IC138 (54, 59, 60). The presence of six dynamic phosphorylation sites within a peptide of IC138 that is situated at its N terminus (Fig. 4C) may reflect the actions of different kinases and phosphatases. An in silico search of kinases for the six sites (amino acids 133, 137, 139, 141, 144, and 148), using NetPhosK (2) and a threshold of 0.4, predicted several kinases, including CK1 (for S133, S137, and T139) and PKA (for S133). In the case of S133, the prediction for CK1 had the highest rate compared to the other kinases. Clearly, such predictions need to be evaluated with experimental data in the future.
Radial spokes relay signals from the central pair of microtubules to the dynein arms (58). They consist of a head, which interacts with the central pair projection, and a thin stalk that is anchored to the doublet microtubule close to the inner dynein arms. The radial spoke of C. reinhardtii is composed of at least 23 proteins, not all of which have been characterized at the molecular level (58). Some of them have been described as phosphorylated by autoradiograms (34). Among the predicted radial spoke phosphoproteins, we could only identify RSP17, which is located in the spoke stalk as a phosphorylated protein.
The generation of wave forms and the beating of cilia and flagella not only depend on multiple dynein isoforms but also involve structures in the central apparatus, which consists of two single microtubules (C1 and C2) and their associated structures, including central pair projection, central pair bridges, and the central pair cap (46). Current hypotheses assume that central pair structures interact with radial spokes, which in turn transmit regulatory signals to the multiple isoforms of dyneins that are attached to outer doublet microtubules (44). Associated with the central projection is the PF6 protein. It appears to serve as a structural scaffold for the assembly of components associated with 1a projection on the C1 microtubule (38). The C1 microtubule is also associated with protein phosphatase 1c (60), which indicates that an interaction of central pair projections and radial spoke heads could be changed based on the phosphorylation of central pair proteins such as PF6.
Tektin filaments are microtubule-associated proteins that are present in the axoneme as stable filaments. They are supposed to have a function in structural properties of the axoneme (28). In this study, we identified tektin as a phosphoprotein with a dynamic pattern of in vivo phosphorylation sites that cluster at its C terminus. Tektin is localized near the area where the B tubule attaches to the A tubule, close to the binding sites for inner dynein arms and radial spokes (28). Tektin has been hypothesized to be phosphorylated in human sperm, since its migration pattern in two-dimensional gel electrophoresis includes several spots (56). The presence of two tektin phosphopeptides, one of which is present with either two, three, or four variable phosphorylation sites, while the other peptide has two variable phosphorylation sites, is in agreement with the presence of several tektin spots by two-dimensional gel electrophoresis.
Flagella and cilia are composed of more than 600 proteins (33) that are all synthesized in the cytoplasm. These components have to find their way from the cytosol to a defined position in the growing axoneme during the assembly of a cilium. This mechanism is realized by the anterograde and retrograde movements of multimeric protein complexes, called IFT particles (37). These particles consist of two subcomplexes, namely, IFTA, associated with retrograde transport and containing at least 6 protein subunits, and IFTB, associated with anterograde transport and consisting of 11 protein subunits. IFT43, which is proposed to be a subunit of IFTA, was identified in our analysis as a phosphorylated protein.
The functional classification of the identified phosphoproteins includes one molecular component that is related to human disease. Mutations in the hydin gene lead to hydrocephalus (20, 45). Hydin was identified as a phosphorylated protein in our analysis as well as in a previous cellular phosphoproteome analysis (51). It is proposed to interact with several proteins of C1P and with KLP1. This suggests that hydin is required for flagellar motility and is involved in the regulation of dynein arm activity by the central pair and the radial spokes (14).
Proteins in other functional categories.
