Abstract
We identified and functionally characterized genes encoding three Gα proteins and one Gβ protein in the dimorphic fungal wheat pathogen Mycosphaerella graminicola, which we designated MgGpa1, MgGpa2, MgGpa3, and MgGpb1, respectively. Sequence comparisons and phylogenetic analyses showed that MgGPA1 and MgGPA3 are most related to the mammalian Gαi and Gαs families, respectively, whereas MgGPA2 is not related to either of these families. On potato dextrose agar (PDA) and in yeast glucose broth (YGB), MgGpa1 mutants produced significantly longer spores than those of the wild type (WT), and these developed into unique fluffy mycelia in the latter medium, indicating that this gene negatively controls filamentation. MgGpa3 mutants showed more pronounced yeast-like growth accompanied with hampered filamentation and secreted a dark-brown pigment into YGB. Germ tubes emerging from spores of MgGpb1 mutants were wavy on water agar and showed a nested type of growth on PDA that was due to hampered filamentation, numerous cell fusions, and increased anastomosis. Intracellular cyclic AMP (cAMP) levels of MgGpb1 and MgGpa3 mutants were decreased, indicating that both genes positively regulate the cAMP pathway, which was confirmed because the WT phenotype was restored by adding cAMP to these mutant cultures. The cAMP levels in MgGpa1 mutants and the WT were not significantly different, suggesting that this gene might be dispensable for cAMP regulation. In planta assays showed that mutants of MgGpa1, MgGpa3, and MgGpb1 are strongly reduced in pathogenicity. We concluded that the heterotrimeric G proteins encoded by MgGpa3 and MgGpb1 regulate the cAMP pathway that is required for development and pathogenicity in M. graminicola.
Signal transduction pathways are important for sensing and responding to different environmental stimuli in both lower and higher eukaryotes. The highly conserved heterotrimeric guanine nucleotide-binding proteins (G proteins) belong to a family of regulatory proteins that are crucial for the transduction of signals, which are perceived by a distinct family of cell surface receptors (4). Heterotrimeric G proteins contain three subunits (α, β, and γ) that are linked in the inactive state. Activation of a Gα subunit by a transmembrane receptor leads to exchange of bound GDP with GTP on the Gα subunit, resulting in dissociation of the Gα and the Gβγ dimeric subunits, which can now interact with downstream effectors that subsequently generate changes in cellular responses (for a review, see reference 10).
Filamentous fungi have one Gβ- and usually three Gα-encoding genes that belong to three major groups. Encoded proteins in groups I and III are related to the mammalian Gαi and Gαs families, respectively, but group II fungal Gα proteins have no mammalian counterpart (1, 4, 14, 22, 33, 53). Interestingly, the corn smut fungus Ustilago maydis contains a unique fourth Gα-encoding gene, and Saccharomyces cerevisiae contains only two Gα proteins (10, 57). Irrespective of the observed numerical variation, Gα proteins regulate a variety of cellular and developmental responses (4). For plant-pathogenic fungi, Gβ-encoding genes have been characterized functionally (9, 14, 22, 27, 31, 48, 52). Apart from the fact that individual Gα-encoding genes and the Gβ-encoding gene have been demonstrated to regulate growth, reproduction, and virulence, comparative functional characterization of all Gα-encoding genes has been reported only for a few plant-pathogenic fungi, including Magnaporthe grisea, Cryphonectria parasitica, and U. maydis (5, 41, 57).
