Abstract
Rbg1 is a previously uncharacterized protein of Saccharomyces cerevisiae belonging to the Obg/CgtA subfamily of GTP-binding proteins whose members are involved in ribosome function in both prokaryotes and eukaryotes. We show here that Rbg1 specifically associates with translating ribosomes. In addition, in this study proteins were identified that interact with Rbg1 by yeast two-hybrid screening and include Tma46, Ygr250c, Yap1, and Gir2. Gir2 contains a GI (Gcn2 and Impact) domain similar to that of Gcn2, an essential factor of the general amino acid control pathway required for overcoming amino acid shortage. Interestingly, we found that Gir2, like Gcn2, interacts with Gcn1 through its GI domain, and overexpression of Gir2, under conditions mimicking amino acid starvation, resulted in inhibition of growth that could be reversed by Gcn2 co-overexpression. Moreover, we found that Gir2 also cofractionated with polyribosomes, and this fractionation pattern was partially dependent on the presence of Gcn1. Based on these findings, we conclude that Rbg1 and its interacting partner Gir2 associate with ribosomes, and their possible biological roles are discussed.
The Obg/CgtA subfamily is a conserved group of monomeric GTP-binding proteins found in the genomes of all organisms sequenced thus far. The evolutionary relationships of the GTPase superfamily suggested a role in ribosome function (6, 38). This prediction appears accurate, since all mitochondrial (8), nuclear (12, 28, 31), and prokaryotic (29, 39, 51, 59-61, 70) Obg/CgtA proteins examined thus far are associated with ribosomes. Moreover, these proteins are also involved in the assembly of the large ribosomal subunit (28, 29, 31, 37, 51, 55). Recently, it has become clear that, in addition to a role in the late assembly of the large ribosomal subunit, the bacterial Obg/CgtA proteins are also directly involved in stress response (30, 48, 58).
In Escherichia coli, amino acid starvation leads to uncharged tRNAs binding to the ribosomal acceptor site (A-site) in a codon-dependent manner (18) which is detected by RelA, a (p)ppGpp synthetase. The increase in (p)ppGpp levels leads to the “stringent response” that provides the cell with the regulatory means to control gene expression and thereby cope with starvation. The levels of (p)ppGpp are kept low in nutrient-rich media by SpoT, a bifunctional enzyme related to RelA that has both (p)ppGpp synthetase and hydrolase activity (19, 20, 71). The hydrolase activity of SpoT is inhibited under nutrient-limiting conditions, allowing intracellular (p)ppGpp levels to increase during the stringent response, as RelA produces (p)ppGpp. In E. coli and Vibrio cholerae, the GTP-binding protein CgtA (also called YhbZ or Obg) interacts with SpoT (48, 70) on the large ribosomal particle (30). Depletion of CgtA results in an increase in (p)ppGpp levels (30, 48), raising the possibility that during exponential growth CgtA directly inhibits the hydrolase activity of SpoT.
It has been proposed that, in Saccharomyces cerevisiae, amino acid starvation also leads to uncharged tRNAs binding to the A-site but that the output is not RelA synthesis of (p)ppGpp (21). The effector protein Gcn1 detects the uncharged tRNA and relays the A-site occupancy information to the protein kinase Gcn2. Gcn2 then phosphorylates the translation initiation factor 2α (eIF-2α), leading to reduced global protein synthesis and increased expression of amino acid biosynthetic enzymes (21). The signal transduction pathway governing Gcn2 is called general amino acid control. Like RelA, Gcn2 and Gcn1 bind to ribosomes, and previous findings support the idea that Gcn1 affects A-site function (53).
In S. cerevisiae there are four Obg/CgtA subfamily members that also appear to play roles in ribosome function: Nog1 (Ypl093w), Mtg2 (Yhr168w), Yal036c, and Ygr173w. Nog1 is a nucleolar protein that plays a key role in assembly of the large ribosomal subunit; depletion of Nog1 leads to a decrease in 60S subunit assembly and formation of halfmer polysomes (12, 28, 31). Mtg2 associates with the large mitochondrial ribosomal subunit, is critical for mitochondrial translation, and is required for the maintenance of proper ribosomal subunit ratios (8).
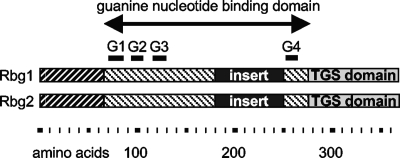
The two remaining S. cerevisiae Obg/CgtA proteins, Yal036c and Ygr173w (hereafter called Rbg1 and Rbg2 for ribosome binding GTPase), belong to the DRG subgroup of Obg/CgtA proteins. Rbg1 and Rbg2 are tripartite proteins that are 52% similar to each other (Fig. 1) and ubiquitously found in all eukaryotes and archaea sequenced to date. These proteins are similar to the other Obg/CgtA proteins only in the guanine nucleotide-binding domain (amino acids [aa] 69 to 275 and aa 67 to 275 for Rbg1 and Rbg2, respectively). Within this conserved GTPase domain, the Rbg proteins also have a 67-aa insertion of unknown function between the conserved G3 and G4 motifs (Fig. 1). This insertion sequence is unique to the eukaryotic and archaeal DRG subfamily (as determined by PSI BLAST searches).
FIG. 1.
Rbg1 and Rbg2 are tripartite proteins, here illustrated as a diagram. Shaded regions represent regions of high similarity. The universally conserved G1 through G4 motifs of the guanine nucleotide binding domain are indicated by overlines. The C-terminal TGS domain is indicated. An amino acid reference scale is provided.
The N and C termini of the Rbg proteins are distinct from those of the nucleolar, mitochondrial, and bacterial Obg/CgtA proteins. The N-terminal amino acids of the Rbg proteins (aa 1 to 68 and aa 1 to 66 for Rbg1 and Rbg2, respectively), predicted to contain two adjacent helices of unknown function, are strongly conserved (45% identical). The C terminus of the Rbg proteins (aa 290 to 368) contains a TGS domain (69), a sequence of ∼50 aa that forms a β-sheet structure (32; Fig. 1). The function of the TGS domain is currently unknown, although a regulatory role has been suggested (3, 69). TGS domains are found in a limited number of proteins including threonyl-tRNA synthetases, DRG-like GTPases and, interestingly, in the (p)ppGpp synthetases SpoT and RelA.
Several lines of evidence suggest that the cytoplasmic Rbg proteins are involved in ribosome function. First, the expression pattern RBG1 displays under various conditions clusters with genes involved in ribosome and rRNA biosynthesis (67). In addition, the RBG1 promoter has a conserved element found in many genes involved in ribosome function (67). Moreover, Rbg1, as well as other proteins involved in translation initiation, was copurified in a complex with eIF4G1-TAP (14, 15). The related gene, RBG2, is synthetically sick in combination with a deletion of the large ribosomal subunit genes rpl22a or rpl6b (N. J. Krogan, unpublished data).
We show here that Rbg1 associates with polyribosomes but not with the 40S or 60S subunits or with 80S monosomes, indicating that Rbg1 specifically associates with translating ribosomes. Interacting partners of Rbg1 were identified by using a yeast two-hybrid screen. Among the interaction partners was a protein of unknown function, Gir2, which has sequence similarity to the N-terminal GI (Gcn2 and Impact) domain of Gcn2 that is involved in Gcn1 binding. We found that Gir2 also associates with ribosomes, and we have several lines of evidence showing that Gir2 binds to Gcn1 via its GI domain. Gir2 overexpression diminishes Gcn2 function, and this could be reverted by Gcn2 overexpression, suggesting that Gir2 competes with Gcn2 for Gcn1 binding. The polyribosome association of Gir2 was not dependent on Rbg1, although its association with polysomes was in part dependent on Gcn1. Based on the connection between Rbg1 and Gir2 and between Gir2 and the components of the general amino acid control pathway, Rbg1 and Gir2 may play a role in adjusting the cell to stress conditions.
MATERIALS AND METHODS
Construction of plasmids.