Protein kinases themselves are regulated by different mechanisms (10). Many kinases consist of additional subunits that may function in response to second messengers such as cAMP or cGMP. Another important mechanism for the regulation of kinases is phosphorylation. We detected one cGMP-dependent protein kinase with one peptide and three in vivo phosphorylation sites. cGMP-dependent kinases are known to have several autophosphorylation sites at the N terminus prior to the cAMP receptor protein (CAP) effector domains (10). Our analysis revealed that the three phosphorylation sites are located in the N terminus of the identified cGMP-dependent protein kinase just in front of the CAP domains (Fig. 5A). The alternation of activation of many kinases can also be regulated by phosphorylation of residues in the activation segment located in the catalytic domain of the kinase (10). An example of such a case is a Ser/Thr protein kinase which was annotated in the JGI database as MAK7. In this protein, the phosphorylation sites are located directly in the catalytic kinase domain (Fig. 5B). It should be noted that MAK7 from C. reinhardtii shows a significantly higher similarity to the human male germ cell-associated kinase, which is present in human cilia, than to MAK7, also present in human cilia. Another example is FAP262, which harbors a Ser/Thr protein kinase domain near its C terminus (Fig. 5C). Again, the phosphorylation sites are situated within the kinase domain. But the location of phosphorylation sites within a functional domain was found only in the two cases of these kinases. In all other cases, they were located outside the conserved domains. The tendency of phosphorylation sites to cluster outside known conserved domains was also found in the eyespot phosphoproteome (52).
Another phosphorylated protein is FAP39. It has a high similarity to a plasma membrane calcium-transporting ATPase. These enzymes are responsible for the removal of excess Ca2+ from the cell in order to maintain the large Ca2+ concentration gradient existing between the cytosol and the extracellular space. Since cilia and flagella can alter their beating patterns through changes in membrane excitation mediated by Ca2+ influx (40), this ATPase could serve as a Ca2+ outward pump to maintain the Ca2+ concentration gradient between the extracellular space and the flagella of C. reinhardtii. As already shown for the PKA-dependent phosphorylation of the plasma membrane Ca2+-ATPase in parotid acinar cells (3), one possible regulatory mechanism could involve phosphorylation.
Uncharacterized FAPs.
In our study, we also identified 20 FAPs. Some FAPs in addition to the above-mentioned FAP262 and FAP39 have predicted domains that provide information about their potential functions. Some have domains involved in protein-protein interaction that can be controlled by phosphorylation. For example, the size of SAM domain protein-containing oligomeric complexes is related to activation of the mitogen-activated protein kinase cascade in yeast (43). Such a scenario could also be envisaged with the identified flagellar SAM protein that is phosphorylated at five clustered sites. An interesting candidate is also FAP288, which has an EF hand and thus may be involved in Ca2+ signaling, which is known to play an important role in signaling in cilia (4, 57).
Some FAPs have no predicted domains. These functional, not yet characterized proteins will also be of specific interest for future analysis because they may represent novel members of signaling cascades within flagella.
Supplementary Material
Acknowledgments
We thank Michael Schroda for giving us anti-VIPP1 antibody and Georg Kreimer for helpful comments on the manuscript. We appreciate the free access to the C. reinhardtii gene models and ESTs provided by the U.S. Department of Energy and by the Kazusa DNA Research Institute, Japan.
Our work was supported by grants from the Deutsche Forschungsgemeinschaft to M.M.