Mycosphaerella graminicola (anamorph Septoria tritici) causes septoria tritici blotch disease in bread and durum wheat in areas with high rainfall during the growing season, particularly in Western Europe, where it is considered to be the most important wheat disease (30). It is a ubiquitous phytopathogen with a lifestyle completely different from that of the aforementioned plant-pathogenic fungi. It is a dimorphic pathogen, and therefore the transition from a yeast-like to a filamentous form is important for initiation of infection (45). M. graminicola does not form appressoria but penetrates the leaves through stomata without forming specific infection structures. Furthermore, as a hemibiotroph, it has a biotrophic phase of about 10 days that is followed by a rapid switch to necrotrophy. The necrotic foliar lesions bear anamorphic and teleomorphic fructifications. M. graminicola is the model fungus for the Mycosphaerellaceae and even for the order Dothideales, an extremely large and diverse class of fungi with over 1,000 named species, including major plant pathogens such as the banana leaf streak fungus Mycosphaerella fijiensis (12, 21). Large expressed sequence tag (EST) libraries and the recently released genome sequence have been instrumental for the identification and characterization of genes involved in the development and pathogenicity of M. graminicola (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html). Recently, we reported that genes encoding mitogen-activated protein kinases (MAPKs) (MgFus3, MgSlt2, and MgHog1) and the catalytic (MgTpk2) and regulatory (MgBcy1) subunits of protein kinase A (PKA) are essential pathogenicity factors and regulate specific steps during the infection process (8, 43-45). To extend our knowledge about the role of G proteins in the development and pathogenicity of M. graminicola, we functionally analyzed three Gα-encoding genes and one Gβ-encoding gene of M. graminicola, which we designated MgGpa1, MgGpa2, MgGpa3, and MgGpb1, respectively. Our results show the requirement of MgGpa1, MgGpa3, and MgGpb1 for pathogenicity, whereas the latter also negatively regulates cell fusion and anastomosis. Among the G protein-encoding genes characterized in this study, MgGpa3 and MgGpb1 positively regulate the cyclic AMP (cAMP) pathway. MgGpa1 seems to be dispensable for cAMP regulation, whereas MgGpa2 appears to be redundant, for none of the assays rendered altered phenotypes. Our results open new perspectives for studying the regulatory machinery of the cAMP pathway in M. graminicola and other plant-pathogenic fungi.
MATERIALS AND METHODS
Fungal strains and culture conditions.
The Dutch field strain M. graminicola IPO323 is highly virulent on winter wheat cv. Obelisk and was used as the wild type (WT) and recipient strain throughout this study. Growing conditions, propagation, and maintenance were as described previously (35, 44).
DNA manipulations and analysis.
Standard protocols were used for DNA manipulation (58). Plasmid DNA was isolated using a QIAprep Spin miniprep kit (Qiagen, Hilden, Germany). Genomic DNA of M. graminicola was extracted using a Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN). A DYEnamic ET dye terminator cycle sequencing kit (Amersham Biosciences, Roosendaal, The Netherlands) was used to sequence DNA on an ABI Prism 3100 capillary automated sequencer according to recommended protocols. Phylogenetic tree generation, DNA and protein sequence alignment, editing, and analysis were performed using MEGALIGN and DNA Star software (Madison, WI). Homology searches were performed with the BLAST program (2).
Construction of plasmid vectors.
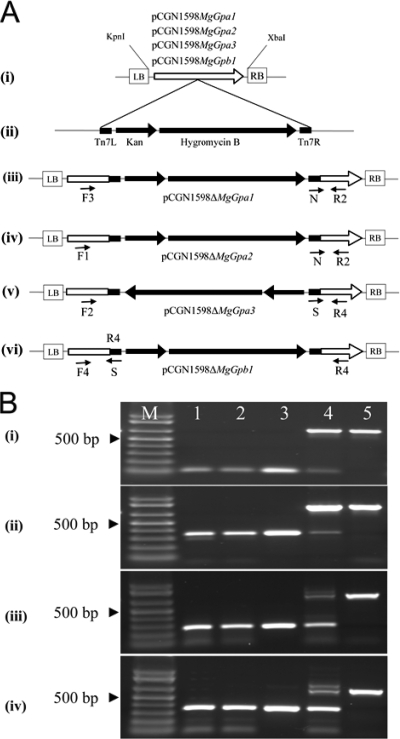
By in silico analyses of EST databases of M. graminicola IPO323 (34), we identified full-length cDNA clones of MgGpa1, MgGpa2, MgGpa3, and MgGpb1. We used the GPS-Mutagenesis system (New England Biolabs, Leusden, The Netherlands) to generate disruption constructs, using a homemade customized donor construct, pGPS3HygKan, that was described previously (44). After transposition, the transposition mixture was cloned into Escherichia coli strain DH10B (Invitrogen, Groningen, The Netherlands) by electroporation, using the recommended protocol of the suppliers, and subsequently plated onto LB containing gentamicin and kanamycin to select for colonies with transposon insertions in the target constructs, which were confirmed by multiplex colony PCR as described previously (44). The exact position of the transposon insertion was determined by sequencing from the right and left borders of the transposon, using primer N and primer S (Table 1), and by comparison with nondisrupted sequences. We selected constructs with transposon insertions in the central part of the WT genes, and these were designated pCGN1589ΔMgGpa1, pCGN1589ΔMgGpa2, pCGN1589ΔMgGpa3, and pCGN1589ΔMgGpb1, respectively (Fig. 1A).