Plasmids used for the present study are listed in Table 1, and the oligonucleotides used in constructing plasmids for the present study are presented in Table 2. pJM1336, a pAS2 bait vector expressing a translational fusion between the GAL4 DNA-binding domain (GAL4-BD) and Rbg1, was constructed as follows. The RBG1 gene was PCR amplified from yeast genomic DNA with primers Fun11*NcoI and 11up. The product was digested with NcoI/SacI and ligated into similarly digested pET28a (Novagen) to create pJM957. An NcoI/SalI RBG1 fragment from pJM957 was then ligated to NcoI/SalI-digested pAS2 to create pJM1336. Sequences encoding full-length GIR2, its N terminus (aa 1 to 170) or C terminus (aa 167 to 265), or the N-terminal GI domain of GCN2 (aa 1 to 125) were amplified by PCR from yeast genomic DNA by using the oligonucleotides YDR152Up/YDR152Down, YDR152Up/YDR152wN, GIR2C/YDR152Down, or GCN2 FORWARD/GCN2 N, respectively. These PCR fragments were cloned into pAS2 by using NdeI and BamHI (restriction sites were designed in the oligonucleotides), creating translational fusions with the GAL4-BD; the resulting plasmids were pAS2-GIR2 (pJM1617), pAS2-GIR2N (pJM2648), pAS2-GIR2C (pJM4550), and pAS2-GCN2 (GI) (pJM2651). Sequences encoding the GI binding domain (aa 2047 to 2383) of GCN1 were amplified by PCR from yeast genomic DNA using the oligonucleotides GCN1 (GCN2 BD) forward and GCN1(GCN2 BD) reverse and cloned into pACT2 using NcoI and BamHI (restriction sites were designed in the oligos), resulting in pACT2-GCN1(GIB) (pJM2646).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pACT2 | GAL4 activation domain; LEU2 | Clontech |
| pAS2 | GAL4 binding domain; TRP1 | Clontech |
| pLAM5′ | Human lamin C fused to the GAL4-BD | Clontech |
| p426GAL1 | 2μ GAL1 URA3 | 45 |
| pES128-9-1 | 2μ GAL1 GST URA3 | 53 |
| pRS423 | 2μ HIS3 | 7 |
| pJM1336 | pAS2-RBG1 | This study |
| pJM1617 | pAS2-GIR2 | This study |
| pJM2648 | pAS2-GIR2N | This study |
| pJM4550 | pAS2-GIR2C | This study |
| pJM2651 | pAS2-GCN2(GI) | This study |
| pJM2646 | pACT2-GCN1(GIB) | This study |
| pJM3958 | pES128-9-1-GIR2 | This study |
| pJM2653 | p426GAL1-GIR2N | This study |
| pJM4563 | p426GAL1-GIR2C | This study |
| pJM2621 | p426GAL1-GIR2 | This study |
| pJM2749 | p426GAL1-GCN2(GI) | This study |
| pAH15 | Yep13-GCN2 | 21 |
| p180 | GCN4-lacZ reporter plasmid | 44 |
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| Fun11*NcoI | GATTGAGCACCATGGCTACTACA |
| 11up | CCTATGATTGCGCACTT |
| Fun11PM1 | CGGGATCCTTAGCCCTTTGGTTTAGT |
| 173*Nco | GCACTACACCATGGGTATTATCG |
| 173down | CGTTAGAGTAGGTTAGC |
| YDR152R1 | ATGCAGAAGAAAATACTTAAATACTTAAGTACACAAGTATGAATTCGAGCTCGTTTAAAC |
| YDR152F2 | GACGTAGCCAAGGGACTTGCCAAGACCGAAATAGCAAATCAACGGATCCCCGGGTTAATTAA |
| YDR152Up | GGATCATATGGATTATAAGGAAGA |
| YDR152wN | GGATCCTTACTGCTTCTCGAGC |
| GIR2C | CATATGCTCGAGAAGCAGTAC |
| Fun11 KO start | GCAGTAGGTGCAAGCGTAGAGTTGTTGATTGAGCAAAATGAGATTGTACTGAGTGCAC |
| Fun11 KO stop | GTTCGAGAATCACTTTTTCAAGATGGTAACAACATCTTCGCTGTGCGGTATTTCACAGCG |
| YDR152Down | TAGGATCCATGCAGAAGAAAATA |
| GCN1(GCN2 BD) forward | CCATGGCGGAAGTTGCTGGCTC |
| GCN1(GCN2 BD) reverse | GGATCCCTACATTTCTTCCTCA |
| GCN2 FORWARD | AAGAATATATACTCC |
| GCN2 N | GGATCCTTACAGTTTTTCTTGA |
| Gir2Xba1UP | ACTTCTAGATACTATGGATTATAA |
| Gir2Sal1DOWN | TTAAGTCGACAAGTATTTATTGAT |
The pAS2-GIR2, pAS2-GIR2N, pAS2-GIR2C, and pAS2-GCN2(GI) plasmids were digested with NheI and BamHI, and the fragments were ligated to SpeI/BamHI-digested p426GAL1 (45), placing the genes under the control of the galactose-inducible GAL1 promoter, thus creating p426GAL1-GIR2 (pJM2621), p426GAL1-GIR2N (pJM2653), p426GAL1-GIR2C (pJM4563), and p426GAL1-GCN2(GI) (pJM2749).
Full-length GIR2 was cloned as a glutathione S-transferase (GST) fusion protein to construct pJM3958 as follows. GIR2 was PCR amplified from yeast genomic DNA by using the primers Gir2Xba1UP and Gir2Sal1DOWN and cloned as a XbaI/SalI fragment into pES128-9-1. The expression of these genes was verified by immunoblotting (data not shown).
Yeast strains.
Yeast strains used for the present study are listed in Table 3. A Gir2 HA chromosomal fusion was generated as previously described (40). Briefly, the hemagglutinin (HA) tag, along with a selectable marker (His3MX6), was PCR amplified (using pFA6-3HA-His3MX6) by using the primers YDR152R1 and YDR152F2 (Table 2), and the PCR product was transformed into the yeast strains BY4704 and BY4705 to generate Gir2-HA (JM3185 and JM3186). JM3370 is a sporulation product from a diploid created by mating the rbg1 deletion strain (370) with JM3186 (GIR2::3HA::HIS3). JM3372 is a sporulation product from a mating between the gcn1 deletion strain (i.e., strain 14562) with JM3185 (GIR2::3HA::HIS3) and sporulating the resulting diploid to obtain haploid cells. The rbg1 rbg2 mutant contains the rbg2::KanMX4 allele from strain 14803 and an rbg1::HIS3 mutant generated by integration of a PCR fragment generated by amplifying HIS3 from pRS423 with oligonucleotides Fun11 KO start and Fun11 KO stop.
TABLE 3.
Strains examined in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| Y190 | MATagal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ LYS2::GAL1-HIS3 cyhR | 17 |
| Y187 | MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ | 17 |
| YW5-1B | MATatrp1 ura3-52 leu2-3,112 | 63 |
| W303 | MATa/MATα trp1/trp1 leu2-3 112/leu2-3 112 his3-11 15/his3-11,15 ura3/ura3 can1-100/can1-100 ade2-1/ade2-1 | 62 |
| BY4704 | MATaade2Δ::hisG his3Δ200 leu2Δ0 met10Δ0 trp1Δ63 ura3Δ0 | ATCC |
| BY4705 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 met10Δ0 trp1Δ63 ura3Δ0 | ATCC |
| CRY1 | MATaura3-52 trp1 leu2-3,112 his3-11 ade2-1 can1-100 | 5 |
| H1511 | MATα ura3-52 trp1-63 leu2-3,112 GAL2+ | 11 |
| H2557 | MATα ura3-52 trp1-63 leu2-3,112 gcn2Δ GAL2+ | 11 |
| BY4741 | MATahis3 leu2 met15 ura3 | Yeast deletion collection |
| BY4742 | MATα his3 leu2 lys2 ura3 | Yeast deletion collection |
| 370 | Same as BY4741 except rbg1::KanMX4 | Yeast deletion collection |
| 14803 | Same as BY4742 except ygr173w::KanMX4 | Yeast deletion collection |
| 14562 | Same as BY4742 except gcn1::KanMX4 | Yeast deletion collection |
| 1350 | Same as BY4742 except gir2::KanMX4 | Yeast deletion collection |
| JM2064 | rbg1::HIS3 rbg2::KanMX4 | This study |
| JM3185 | MATaGIR2::3HA::HIS3 | This study |
| JM3186 | MATα GIR2::3HA::HIS3 | This study |
| JM3370 | rbg1::KanMX4 GIR2::3HA::HIS3 | This study |
| JM3372 | gcn1::KanMX4 GIR2::3HA::HIS3 | This study |
Yeast polysome analysis.