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blom, N., T. Sicheritz-Ponten, R. Gupta, S. Gammeltoft, and S. Brunak. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 41633-1649. [DOI] [PubMed] [Google Scholar]
- 3.Bruce, J. I. E., D. I. Yule, and T. J. Shuttleworth. 2002. Ca2+-dependent protein kinase-A modulation of the plasma membrane Ca2+-ATPase in parotid acinar cells. J. Biol. Chem. 27748172-48181. [DOI] [PubMed] [Google Scholar]
- 4.Dymek, E. E., and E. F. Smith. 2007. A conserved CaM- and radial spoke-associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 179515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng, J., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5976-989. [DOI] [PubMed] [Google Scholar]
- 6.Fliegauf, M., T. Benzing, and H. Omran. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell. Biol. 8880-893. [DOI] [PubMed] [Google Scholar]
- 7.Habermacher, G., and W. S. Sale. 1997. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 136167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, E. H. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 9.Hendrickson, T. W., C. A. Perrone, P. Grifin, K. Wuichet, J. Mueller, P. Yang, M. E. Porter, and W. S. Sale. 2004. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell 155431-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, L. N., M. E. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85149-158. [DOI] [PubMed] [Google Scholar]
- 11.Keller, L. C., E. P. Romijn, I. Zamora, J. R. Yates III, and W. F. Marshall. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 151090-1098. [DOI] [PubMed] [Google Scholar]
- 12.Keller, L. C., S. Geimer, E. Romijn, J. R. Yates III, I. Zamora, and W. F. Marshall. 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 201150-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King, S. M., and G. B. Witman. 1994. Multiple sites of phosphorylation within the alpha heavy chain of Chlamydomonas outer arm dynein. J. Biol. Chem. 2695452-5457. [PubMed] [Google Scholar]
- 14.Lechtreck, K. F., and G. B. Witman. 2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine, R. P., and W. T. Ebersold. 1958. The relation of calcium and magnesium to crossing over in Chlamydomonas reinhardtii. Z. Vererbungsl. 89631-635. [DOI] [PubMed] [Google Scholar]
- 16.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17676-682. [DOI] [PubMed] [Google Scholar]
- 17.Liu, C., F. Willmund, P. Whitelegge, S. Hawat, P. Knapp, M. Lodha, and M. Schroda. 2005. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 161165-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Q., G. Tan, N. Levenkova, T. Li, P. N. Pugh, Jr., J. J. Rux, D. W. Speicher, and E. A. Pierce. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 61299-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann, M., S. E. Ong, M. Grønborg, H. Stehen, O. N. Jensen, and A. Pandey. 2002. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 20261-268. [DOI] [PubMed] [Google Scholar]
- 20.Marshall, W. F. 2008. The cell biological basis of ciliary disease. J. Cell Biol. 18017-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzihradszky, K. F., Z. Darula, E. Perlson, M. Fainzilber, R. J. Chalkley, H. Ball, D. Greenbaum, M. Bogyo, D. R. Tyson, R. A. Bradshaw, and A. L. Burlingame. 2004. O-sulfonation of serine and threonine. Mol. Cell. Proteomics 3429-440. [DOI] [PubMed] [Google Scholar]
- 22.Medzihradszky, K. F., S. Guan, D. A. Maltby, and A. L. Burlingame. 2007. Sulfopeptide fragmentation in electron-capture and electron-transfer dissociation. J. Am. Mass Spectrom. 181617-1624. [DOI] [PubMed] [Google Scholar]