TABLE 1.
Generated Mycosphaerella graminicola G protein transformants and primers used in this study
| Gene or position | M. graminicola transformant | Type | Primer | Primer sequence |
|---|---|---|---|---|
| MgGpa1 | IPO323 ΔMgGpa1-14 | Disruption strain | MgGpa1-F3 | TTCAGCGAAGCCGAGAATATC |
| IPO323 ΔMgGpa1-15 | Disruption strain | MgGpa1-R2 | CACATGCGCAGGTTCTCTTG | |
| IPO323 E-MgGpa1-13 | Ectopic transformant | |||
| MgGpa2 | IPO323 ΔMgGpa2-2 | Disruption strain | MgGpa2-F1 | GGCGTACAGCTTGCGAT |
| IPO323 ΔMgGpa2-6 | Disruption strain | MgGpa2-R2 | CATCAAGTTATGCAAGTTCCGC | |
| IPO323 E-MgGpa2-10 | Ectopic transformant | |||
| MgGpa3 | IPO323 ΔMgGpa3-21 | Disruption strain | MgGpa3-F2 | AGCAGTGGGAACAATGAGGAG |
| IPO323 ΔMgGpa3-33 | Disruption strain | MgGpa3-R4 | TGAATACTGAGCTGGCCCAT | |
| IPO323 E-MgGpa3-2 | Ectopic transformant | |||
| MgGpb1 | IPO323 ΔMgGpb1-3 | Disruption strain | MgGpb1-F4 | ACCACGAACAAAGTACACGCC |
| IPO323 ΔMgGpb1-22 | Disruption strain | MgGpb1-R4 | AGTCGACCTGAGACGGAGAAAG | |
| IPO323 E-MgGpb1-1 | Ectopic transformant | |||
| Left border of transposon (Tn7L) | S | ATAATCCTTAAAAACTCCATTTCCACCCCT | ||
| Right border of transposon (Tn7R) | N | ACTTTATTGTCATAGTTTAGATCTATTTTG |
FIG. 1.
Generation of disruption constructs and subsequent identification of mutants of Mycosphaerella graminicola. (A) (i) The cDNA fragments of MgGpa1, MgGpa2, MgGpa3, and MgGpb1 were excised from the pSport1 vector, using KpnI/XbaI restriction enzymes, and inserted into the same site of the binary vector of Agrobacterium tumefaciens (pCGN1589), generating the target constructs pCGN1589MgGpa1, pCGN1589MgGpa2, pCGN1589-MgGpa3, and pCGN1589MgGpb1, respectively. The target constructs were transposed using the customized donor construct pGPS3HygKan (ii), resulting in the disruption constructs pCGN-1598ΔMgGpa1 (iii), pCGN1598ΔMgGpa2 (iv), pCGN1598 ΔMgGpa3 (v), and pCGN1598ΔMgGpb1 (vi). (B) Multiplex PCR screening to identify homologous recombinants in MgGpa1 (i), MgGpa2 (ii), MgGpa3 (iii), and MgGpb1 (iv), using two gene-specific primers (indicated in panel A) together with either primer N or primer S, located at the right or left border of the transposon, respectively. Homologous recombinants (lanes 1 and 2) amplified a small amplicon identical in size to the amplicon generated from the disruption construct (lane 3), whereas in ectopic transformants (lane 4) two bands were amplified, including a small band corresponding to the disruptant strains and a larger band of equal size to the amplicon generated from the WT strain M. graminicola IPO323 (lane 5). Lane M shows One Kb Plus markers.