Cells grown in 200 ml of yeast extract-peptone-adenine-dextrose (YPAD) medium at 30°C to an optical density at 600 nm (OD600) of 0.5 to 1.0 were transferred to a 250-ml centrifuge tube, to which 1 ml of a 10-mg/ml concentration of cycloheximide and ice (to fill the tube) were added. Cells were incubated on ice with periodic swirling for 10 min, harvested at 7,000 rpm for 5 min in an SLA 1500 rotor, and washed with 20 ml of ice-cold lysis buffer (LBS; 40 mM Tris-HCl [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 50 μg of cycloheximide/ml). The cell pellet was resuspended in 2.5 ml of LBS and transferred to a 15-ml Corex tube. Then, 1 ml of glass beads was added, and the cell suspension was vortexed six times for 30 s with 30-s rests on ice between bursts. An additional 2.5 ml of LBS was added, and the cell suspension was centrifuged at 6,500 rpm in a SA600 rotor to remove any unbroken cells. The resulting supernatant was further clarified at 9,200 rpm for 10 min. Where indicated, the 9,200 rpm supernatant was incubated on ice for 30 min in the presence of 5 U/μl of micrococcal nuclease and 3 mM CaCl2 before loading onto sucrose gradients. We loaded 10 OD260 units of cell extract in LBS and 1 mM dithiothreitol onto 10-ml 7 to 47% sucrose gradients that were centrifuged at 28,000 rpm for 4 h in an SW41 Ti rotor. Then, 500-μl fractions were collected manually using an ISCO UV monitor with a 254-nm filter. Samples were concentrated, and the sucrose was removed by the addition of water to 1 ml, followed by incubation on ice with 30 μl of 1% deoxycholate for 10 min. Next, 150 μl of 100% trichloroacetic acid was added, and the samples were incubated on ice overnight. Samples were centrifuged at 13,000 rpm for 10 min, vacuum dried, and resuspended in 1 M Tris and 6× sodium dodecyl sulfate (SDS) sample loading buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and subsequently transferred to nitrocellulose membrane. Immunoblot analysis was performed as described below.
Detection of protein-protein interaction by yeast two-hybrid analysis.
pJM1336 was transformed into the yeast strain Y190, and the resulting strain was transformed with a yeast genomic library cloned into pGAD-C1 (27). Approximately 200,000 transformants were selected on SD plates lacking histidine, leucine, and tryptophan (SD−His−Leu−Trp) supplemented with 60 mM 3-aminotriazole (3-AT). Each transformant was screened for lacZ activity by an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) colony filter lift (4). Transformants that resulted in expression (above background levels) from both the HIS3 and the lacZ reporter genes were further assayed for lack of reporter gene activation (i) in the absence of pJM1336, (ii) in the presence of pAS2 alone, and (iii) in the presence of pLAM5′ (human lamin C protein fused to the GAL4-BD). Transformants that met all three criteria were mated to a pJM1336 containing Y187 strain to reconfirm interaction, and their library clones were sequenced to identify the genes encoded.
All pAS2 clones were transformed into Y190, pACT2-GCN1(GIB) was transformed into Y187, the strains were mated, and the resulting diploids were assayed for HIS3 reporter gene activation by monitoring the growth of serial dilutions of liquid cultures on SD−His−Leu−Trp plates supplemented with 60 mM 3-AT. Expression of all fusion proteins was verified by immunoblot analysis as described below.
GST pulldown assays.
To prepare extracts from yeast cells expressing GST (pES128-9-1) or GST-Gir2 (pJM3958) under a galactose-inducible promoter, cells were grown to mid log phase in SD lacking uracil (SD-Ura) with 4% raffinose, galactose was added to a final concentration of 2%, and the cultures were further incubated at 30°C for 4 h. Cells were harvested, washed once, and resuspended in HEPES buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 25 mM β-glycerophosphate, 0.1 mM Na3VO4, and 1 mM dithiothreitol containing the protease inhibitors 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupepstatin/ml, 2 μg of aprotinin/ml, 1.25 mM benzamidine, and 0.5 μg of pepstatin A/ml). Cells were lysed in the presence of glass beads, and Triton X-100 was added to a final concentration of 1%. The lysates were centrifuged at 20,800 × g for 15 min to remove cell debris and, if ribosomes were to be removed, the resulting supernatants were further clarified by centrifugation at 100,000 × g for 10 min. Then, 125 μg of RNase A was added for 30 min at 4°C to samples in which RNA species were digested. A total of 2 mg of cell lysate was mixed with 20 μl of a 75% slurry of glutathione-Sepharose beads, followed by incubation at 4°C for 2 h with constant agitation. The glutathione beads were collected and then washed twice with HEPES buffer plus 1% Triton X-100, and bound proteins were eluted by boiling for 5 min in the presence of Laemmli SDS-PAGE loading buffer. Samples were subjected to SDS-PAGE, and proteins were detected by immunoblotting as described below.
Assaying β-galactosidase reporter gene activity.
Yeast strains were transformed with plasmid p180 (44) harboring the GCN4-lacZ reporter gene. For each plasmid, two independent transformants were grown to saturation and diluted to an OD600 of ∼0.2 in two identical cultures. After 2 h of growth, one set of cultures was supplemented with 0.5 μg of Sulfometuron/ml to induce starvation for branched amino acids. Unstarved cultures were grown for a total of 6 h, and starved cultures were grown in the presence of Sulfometuron for 6 h. Enzyme assays were performed as previously described (43).
Overexpression of GIR2 under derepressing conditions.
p426GAL1-GIR2 (pJM2621), p426GAL1-GIR2N (pJM2653), p426GAL1-GIR2C (pJM4563), and p426GAL1-GCN2(GI) (pJM2749), as well as the p426GAL1 vector itself, were transformed into yeast strain YW5-1B or YW5-1B containing pAH15 for expression of Gcn2 (22). Transformants were grown in SD−Ura−His liquid medium at 30°C to an OD600 of 1. Serial dilutions of the cultures were spotted onto SD-Ura or SD-Ura-His plates supplemented with 10 mM 3-AT in the presence of 2% glucose or 10% galactose. The plates were incubated at 30°C for 2 to 5 days.
Analysis of in vivo eIF-2α phosphorylation levels in cells overexpressing Gir2.
Yeast strains (H1511) expressing GST (pES128-9-1) or GST-Gir2 (pJM3958) and isogenic gcn2Δ strain H2556 expressing GST (pES128-9-2) were grown to exponential phase in medium containing 2% galactose, subjected to starvation, and analyzed for eIF-2α phosphorylation as described previously (52).
Immunoblot analysis.