- 23.Merchant, S. S., S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz, G. B. Witman, A. Terry, A. Salamov, L. K. Fritz-Laylin, L. Maréchal-Drouard, W. F. Marshall, L. H. Qu, D. R. Nelson, A. A. Sanderfoot, M. H. Spalding, V. V. Kapitonov, Q. Ren, P. Ferris, E. Lindquist, H. Shapiro, S. M. Lucas, J. Grimwood, J. Schmutz, P. Cardol, H. Cerutti, G. Chanfreau, C. L. Chen, V. Cognat, M. T. Croft, R. Dent, S. Dutcher, E. Fernández, H. Fukuzawa, D. González-Ballester, D. González-Halphen, A. Hallmann, M. Hanikenne, M. Hippler, W. Inwood, K. Jabbari, M. Kalanon, R. Kuras, P. A. Lefebvre, S. D. Lemaire, A. V. Lobanov, M. Lohr, A. Manuell, I. Meier, L. Mets, M. Mittag, T. Mittelmeier, J. V. Moroney, J. Moseley, C. Napoli, A. M. Nedelcu, K. Niyogi, S. V. Novoselov, I. T. Paulsen, G. Pazour, S. Purton, J. P. Ral, D. M. Riaño-Pachón, W. Riekhof, L. Rymarquis, M. Schroda, D. Stern, J. Umen, R. Willows, N. Wilson, S. L. Zimmer, J. Allmer, J. Balk, K. Bisova, C. J. Chen, M. Elias, K. Gendler, C. Hauser, M. R. Lamb, H. Ledford, J. C. Long, J. Minagawa, M. D. Page, J. Pan, W. Pootakham, S. Roje, A. Rose, E. Stahlberg, A. M. Terauchi, P. Yang, S. Ball, C. Bowler, C. L. Dieckmann, V. N. Gladyshev, P. Green, R. Jorgensen, S. Mayfield, B. Mueller-Roeber, S. Rajamani, R. T. Sayre, P. Brokstein, I. Dubchak, D. Goodstein, L. Hornick, Y. W. Huang, J. Jhaveri, Y. Luo, D. Martínez, W. C. Ngau, B. Otillar, A. Poliakov, A. Porter, L. Szajkowski, G. Werner, K. Zhou, I. V. Grigoriev, D. S. Rokhsar, and A. R. Grossman. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mineyuki, Y. 2007. Plant microtubule studies: past and present. J. Plant Res. 12045-51. [DOI] [PubMed] [Google Scholar]
- 25.Monigatti, F., B. Hekkin, and H. Steen. 2006. Protein sulfation analysis—a primer. Biochim. Biophys. Acta 17641904-1913. [DOI] [PubMed] [Google Scholar]
- 26.Neuhoff, V., K. Philipp, H. G. Zimmer, and S. Mesecke. 1979. A simple, versatile, sensitive and volume-independent method for quantitative protein determination, which is independent of other external influences. Hoppe-Seyler's Z. Physiol. Chem. 3601657-1670. [DOI] [PubMed] [Google Scholar]
- 27.Nicastro, D., C. Schwartz, J. Pierson, R. Gaudette, M. E. Porter, and J. R. McIntosh. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313944-948. [DOI] [PubMed] [Google Scholar]
- 28.Nojima, D., R. W. Linck, and E. H. Egelman. 1995. At least one of the protofilaments in flagellar microtubules is not composed of tubulin. Curr. Biol. 5158-167. [DOI] [PubMed] [Google Scholar]
- 29.Ostrowski, L. E., K. Blackburn, K. M. Radde, M. B. Moyer, D. M. Schlatzer, A. Moseley, and R. C. Boucher. 2002. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics 1451-456. [DOI] [PubMed] [Google Scholar]
- 30.Pan, J., and W. J. Snell. 2000. Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 3596-602. [DOI] [PubMed] [Google Scholar]
- 31.Pazour, G. J., and G. B. Witman. 2003. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15105-110. [DOI] [PubMed] [Google Scholar]
- 32.Pazour, G. J. 2004. Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J. Am. Soc. Nephrol. 152528-2536. [DOI] [PubMed] [Google Scholar]
- 33.Pazour, G. J., N. Agrin, J. Leszyk, and G. B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piperno, G., B. Huang, Z. Ramanis, and D. J. Luck. 1981. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 8873-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter, M. E., and W. S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 15137-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinders, J., and A. Sickmann. 2005. State-of-the-art in phosphoproteomics. Proteomics 54052-4061. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbaum, J. L., and G. B. Witman. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3813-825. [DOI] [PubMed] [Google Scholar]
- 38.Rupp, G., E. O'Toole, and M. E. Porter. 2001. The Chlamydomonas pf6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salathe, M. 2007. Regulation of mammalian ciliary beating. Annu. Rev. Physiol. 69401-422. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, J. A., and R. Eckert. 1976. Calcium couples flagellar reversal to photostimulation in Chlamydomonas reinhardtii. Nature 262713-715. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, M., G. Gessner, M. Luff, I. Heiland, V. Wagner, M. Kaminski, S. Geimer, N. Eitzinger, T. Reissenweber, O. Voytsekh, M. Fiedler, M. Mittag, and G. Kreimer. 2006. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 181908-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silflow, C. D., and P. A. Lefebvre. 2001. Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol. 1271500-1507. [PMC free article] [PubMed] [Google Scholar]