Fungal transformation.
The disruption constructs pCGN1589ΔMgGpa1, pCGN1589ΔMgGpa2, pCGN1589ΔMgGpa3, and pCGN1589ΔMgGpb1 were cloned into Agrobacterium tumefaciens strain LBA1100 by electroporation. A. tumefaciens-mediated transformation was performed according to a standard protocol (44, 72). Homologous recombinants were identified by PCR screening (Fig. 1B), using primers that were designed upstream and downstream of the transposon insertions together with either primer S or primer N, located in the left or right border of the transposon, respectively (Table 1). Eventually, we selected two mutant strains and one ectopic transformant for each gene for phenotypic analyses (Table 1). To ensure the independence of the transformants, we performed transformation experiments over time. In cases where transformants were generated in the same experiment, they were collected mostly from different plates (at least four plates for each transformation) to prevent the selection of clones. Moreover, in contrast to polyethylene glycol transformation, A. tumefaciens-mediated transformation precludes the selection of clones, as cocultivation of the fungus and Agrobacterium is done on solid nitrocellulose filters, and hence each colony represents an independent transformation event.
Phenotyping.
We phenotyped all disruptant strains along with ectopic transformants and the WT as control strains.
In vitro assays were performed on agar plates to study colony development by spotting approximately 1 μl of spore suspension (107 spores ml−1) on potato dextrose agar (PDA), which was subsequently cultured for 10 days at 20°C under continuous light, with and without 13 mM exogenous cAMP, or at 28°C in darkness, as well as in liquid yeast glucose broth (YGB) at 20°C under daylight settings. Spore length measurements were performed on 100 randomly selected spores per strain from YGB cultures.
Histological analyses were performed on water agar (WA) and PDA, and the effect of exogenous cAMP (15 mM) on the early development of the strains was examined. Samples were monitored every 24 h until 80 h after initiation of the experiment, using a Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany) equipped with a Carl Zeiss AxioCam 6.08.0 digital camera connected to an IBM computer. Pictures were saved with Axio Vision 4.1 software (Carl Zeiss, Jena, Germany), and composed pictures were prepared with Adobe Photoshop 7.0.
In planta phenotyping assays were performed according to a previously described protocol (44, 45). Disease progress was monitored at various time points after inoculation, and final observations were performed at 20 days postinoculation (dpi).
cAMP measurements.
Quantification of intracellular cAMP was performed on 12 isolates (Table 1), including the WT, according to the protocol suggested by the manufacturer (Amersham Life Science Inc., Arlington Heights, IL). Spores were collected and lyophilized after being cultured for 5 days in YGB. Lyophilized spores (100 mg) were ground to a fine powder in a 2-ml Eppendorf tube by use of a bead beater (Ribolyser; Hybaid, Heidelberg, Germany). Powdered mycelia were resuspended in distilled water, and proteins were precipitated by adding 700 μl of 6% tricarboxylic acid (TCA). After centrifugation, the supernatant was extracted five times with water-saturated ether to remove the TCA and subsequently lyophilized. The lyophilized sample was dissolved in phosphate buffer (10 mM, pH 7.4) and used for the cAMP assay. Before protein precipitation with TCA, two replicate samples of 100 μl of the supernatant were taken for protein quantification using a Coomassie Plus protein assay reagent kit (Pierce Biotechnology, Inc.), with bovine serum albumin as a reference. Before cAMP quantification, the pH was measured, and if necessary, 50% KHCO3 was added until a pH of 6 to 8 was obtained. In total, three biological replicates of each strain were assayed in duplicate by a cAMP (3H) assay system following the manufacturer's instructions (Amersham Life Science Inc., Arlington Heights, IL). After quantification, the cAMP values were normalized to the protein level for each strain (pmol cAMP/mg total protein).