Rbg1 antibody (Sigma Genosys) was raised in rabbits against a truncated Rbg1 lacking the putative amphipathic helices (aa 1 to 302). Rbg1 was detected by using purified antibody, which had been treated with acetone powders derived from ATCC 4000370 at a dilution of 1:250 or 1:500. Antibody against Rbg2 (Sigma Genosys) was raised in rabbits against full-length Rbg2, affinity purified, and used at a dilution of 1:500. The specificity of these antibodies was verified in Western blots using wild-type and RBG1 and RBG2 deletion strains (data not shown). Gir2-HA fusions were detected using monoclonal anti-HA antibody (Covance) at a 1:1,000 dilution. The S2 antibody was a gift from the laboratory of Jonathon R. Warner and was used at a 1:2,000 dilution (66). Antibodies to eIF-2α and Gcn1 were a gift from the laboratory of Alan G. Hinnebusch and were used at dilutions of 1:2,000 (9) or 1:1,000 (65), respectively. eIF-2α phosphorylated on Ser-51 was detected by using antibodies from BioSource International, Inc., at 1:5,000. All antibody reactions were performed in the presence of 5% nonfat dried milk as a blocking agent. Immunoblots were visualized using an enhanced chemiluminescence detection system (Amersham Biosciences) as directed by the manufacturer.
RESULTS
Rbg1 cofractionates with polyribosomes.
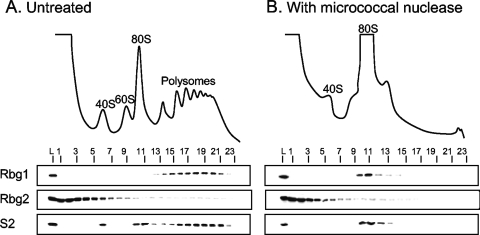
All of the Obg/CgtA proteins examined thus far associate with the large ribosomal subunit (8, 12, 31, 39, 51, 59, 60, 70). To test whether this was also the case for Rbg1 and Rbg2, cell lysates were subjected to sucrose density gradient centrifugation and fractionated, and the cosedimentation of Rbg1 and Rbg2 with ribosomes was determined by immunoblotting. Antibodies to the small ribosomal subunit protein S2 were used as reference to monitor the migration and/or fractionation of the 40S ribosomal subunit, the 80S monosomes, and the polyribosomes. Rbg1 was found exclusively in the polyribosome fractions (Fig. 2A, fractions 11 to 20), a localization pattern distinctly different from that of the nucleolar, mitochondrial, and bacterial Obg/CgtA proteins. The polysome association detected here for Rbg1 would be consistent, however, with a cytoplasmic localization for the Rbg1 protein. The association between Rbg2 and ribosomes is unclear since Rbg2 fractionated throughout the gradient, with the majority of Rbg2 being in the lighter fractions. To confirm that the observed migration of Rbg1 was due to an association with polyribosomes and not due to association with another large protein complex, migration on sucrose gradients was examined after disruption of the polyribosomes with micrococcal nuclease. The addition of micrococcal nuclease to cell lysates results in cleavage of the mRNA between translating ribosomes which, in turn, results in a significant reduction in polyribosomes and a concomitant increase in 80S monosomes (Fig. 2B). Under these conditions, Rbg1 specifically cofractionated with the 80S monosomes (Fig. 2B, fractions 10 to 14). After mRNA digestion the distribution of Rbg2 in the polyribosome fractions, however, was unchanged. We conclude that Rbg1 interacts specifically with translating ribosomes and that this interaction is stable after digestion of the intervening mRNA. Rbg2, in contrast, does not appear to be associated specifically with ribosomes under the conditions assayed (Fig. 2).
FIG. 2.
Rbg1 associates with polyribosomes. (A) Whole-cell extracts from strain W303 were resolved by density sedimentation in 7 to 47% sucrose gradients. (B) Polyribosomes were disrupted by the addition of micrococcal nuclease (5 U/μl) prior to loading of extracts on gradients. The UV absorbance trace (254 nm) obtained during fractionation is shown with the positions of the 40S, 60S, 80S, and polyribosomes indicated. Fractions (numbered) were analyzed by immunoblotting with antibodies against Rbg1, Rbg2 and S2, a small ribosomal subunit protein, as indicated. L, 1/100 cell extract loaded on gradient.
It is not known whether Rbg2 and Rbg1 are functionally redundant, since no phenotypes have been assigned to either mutant (Saccharomyces Genome Database and data not shown). Furthermore, we did not observe any obvious phenotypes when working with either the single or double deletion mutants (e.g., growth rate), suggesting that the protein synthesis rate is not severely affected. When scoring for sensitivity to some antibiotics we found that hygromycin B sensitivity was unchanged in rbg1Δ, rbg2Δ, and rbg1Δrbg2Δ strains. The sensitivity among these single- and double-deletion strains toward cycloheximide and anisomycin was similar and only marginally higher than that of wild type (data not shown). This finding may support the idea that the corresponding proteins have a function on the ribosome.
Identification of Rbg1 interacting partners by yeast two-hybrid screening.
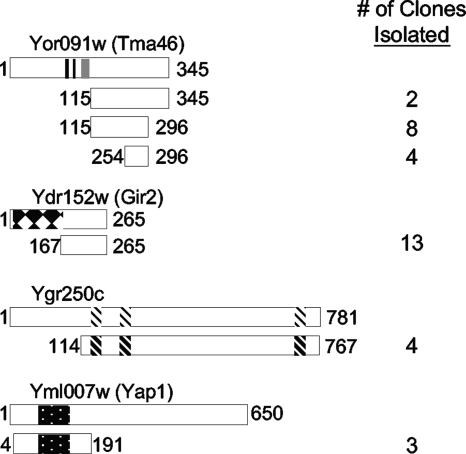
In order to further investigate the cellular function of Rbg1, we attempted to identify potential interacting partners of Rbg1 through a yeast two-hybrid screen using Rbg1 fused to the GAL4-BD as the bait protein. Approximately 200,000 Gal4-activating clones (GAL4-AD) were screened from a random library, and three distinct gene products—Tma46 (Yor091w), Ygr250c, and Gir2 (Ydr152w)—were identified a minimum of four times each as interacting partners of Rbg1 (Fig. 3). Furthermore, Yap1 (Yml007w) was identified three times. Yap1 is a basic leucine zipper transcription factor that is essential for the oxidative stress tolerance of cells (16, 35, 36). Amino acids 4 to 191 containing the leucine zipper domain were sufficient for Rbg1 binding, suggesting that this domain may be involved in the interaction with Rbg1 (Fig. 3).
FIG. 3.
Interacting partners of Rbg1. A yeast two-hybrid screen identified Tma46, Gir2, Ygr250c, and Yap1 as potential interacting partners of Rbg1. The encoded protein fragments are depicted under a diagram of each full-length protein. Beginning and ending amino acids are indicated, as well as the number of clones for each fragment. Tma46 encodes a potential nuclear localization sequence and a zinc finger domain as indicated by the black and gray bars, respectively. Gir2 contains an N-terminal GI domain (black and white diamonds), as discussed in the text. Ygr250c possesses three potential RNA-binding motifs (diagonals), where indicated. The well-characterized Yap1 contains a basic-leucine zipper domain (black with dots).
In contrast to Yap1, the biological role of the other identified Rbg1 interaction proteins is unclear. Four identical YGR250c clones encoding the majority of the protein (aa 114 to 767) were isolated. Interaction between Rbg1 and Ygr250c was also reported in a genomewide two-hybrid study (26). Ygr250c is a cytoplasmic protein (23) that possesses three RNA-binding motifs. The function of Ygr250c is unknown and deletion of YGR250c resulted in no detectable phenotype (50; data not shown).
We obtained 14 overlapping fragments of Tma46 (Fig. 3), a protein of unknown function that was previously reported to be in a complex with Rbg1 (10, 33). The smallest Rbg1-interaction domain of Tma46 consisted of 42 aa (i.e., aa 254 to 296), suggesting that this portion contains the Rbg1 binding site. Tma46 has a putative zinc finger (aa 88 to 113) and has been identified as a protein associated with the nuclear pore complex (49, 64). Tma46 is was also reported to associate with polysomes (10).