- 43.Slaughter, B. D., J. M. Huff, W. Wiegraebe, J. W. Schwartz, and R. Li. 2008. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS ONE 3e1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, E. F. 2002. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskelet. 5233-42. [DOI] [PubMed] [Google Scholar]
- 45.Smith, E. F. 2007. Hydin seek: finding a function in ciliary motility. J. Cell Biol. 176403-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, E. F., and P. A. Lefebvre. 1997. The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil. Cytoskelet. 3811-18. [DOI] [PubMed] [Google Scholar]
- 47.Sopko, R., and B. J. Andrews. 2008. Linking the kinome and phosphorylome—a comprehensive review of approaches to find kinase targets. Mol. Biosyst. 4920-933. [DOI] [PubMed] [Google Scholar]
- 48.Takada, S., C. G. Wilkerson, K. Wakabayashi, R. Kamiya, and G. B. Witman. 2002. The outer dynein arm-docking complex: composition and characterization of a subunit (Oda1) necessary for outer arm assembly. Mol. Biol. Cell 131015-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turkina, M. V., J. Kargul, A. Blanco-Rivero, A. Villarejo, J. Barber, and A. V. Vener. 2006. Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 51412-1425. [DOI] [PubMed] [Google Scholar]
- 50.Turkina, M. V., A. Blanco-Rivero, J. P. Vainonen, A. V. Vener, and A. Villarejo. 2006. CO2 limitation induces specific redox-dependent protein phosphorylation in Chlamydomonas reinhardtii. Proteomics 62693-2704. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, V., G. Gessner, I. Heiland, M. Kaminski, S. Hawat, K. Scheffler, and M. Mittag. 2006. Analysis of the phosphoproteome of Chlamydomonas reinhardtii provides new insights into various cellular pathways. Eukaryot. Cell 5457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, V., K. Ullmann, A. Mollwo, M. Kaminski, M. Mittag, and G. Kreimer. 2008. The phosphoproteome of a Chlamydomonas reinhardtii eyespot fraction includes key proteins of the light signaling pathway. Plant Physiol. 146772-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, N. F., and P. A. Lefebvre. 2004. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 31307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirschell, M., T. Hendrickson, and W. S. Sale. 2007. Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil. Cytoskelet. 64569-579. [DOI] [PubMed] [Google Scholar]
- 55.Witman, G. B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134280-290. [DOI] [PubMed] [Google Scholar]
- 56.Wolkowicz, M. J., S. Naaby-Hansen, A. R. Gamble, P. P. Reddi, C. J. Flickinger, and J. C. Herr. 2002. Tektin B1 demonstrates flagellar localization in human sperm. Biol. Reprod. 66241-250. [DOI] [PubMed] [Google Scholar]
- 57.Wood, C. D., A. Darszon, and M. Whitaker. 2003. Speract induces calcium oscillations in the sperm tail. J. Cell Biol. 16189-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, P., D. R. Diener, C. Yang, T. Kohno, G. J. Pazour, J. M. Dienes, N. S. Agrin, S. M. King, W. S. Sale, R. Kamiya, J. L. Rosenbaum, and G. B. Witman. 2005. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 1191165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, P., and W. S. Sale. 2000. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J. Biol. Chem. 27518905-18912. [DOI] [PubMed] [Google Scholar]
- 60.Yang, P., L. Fox, R. J. Colbran, and W. S. Sale. 2000. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J. Cell Sci. 11391-102. [DOI] [PubMed] [Google Scholar]
- 61.Yokoyama, R., E. O'Toole, S. Ghosh, and D. R. Mitchell. 2004. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 10117398-17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., A. Wolf-Yadlin, P. L. Ross, D. J. Pappin, J. Rush, D. A. Lauffenburger, and F. M. White. 2005. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 41240-1250. [DOI] [PubMed] [Google Scholar]
- 63.Zhao, B., C. Schneid, D. Iliev, E.-M. Schmidt, V. Wagner, F. Wollnik, and M. Mittag. 2004. The circadian RNA-binding protein CHLAMY1 represents a novel type heteromer of RNA recognition motif and lysine homology-containing subunits. Eukaryot. Cell 3815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.