Statistical analyses.
cAMP (pmol cAMP/mg total protein) and spore length (μm) data were natural log transformed and further analyzed by analysis of variance followed by pairwise comparisons among treatment means (based on t tests with a P value of 0.05) to test for significant differences between experiments, using the statistical software package Genstat (54) extended with Biometris Procedure library applications (20). The analyses did not show significant differences between control strains (WT and ectopic transformants) and between the independent mutants of the same gene, which allowed us to lump these data into groups for final analyses (controls and MgGpa1, MgGpa3, and MgGpb1 mutants).
Nucleotide sequence accession numbers.
Sequence data from this article were deposited with the EMBL/GenBank data libraries under accession numbers DQ458049, DQ458050, DQ458051, and DQ458052, for MgGpa1, MgGpa2, MgGpa3, and MgGpb1, respectively.
RESULTS
Sequence comparison and phylogenetic analyses of MgGpa1, MgGpa2, MgGpa3, and MgGpb1.
Screening of a large EST resource comprising 27,007 M. graminicola ESTs (34) enabled us to identify three Gα-encoding genes and one Gβ-encoding gene. Assembly of individual reads of these ESTs combined with sequencing of full-length clones resulted in identification of the genes MgGpa1, MgGpa2, MgGpa3, and MgGpb1, with 1,059-, 1,065-, 1,068-, and 1,050-bp open reading frames encoding proteins consisting of 353, 355, 356, and 350 amino acids, respectively. Screening of the recently released genome sequence of M. graminicola IPO323 (U.S. Department of Energy Joint Genome Institute [http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html]) revealed no additional Gα- and Gβ-encoding genes, but we identified an expected putative Gγ protein-encoding gene, showing 60% identity with the Gγ subunit of Aspergillus fumigatus (GenBank accession number XP_750270), which was not present in the EST database (34) and hence was not considered in this project.
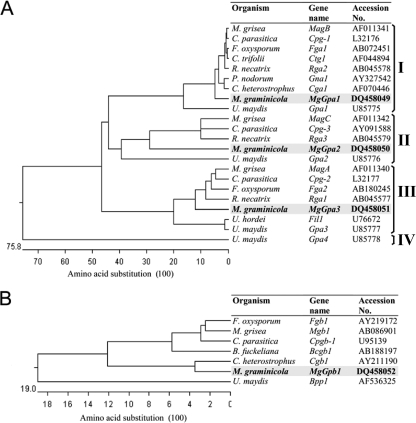
Comparison of proteins encoded by these genes with orthologs in M. grisea showed that MgGPA1, MgGPA2, MgGPA3, and MgGPB1 are the orthologs of MAGB, MAGC, MAGA, and MGB1, with 89, 76, 87, and 88% similarity, respectively. Analyses of the genomic DNA sequences and the EST contigs of these genes revealed that MgGpa1, MgGpa2, MgGpa3, and MgGpb1 contain one, four, five, and four introns, respectively. Multiple sequence alignments of the fungal Gα protein sequences resulted in a phylogenetic tree with three major groups within the superfamily of the Gα proteins (Fig. 2A). All Gα proteins (MgGPA1, MgGPA2, and MgGPA3) contain the characteristic P-loop motif, which is one of the ATP/GTP binding regions (G/AXXXXGKT/S), and a G motif that stabilizes the guanine ring (24, 55). MgGPA1 belongs to major group I and contains the consensus myristoylation site, indicated as MGX (an uncharged residue), XX (small uncharged residues), S at the N terminus, and a consensus CXXX sequence at the C terminus, which is the fingerprint of the pertussis toxin labeling site (17, 60). Multiple sequence alignments showed that MgGPA2 has no mammalian counterpart and belongs to group II, which is a distinct group lacking the consensus myristoylation site (49). MgGPA3 belongs to major group III, which has only the consensus myristoylation sequence, which is characteristic for adenylyl cyclase-stimulating G proteins (Gαs).
FIG. 2.