Thirteen identical YDR152w clones (aa 167 to 265) were isolated as encoding Rbg1 interacting partners (Fig. 3), suggesting that Rbg1 interacts with the C terminus of Ydr152w. Ydr152w was previously identified as an interacting partner of both yeast DRG proteins, Rbg1 and Rbg2, in genomewide two-hybrid and protein-protein interaction studies (25, 33, 64). It was demonstrated that YDR152w has a synthetic interaction with ribosomal genes and, therefore, YDR152w was renamed GIR2 for “genetically interacts with ribosomal genes” (Krogan, unpublished). Although the function of Gir2 is unknown, the N terminus (aa 5 to 156) shares sequence similarity to the N-terminal GI domain of Gcn2 (34), a polyribosome-associated protein (47). Interestingly, native Gir2 seems to be highly unstructured, perhaps only adopting a folded conformation in the presence of binding partners (1, 2).
Gir2 interacts with Rbg1 and Gcn1.
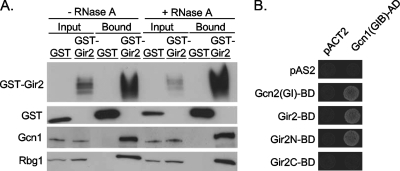
Of the proteins identified in the yeast two-hybrid screen, we focused on the interaction between Rbg1 and Gir2. To confirm the ability of Gir2 to interact with Rbg1, GST-tagged Gir2 was overexpressed in wild-type cells and precipitated using glutathione-Sepharose, and the precipitates were analyzed by immunoblotting. Rbg1 was easily detectable as a copurifying protein in the extracts containing GST-Gir2 but not with the GST-negative control (Fig. 4A). To determine whether an RNA species was necessary for the interaction between Gir2 and Rbg1, the pulldown analyses were repeated in the presence of RNase A. The interaction between Gir2 and Rbg1 was not altered upon digestion of RNA (Fig. 4A). In addition, the interaction between Gir2 and Rbg1 was analyzed upon removal of ribosomes by centrifugation of the samples at 100,000 × g. Under these conditions the interaction between Gir2 and Rbg1 persisted (data not shown). We conclude that Gir2 and Rbg1 interact in vivo.
FIG. 4.
Association of Gir2 with Gcn1 and Rbg1. (A) Copurification of Rbg1 and Gcn1 with GST-Gir2. GST and GST-Gir2 were affinity-purified with glutathione-Sepharose beads from yeast strains (H1511) expressing plasmid-borne GST alone or GST-Gir2 from a galactose-inducible promoter (pES128-9-1 and pJM3958, respectively). Aliquots of cellular extract, treated with or without RNase A, loaded onto the beads (input) or from the affinity-purified (bound) fractions, were analyzed by immunoblotting with antibodies to GST, Rbg1, or Gcn1, as indicated. (B) Cells expressing the GAL4 activation domain alone (pACT2) or fused to Gcn1(GIB)-AD [pACT2-GCN1(GIB)] and either the GAL4-binding domain alone (pAS2) or fused to Gcn2(GI), Gir2, Gir2N, or Gir2C [pAS-GCN2(GI), pAS-GIR2, pAS-GIR2N, and pAS-GIR2C, respectively] were analyzed for expression of HIS3 reporter gene by growth on SD−His−Trp−Leu plus 3-AT.
The GI domain of Gcn2 is necessary and sufficient for in vivo binding to Gcn1, a key regulatory protein in general amino acid control pathway(34, 53). Since the N terminus of Gir2 shares sequence similarity with the GI domain of Gcn2 (34), we tested whether Gir2 also interacts with Gcn1 in vivo using GST-Gir2 mediated glutathione precipitation. We found that Gcn1 was specifically detected in the GST precipitation of cell extracts harboring GST-Gir2 (Fig. 4A), suggesting that, in addition to Rbg1, Gir2 also interacts with Gcn1 in vivo. The interaction between Gir2-Gcn1 was stable in either the presence of RNase (Fig. 4A) or the absence of ribosomes (data not shown).
We next analyzed the interaction between Gir2 and Gcn1 by using the yeast two-hybrid analysis. The Gcn2 binding domain of Gcn1 (aa 2048 to 2383), called the GIB domain (for GI binding), has been previously defined (34, 53). As expected, therefore, cells expressing Gcn1(GIB)-AD and Gcn2(GI)-BD on the appropriate selective medium grew, whereas cells harboring Gcn1(GIB)-AD and the vector control pAS2 did not (Fig. 4B). Consistent with the ability of GST-Gir2 to precipitate Gcn1, we observed growth for cells expressing Gcn1(GIB)-AD and Gir2-BD on the appropriate selective medium. We conclude that full-length Gir2 interacts with Gcn1.
To determine whether the N-terminal GI domain of Gir2 was sufficient for its interaction with Gcn1(GIB)-AD, the N terminus (aa 1 to 170) or C terminus (aa 167 to 265) of Gir2 was expressed as a fusion protein with the GAL4 DNA-binding domain (Gir2N-BD and Gir2C-BD, respectively). Expression of these truncated fusions was verified by immunoblotting (data not shown). Cells expressing Gcn1(GIB)-AD and Gir2N-BD showed growth comparable to that of the positive control, whereas cells expressing Gcn1(GIB)-AD and Gir2C-BD did not grow, suggesting that the GI domain of Gir2 is necessary and sufficient for its interaction with Gcn1 (Fig. 4B).
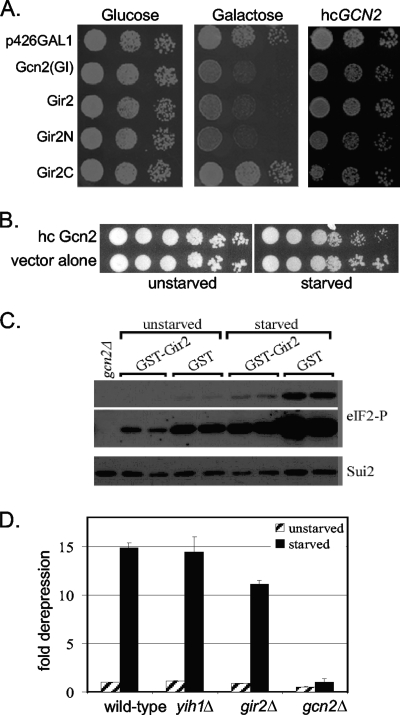
To provide evidence that Gir2 and Gcn1 interact in vivo, we exploited the well-characterized general amino acid control pathway. This signal transduction pathway enables cells to overcome amino acid starvation. In particular, direct physical interaction between Gcn1 and Gcn2 is essential for activating Gcn2 and thus the general amino acid control (21, 53). The function of this pathway can be assayed by growing cells on medium containing 3-AT, an inhibitor of the histidine biosynthetic enzyme encoded by H1S3. Overproduction of the GI domain of Gcn2 results in a dominant-negative phenotype, observable as the lack of growth on 3-AT containing medium (13, 34; Fig. 5A). The lack of growth is due to the GI domain competing with native Gcn2 for Gcn1 binding, thereby dampening Gcn2 activation and consequently diminishing the ability of cells to overcome amino acid starvation imposed by 3-AT (53). Simultaneous coexpression of Gcn2 overcomes the dominant-negative phenotype (Fig. 5A) (53). We posited that if Gir2 binds to the same domain in Gcn1 as Gcn2 does, then overexpression of Gir2 would also titrate Gcn1 and lead to a dominant-negative phenotype. To test this, we induced expression of full-length Gir2 (under the control of the galactose inducible GAL1 promoter) in an otherwise wild-type strain. We observed a severe growth inhibition on medium containing 3-AT (Fig. 5A), while co-overexpression of Gcn2 restored growth. Furthermore, suppression of the 3-AT sensitivity phenotype by Gcn2 was not simply due to a growth advantage conferred to cells overexpressing Gcn2, since wild-type cells overexpressing Gcn2 grew less well than cells harboring a vector alone under starved conditions (Fig. 5B). Taken together, these data support the model that overexpressed Gir2 competes with Gcn2 for Gcn1 binding by utilizing the same or overlapping binding domains in Gcn1 and thereby dampens signaling of Gcn1 to Gcn2 under amino acid limiting conditions.
FIG. 5.