Phylogenetic comparison of orthologs of Gα proteins (A) and Gβ proteins (B), based on amino acid sequence alignments. The protein sequences encoded by MgGpa1 (GenBank accession number DQ458049), MgGpa2 (GenBank accession number DQ458050), MgGpa3 (GenBank accession number DQ458051), and MgGpb1 (GenBank accession number DQ458052) from Mycosphaerella graminicola were compared with orthologs present in other fungi. Multiple sequence alignments and the phylogenetic tree were constructed using the MEGALIGN program (DNAStar, Madison, WI).
Protein comparisons via an NCBI conserved domain search showed that MgGPB1 contains seven WD40 domains, which are characteristic of a family of ancient regulatory proteins (51). WD40 repeats in M. graminicola generally contain a C-terminal GH dipeptide, an inter-WD40 region of 13 to 29 amino acids, and an N-terminal WD dipeptide, representing the WD40 signature. However, there is variation in either one or both GH/WD dipeptides. The second WD40 repeat (residues 104 to 134) in MgGPB1 is the most variable domain among fungi and contains LR/YN at the C/N termini in M. graminicola instead of GH/WD. The other WD40 domains are highly conserved in one or both of the C/N termini.
Transposon-mediated vector construction and targeted gene disruption.
The disruption constructs for MgGpa1, MgGpa2, MgGpa3, and MgGpb1 were generated using an in vitro transposition system as described previously (44). PCR screening and sequencing enabled the selection of constructs with transposon insertions downstream of the start codon in the open reading frames of MgGpa1, MgGpa2, MgGpa3, and MgGpb1, at 666, 772, 393, and 586 bp, respectively (Fig. 1A). We used these constructs to disrupt MgGpa1, MgGpa2, MgGpa3, and MgGpb1 in the WT through A. tumefaciens-mediated transformation (44, 72). After transformation, homologous recombinants were identified by PCR screening (Fig. 1B). Among 28, 25, 92, and 47 hygromycin-resistant transformants for MgGpa1, MgGpa2, MgGpa3, and MgGpb1, we identified 3, 14, 3, and 2 disruptant strains, respectively, and 12 transformants were selected for further studies (Table 1).
Phenotypic characteristics.
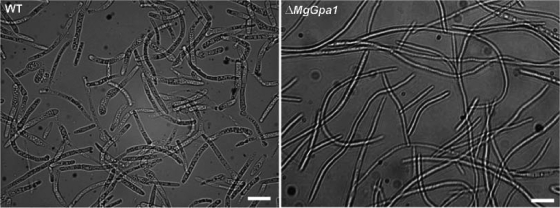
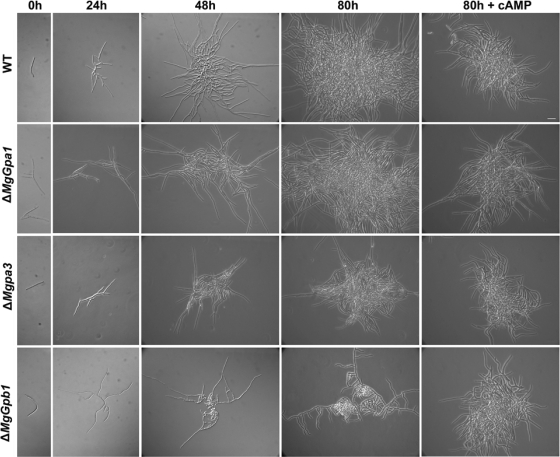
The MgGpa1, MgGpa3, and MgGpb1 mutants showed altered phenotypes on PDA at 20°C under continuous light (Fig. 3A) and darkness (not shown), whereas the phenotypes of MgGpa2 mutants were identical to those of the controls under all conditions tested (not shown). The colonies of the WT and ectopic transformants started to melanize after incubation for 4 to 5 days and were completely melanized and covered with a thin layer of white aerial mycelium 10 days after incubation, whereas the colonies of MgGpa1 and MgGpb1 mutants were nonmelanized and showed fimbriated borders (Fig. 3A). Spore size measurements showed that the MgGpa1 disruptants produced significantly longer spores in YGB than those of the WT and ectopic controls (Table 2; Fig. 4) (WT and ectopic control spores were 32.2 μm long, whereas transformant spores were ≥62.8 μm long; P < 0.05). The growth rate of the MgGpb1 mutant was significantly reduced (∼25% colony area, with fimbriated borders 56% of those of the WT after 10 days), whereas the MgGpa3 mutant showed only a slight growth reduction and delayed melanization. In addition, the MgGpa3 mutant exhibited an extensive yeast-like growth in the center of the