Gir2 expression and amino acid control. (A) Overexpression of Gir2 leads to growth inhibition on 3-AT. Tenfold serial dilutions of cells harboring vector alone (p426-GAL1) or expressing GCN2(GI), GIR2, GIR2N, or GIR2C under the GAL1 promoter were spotted on plates containing either glucose or galactose, as indicated, in the presence of 10 mM 3-AT and incubated at 30°C. Similar strains that also express Gcn2 from a high-copy plasmid (hcGCN2; pAH15) were grown in the presence of galactose and 10 mM 3-AT. (B) Overexpression of hcGCN2 slightly perturbs the growth of starved cells. Tenfold serial dilutions of wild-type cells harboring hcGCN2 or vector alone in the absence (unstarved) or presence (starved) of 10 mM 3-AT. (C) Yeast strains (H1511) expressing plasmid-borne GST alone or GST-Gir2 and the isogenic gcn2Δ strain H2557 were grown to exponential phase and harvested, followed by immunoblot analysis with antibodies to eIF-2α and phosphorylated eIF-2α. Two different exposure times for phosphorylated eIF-2α are shown (two panels linked together by a gray bar on the right-hand side, with the upper panel showing short exposure and the lower panel showing long exposure). (D) Expression of GCN4-lacZ. Expression of GCN4 was determined in the strains indicated using a lacZ-based reported construct (p180), and the fold derepression was determined as the ratio of the wild-type values under nonstarvation conditions. For each strain, at least two independent transformants were assayed, in duplicate (gcn2Δ strain) or at least in triplicate, and the average values as well as the standard error values are shown in the graph.
We next sought to determine whether the GI domain of Gir2 alone was sufficient to produce the dominant-negative phenotype. Overexpression of Gir2N in the presence of 3-AT resulted in a dominant-negative phenotype, whereas overexpression of Gir2C had no effect on cell growth (Fig. 5A). Thus, we conclude that the N-terminal GI domain of Gir2 is both necessary and sufficient for binding Gcn1 in vivo.
The basal Gcn2 enzyme activity, as well as the activation of Gcn2 under amino acid starvation, is dependent on direct Gcn1-Gcn2 interaction. If the dominant-negative phenotype of Gir2 overexpression is due to Gir2 disrupting Gcn1-Gcn2 interaction, then this should result in reduced phosphorylation of the Gcn2 substrate, eIF-2α, even under amino acid-replete conditions. To test our prediction, we examined the phosphorylation state of eIF-2α in strains overexpressing GST-Gir2. As expected, the levels of eIF-2α phosphorylation were dramatically decreased in comparison to cells overexpressing GST alone (Fig. 5C). These data further support the conclusion that Gir2 binds Gcn1 and, at least when Gir2 is overexpressed, competes with Gcn2 for Gcn1 binding and thereby inhibits the activation of Gcn2 by Gcn1.
Although our results support the idea that Gir2 is a binding partner for Gcn1, it is not clear that the main role of Gir2 is in regulating the general amino acid control pathway. Therefore, we examined whether the gir2Δ mutant had an effect on the output of a Gcn2-regulated gene, GCN4. Isogenic wild-type, yih1Δ, gir2Δ, and gcn2Δ strains were transformed with a plasmid (p180) harboring a GCN4-lacZ reporter construct in which the translation of β-galactosidase is under the control of the GCN4 5′ untranslated region containing four upstream open reading frames, and β-galactosidase activity was assayed under nonstarvation and starvation conditions. In contrast to a GCN2 deletion, a GIR2 deletion did not alter the level of GCN4 expression (Fig. 5D). Thus, in vivo, it is unlikely that Gir2 plays a general role in controlling Gcn2 activity. Similarly, deletion of another gene, YIH1, also encoding a GI domain known to bind Gcn1 (54), also resulted in no increase in GCN4 expression (Fig. 5D), in agreement with previous findings that deletion of YIH1 did not lead to increased eIF-2α phosphorylation (54).
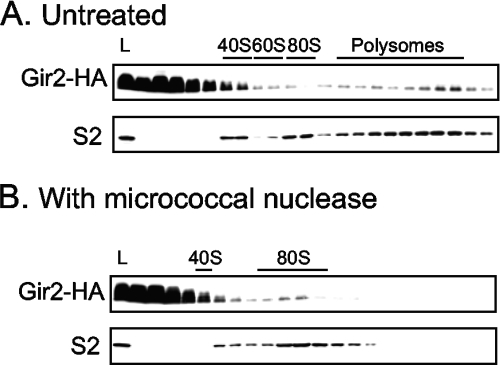
Gir2 comigrates with ribosomes.
Thus far, we have shown that Gir2 interacts with Rbg1 (Fig. 3 and 4) and Gcn1 (Fig. 4). Moreover, both Rbg1 and Gcn1 are associated with polyribosomes (Fig. 2; 41). Therefore, we sought to determine whether Gir2 was also ribosome associated. We epitope tagged the chromosomal GIR2 gene and subjected the resulting strain to velocity sedimentation through a sucrose gradient to resolve ribosomes. Under the conditions assayed, the vast majority of Gir2-3HA migrated at the top of the sucrose gradient (Fig. 6A), indicating that the majority of Gir2-3HA is either not associated with larger complexes or becomes dissociated from larger complexes during centrifugation. A significant amount of Gir2-3HA, however, was found in fractions corresponding to polyribosomes (Fig. 6A). Interestingly, this pattern of fractionation is similar to that previously reported for Gcn1 (42). Furthermore, the Gir2-3HA levels appear to increase with the size of the polyribosomes, a profile similar to that seen for the small ribosomal subunit protein S2 (Fig. 6A). Upon disruption of the elongating ribosomes by the addition of micrococcal nuclease, the amount of Gir2 found in the polyribosome fractions decreased, and there was an increase in the signal corresponding to the monosome peak (Fig. 6B). Thus, at least a proportion of the population of Gir2 is associated with the polyribosomes.
FIG. 6.
Gir2 partially cofractionates with translating ribosomes. Extracts from cells expressing Gir2-HA from the chromosome without (A) or with (B) the addition of micrococcal nuclease (5 U/μl) prior to fractionation on sucrose gradients. Immunoblots of indicated fractions reveal the positions of Gir2-HA and ribosomal protein S2. The positions of the 40S, 60S, and 80S monosomes and the polysomes are labeled. L, 1/100 extract loaded on gradient.
The association of Gir2 with polyribosomes is partially dependent on Gcn1.
Gir2, Rbg1, and Gcn1 each associate with polyribosomes (Fig. 2 and 6; 42), and Gir2 interacts with both Rbg1 and Gcn1 (Fig. 3 and 4). It is therefore possible that, on the translating ribosome, Gir2 interacts with both Rbg1 and Gcn1 simultaneously, with its N-terminal GI domain tethered to Gcn1 and its C terminus tethered to Rbg1. To determine whether the association of Gir2 with the polysomes was a direct interaction or was dependent on either of its binding partners, we examined the fractionation pattern of Gir2-3HA on sucrose density gradients in strains lacking either RBG1 or GCN1.
In the absence of Rbg1, the distribution of Gir2-3HA in sucrose density gradients does not significantly differ from that seen in wild-type extracts (Fig. 7B compared to Fig. 7A), suggesting that Gir2-3HA associates with the polysomes independently of Rbg1. In the absence of Gcn1, however, there was a dramatic decrease in the amount of Gir2-3HA associated with polyribosomes (Fig. 7C). Furthermore, the polyribosome association can be restored by expression of Gcn1 from a plasmid (data not shown). Thus, the polysome association of Gir2 is at least partially dependent on Gcn1 but not on Rbg1.
FIG. 7.
Gir2 association with polysomes is partially dependent on Gcn1. Cell extracts from a wild-type strain (A), an rbg1Δ strain (B), or a gcn1Δ strain (C) were resolved by velocity sedimentation in sucrose gradients, and the positions of Gir2-HA and Rbg1 were detected by immunoblot analysis, as indicated. The positions of the 40S, 60S, and 80S monosomes and the polysomes are indicated. L, 1/100 extract loaded on gradient.
We also examined the requirements of Rbg1 for polysome association. The Rbg1 fractionation pattern in cells lacking Gcn1 was identical to that of wild type (Fig. 7C compared to Fig. 7A), indicating that the ribosome association of Rbg1 is independent of Gcn1. Likewise, the fractionation pattern of Rbg1 in cells lacking Gir2 is indistinguishable from that of wild type (data not shown). Thus, the polysome association of Rbg1 is independent of Gir2 and Gcn1.
DISCUSSION
Here we show that Rbg1, a member of the Obg/CgtA subfamily of GTPases, is associated with polyribosomes. This finding is in agreement with the prediction that all Obg/CgtA proteins are involved in ribosome function (6, 38). Unlike the nucleolar, mitochondrial, and bacterial Obg/CgtA proteins examined that specifically interact with the large ribosomal subunit, however, Rbg1 cofractionates with polyribosomes (Fig. 2). Thus, it is likely that Rbg1 plays a cellular role distinct from the large subunit assembly role filled by the Nog1 and Mtg2 members of the Obg/CgtA family (8, 12, 28, 31).
The nature of the polysome association of Rbg1 is unclear. It is possible that a specific translational factor(s) recruits Rbg1 to the ribosome only after initiation of translation or that Rbg1 binds to a specific ribosome conformational state that only occurs during translation elongation. Alternatively, Rbg1 could interact directly with tRNA and remain associated with the translating ribosome due to a dynamic tRNA association. Finally, it is possible that Rbg1 associates with a specific subset of polysomes, such as the subpopulation of ribosomes that are stalled because the required aminoacyl-tRNA is not available or because the ribosome has accepted an uncharged tRNA rather than a charged one. This latter possibility is particularly intriguing given that uncharged tRNAs can activate both the TGS-containing RelA protein in bacteria and the Gcn1 interacting partner, Gcn2, in yeast (21, 57).
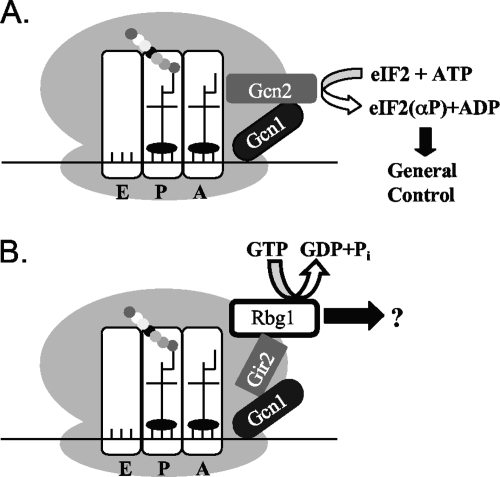
We identified potential interacting partners of Rbg1: Yap1, Ygr250c, Tma46, and Gir2. We focused here on the interactions between Rbg1 and Gir2, Gir2 and Gcn1, and all three of these proteins with polyribosomes. We show that Gir2 interacts with Rbg1 and Gcn1 in physically distinct areas with the Gir2 C terminus binding to Rbg1 (Fig. 3) and the N-terminal GI domain binding Gcn1 (Fig. 5). Thus, Gir2 could interact with both Rbg1 and Gcn1 simultaneously. Moreover, we show that in ribosome cosedimentation assays, Gir2 bound to ribosomes in a manner that is at least in part dependent on Gcn1, whereas the Gir2-ribosome interaction was independent of Rbg1 and the Rbg1-ribosome interaction was independent of Gcn1. We therefore propose that Gir2 acts as a linker between Rbg1 and Gcn1, perhaps to mediate signals from Gcn1 to Rbg1. One attractive possibility is that hydrolysis of GTP by Rbg1 could be the output of this signaling cascade (Fig. 8B).
FIG. 8.
Modeling the interaction between translating ribosomes and Gcn2, Gcn1, Rbg1, and Gir2. (A) Gcn1 interacts with Gcn2 on the ribosome for detecting uncharged tRNAs under amino acid starvation. This leads to the stimulation of the Gcn2 kinase domain, subsequent phosphorylation of its substrate eIF-2α, and finally to the activation of the general amino acid control (see references 42 and 53). (B) On the ribosome, Gir2 binds simultaneously Rbg1 via its C-terminal domain, and Gcn1 binds via its N-terminal domain. In a manner analogous to the general amino control pathway described in panel A, the Rbg1 pathway may receive a signal from Gcn1 that is relayed through Gir2. It is possible that this signal alters the guanine nucleotide bound state of Rbg1 to elicit an as-yet-undefined biological response.
We propose that Gcn1, Gir2, and Rbg1 may represent another polysome-associated, response system (Fig. 8B). One exciting possibility is that, in the course of evolution, there was a shuffling of protein domains between the two interacting bacterial proteins (Obg/CgtA and SpoT) that resulted in the archaeal and eukaryotic hybrid Rbg1-like protein consisting of the GTP-binding domain of Obg/CgtA and the TGS domain of SpoT. Although mechanistically different, it has been postulated that the RelA-mediated response to stress (i.e., amino acid starvation) is analogous to the Gcn1/Gcn2 response in S. cerevisiae (42, 53). RelA, a polysome-associated protein, is activated by the binding of uncharged tRNAs (46). The SpoT protein is 50% similar to RelA (19). E. coli CgtA, interacts with and controls the activity of SpoT, on the ribosome (30, 70), clearly implicating this bacterial GTPase in stress response. In S. cerevisiae, during amino acid starvation, induction of the Gcn2 kinase activity requires association of Gcn2 with uncharged tRNA and Gcn1, which is proposed to be directly involved in relaying this signal to Gcn2 (Fig. 8A; 21). Gcn2, contains a domain similar to that of histidyl-tRNA synthetases that is important for binding to uncharged tRNA (68). Conceivably, the TGS domain of Rbg1 could also bind uncharged tRNA, specifically threonyl-tRNA, on a subset of ribosomes. In a manner analogous to the activation of Gcn2, Rbg1 binding to uncharged tRNA, coupled with a specific signal (e.g., an as-yet-unknown stress condition) relayed by Gcn1 through Gir2, could result in an as-yet-undefined Rbg1-mediated biological response (Fig. 8).
The high sequence conservation of Rbg1 and Rbg2 orthologs suggests that these proteins are involved in fundamental pathways and perhaps their binding partners are also conserved. In fact, the mammalian Rbg proteins interact with protein orthologs we identified in the present study, with some interesting differences: mammalian DRG1 (ortholog to yeast Rbg1) binds to DFRP1 (Tma46), and DRG2 (Rbg2) binds to DFRP2 (Gir2) (24). These DRG proteins are susceptible to degradation through the ubiquitin-mediated pathway, and their degradation is prevented by their interaction with DFRP proteins. Thus, the DFRP proteins provide a means of regulating DRG protein levels, which may be important for their function in tissue specific and developmentally specific expression (56). DFRP1 and DFRP2 share sequence similarity in a small region that is involved in DRG binding, but otherwise they are very different in domain structure. The Tma46 and Gir2 fragments we identified as being sufficient for a two-hybrid interaction with Rbg1 coincided largely with the DRG binding sites of the mammalian proteins, a finding consistent with the idea that their functions are evolutionary conserved.
If Gir2 can compete with Gcn2 for the binding of Gcn1 in vivo, Gir2 could be expected to act as a negative regulator of Gcn1, maintaining Gcn1 in an inactive state until an environmental stress signal is present. This appears not to be the case, however, since deletion of GIR2 did not lead to increased derepression of GCN4 as determined by GCN4-lacZ expression studies (Fig. 5D). On the contrary, it appears that Gir2 may have a slight positive regulatory role in Gcn2 activation, but only when cells are starved. Although further verification of such an additional positive regulatory role would be necessary, our findings suggest that, as proposed for Yih1 (54), Gir2 may only inhibit Gcn2 activation under specific circumstances or in specific cellular compartments when or where Gcn2 activation is disadvantageous to the cell.
Acknowledgments
This study was supported by NSF grant MCB0316357 and NIH grant GM077628 to J.R.M., a University of Michigan Rackham Merit Fellowship to P.K.W., and by the Royal Society of New Zealand Marsden fund (MAU0607) and Massey University Research Fund to E.S.
We are grateful to Kaustuv Datta, Jonathan Warner, and Alan Hinnebusch for clones and antibodies; Suzanne Lybarger and Raji Janakiraman for clones; Nidal Ayoub for technical assistance; and Usha Nair for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 15 May 2009.
REFERENCES
- 1.Alves, V. S., and B. A. Castilho. 2005. Gir2 is an intrinsically unstructured protein that is present in Saccharomyces cerevisiae as a group of heterogeneously electrophoretic migrating forms. Biochem. Biophys. Res. Commun. 332450-455. [DOI] [PubMed] [Google Scholar]
- 2.Alves, V. S., D. C. Pimenta, E. Sattlegger, and B. A. Castilho. 2004. Biophysical characterization of Gir2, a highly acidic protein of Saccharomyces cerevisiae with anomalous electrophoretic behavior. Biochem. Biophys. Res. Commun. 314229-234. [DOI] [PubMed] [Google Scholar]
- 3.Battesti, A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 621048-1063. [DOI] [PubMed] [Google Scholar]
- 4.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50643-650. [DOI] [PubMed] [Google Scholar]
- 5.Brickner, J. H., and R. S. Fuller. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 13923-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6135-139. [DOI] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 8.Datta, K., J. L. Fuentes, and J. R. Maddock. 2005. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell 16954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68585-596. [DOI] [PubMed] [Google Scholar]
- 10.Fleischer, T. C., C. M. Weaver, K. J. McAfee, J. L. Jennings, and A. J. Link. 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 201294-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foiani, M., A. M. Cigan, C. J. Paddon, S. Harashima, and A. G. Hinnebusch. 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 113203-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuentes, J. L., K. Datta, S. M. Sullivan, A. Walker, and J. R. Maddock. 2007. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics 278105-123. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Barrio, M., J. Dong, S. Ufano, and A. G. Hinnebusch. 2000. Association of GCN1-GCN20 regulatory complex with the N terminus of eIF2α kinase GCN2 is required for GCN2 activation. EMBO J. 191887-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440631-636. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415141-147. [DOI] [PubMed] [Google Scholar]
- 16.Gulshan, K., S. A. Rovinsky, S. T. Coleman, and W. S. Moye-Rowley. 2005. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J. Biol. Chem. 28040524-40533. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75805-816. [DOI] [PubMed] [Google Scholar]
- 18.Haseltine, W. A., and R. Block. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. USA 701564-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinemeyer, E. A., and D. Richter. 1977. In vitro degradation of guanosine tetraphosphate (ppGpp) by an enzyme associated with the ribosomal fraction from Escherichia coli. FEBS Lett. 84357-361. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, V. J., and H. Bremer. 1991. Escherichia coli ppGpp synthetase II activity requires spoT. J. Biol. Chem. 2665991-5999. [PubMed] [Google Scholar]
- 21.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59407-450. [DOI] [PubMed] [Google Scholar]
- 22.Hinnebusch, A. G., and G. R. Fink. 1983. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 805374-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa, K., S. Azuma, S. Ikawa, K. Semba, and J. Inoue. 2005. Identification of DRG family regulatory proteins (DFRPs): specific regulation of DRG1 and DRG2. Genes Cells 10139-150. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 984569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 971143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen, B. C., Q. Wang, C. T. Kifer, and M. Parsons. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J. Biol. Chem. 27832204-32211. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, M., K. Datta, A. Walker, J. Strahler, P. Bagamasbad, P. C. Andrews, and J. R. Maddock. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 1886757-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, M., S. M. Sullivan, P. K. Wout, and J. R. Maddock. 2007. G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 1896140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 234344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koller-Eichhorn, R., T. Marquardt, R. Gail, A. Wittinghofer, D. Kostrewa, U. Kutay, and C. Kambach. 2007. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 28219928-19937. [DOI] [PubMed] [Google Scholar]
- 33.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440637-643. [DOI] [PubMed] [Google Scholar]
- 34.Kubota, H., Y. Sakaki, and T. Ito. 2000. GI domain-mediated association of the eukaryotic initiation factor 2α kinase GCN2 with its activator GCN1 is required for general amino acid control in budding yeast. J. Biol. Chem. 27520243-20246. [DOI] [PubMed] [Google Scholar]
- 35.Kuge, S., and N. Jones. 1994. YAP1-dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 161710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapik, Y. R., J. M. Misra, L. F. Lau, and D. G. Pestov. 2007. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol. Cell. Biol. 277735-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 31741-72. [DOI] [PubMed] [Google Scholar]
- 39.Lin, B., D. A. Thayer, and J. R. Maddock. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 41.Marton, M. J., D. Crouch, and A. G. Hinnebusch. 1993. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 133541-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marton, M. J., C. R. Vazquez de Aldana, H. Qiu, K. Chakraburtty, and A. G. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell. Biol. 174474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moehle, C. M., and A. G. Hinnebusch. 1991. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 112723-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller, P. P., and A. G. Hinnebusch. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45201-207. [DOI] [PubMed] [Google Scholar]
- 45.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 225767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramagopal, S., and B. D. Davis. 1974. Localization of the stringent protein of Escherichia coli on the 50S ribosomal subunit. Proc. Natl. Acad. Sci. USA 71820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramirez, M., R. C. Wek, and A. G. Hinnebusch. 1991. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 113027-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskin, D. M., N. Judson, and J. J. Mekalanos. 2007. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1044636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sartori, G., G. Mazzotta, S. Stocchetto, A. Pavanello, and G. Carignani. 2000. Inactivation of six genes from chromosomes VII and XIV of Saccharomyces cerevisiae and basic phenotypic analysis of the mutant strains. Yeast 16255-265. [DOI] [PubMed] [Google Scholar]
- 51.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10393-408. [DOI] [PubMed] [Google Scholar]
- 52.Sattlegger, E., and A. G. Hinnebusch. 2005. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 28016514-16521. [DOI] [PubMed] [Google Scholar]
- 53.Sattlegger, E., and A. G. Hinnebusch. 2000. Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 196622-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sattlegger, E., M. J. Swanson, E. A. Ashcraft, J. L. Jennings, R. A. Fekete, A. J. Link, and A. G. Hinnebusch. 2004. YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 27929952-29962. [DOI] [PubMed] [Google Scholar]
- 55.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 234449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sazuka, T., Y. Tomooka, Y. Ikawa, M. Noda, and S. Kumar. 1992. DRG: a novel developmentally regulated GTP-binding protein. Biochem. Biophys. Res. Commun. 189363-370. [DOI] [PubMed] [Google Scholar]
- 57.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor sigma(B). J. Bacteriol. 1814653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott, J. M., J. Ju, T. Mitchell, and W. G. Haldenwang. 2000. The Bacillus subtilis GTP binding protein Obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 1822771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikora, A. E., R. Zielke, K. Datta, and J. R. Maddock. 2006. The Vibrio harveyi GTPase CgtAV is essential and is associated with the 50S ribosomal subunit. J. Bacteriol. 1881205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, J., U. Jakob, and J. C. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 1842692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 63.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333169-174. [DOI] [PubMed] [Google Scholar]
- 64.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403623-627. [DOI] [PubMed] [Google Scholar]
- 65.Vazquez de Aldana, C. R., M. J. Marton, and A. G. Hinnebusch. 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 143184-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vilardell, J., and J. R. Warner. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 171959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wade, C., K. A. Shea, R. V. Jensen, and M. A. McAlear. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 218638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 154497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases: analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9689-710. [PubMed] [Google Scholar]
- 70.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 1865249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]