colonies, which was characterized by an intensive microconidiation and a concatenation of budding cells (Fig. 3A). Interestingly, such a tendency to yeast-like growth and reduced filamentation was also observed in the MgTpk2 mutant, in which the catalytic subunit of PKA was disrupted (43). At 28°C, the growth patterns and phenotypes of the WT and ectopic transformant controls and the mutants were nearly similar and were characterized by a fluffy white aerial mycelium that covered the colony (not shown). At temperatures of >29°C, all colonies stopped growing, including the WT and ectopic transformant controls (not shown). In YGB, controls and all mutants merely produced yeast-like cells, except for the MgGpa1 mutants, which produced fluffy mycelia (Fig. 3B). MgGpa3 mutants secreted a dark brown pigment into YGB (Fig. 3C), similar to our previous observation with the MgTpk2 mutant (43).
FIG. 3.
Phenotypic characterization of Gα- and Gβ-encoding gene mutants, strain Mycosphaerella graminicola IPO323 (WT), and ectopic transformants (named with an E prefix) grown in vitro under different culture conditions. (A) Phenotypes of 10-day-old cultures grown on PDA without and with exogenous cAMP (13 mM) under continuous light at 20°C. (B) Highly increased filamentous growth of MgGpa1 mutants in YGB medium at 18°C after 5 days. (C) MgGpa3 mutants secrete dark brown pigments, as shown for 5-day-old YGB cultures grown at 18°C.
TABLE 2.
Comparison of microscopic phenotypes (24 to 80 h) and intracellular cAMP levels in Mycosphaerella graminicola mutants of G protein-encoding genes versus the M. graminicola IPO323 WT and ectopic controls
| Mycosphaerella graminicola isolate | Intracellular cAMP value (pmol cAMP/ mg total protein)a | Phenotype on PDA | Phenotype on PDA plus cAMP |
|---|---|---|---|
| Controls | |||
| IPO323 | Short germ tubes | Short germ tubes | |
| IPO323 E-MgGpa | 10.17c | Budding on apical and intermediate cells of spores and secondary filaments (24 h) | Budding on apical and intermediate cells of spores and secondary filaments (24 h) |
| IPO323 E-MgGpb | Filamentation (48 h) | Reduced filamentation (48 h) | |
| Microconidiation in center of colony (80 h) | Microconidiation in center of colony (80 h) | ||
| Mutants | |||
| IPO323 ΔMgGpa1 | 9.45bc | Long germ tubes | Reduced filamentation |
| Increased filamentation, reduced branching (48 to 80 h) | |||
| IPO323 ΔMgGpb1 | 5.48b | Fused germ tubes (48 h) | Restored to control phenotype |
| Uncontrolled anastomosis (80 h) | |||
| IPO323 ΔMgGpa3 | 1.68a | Short germ tubes | Restored to control phenotype |
| No secondary and tertiary filaments | |||
| Extensive budding |
Back-transformed values. Values with the same letter are not significantly different at a P level of 0.05.
FIG. 4.
Comparative phenotyping of Mycosphaerella graminicola mutants of the Gα-encoding gene MgGpa1 and of M. graminicola IPO323 (WT) in YGB at 20°C under daylight settings. The MgGpa1 disruptants produced significantly (P < 0.05) longer spores in YGB. Bar = 30 μm (also see the text for further details).
Complementation of mutants with cAMP.
To test whether the G proteins are involved in the cAMP-dependent signaling pathway, we supplemented PDA with exogenous cAMP (Fig. 3A). This had pleiotropic effects on the WT and all mutants. In the WT, cAMP complementation had multiple effects on appearance, filamentation, and growth rate, resulting in a smaller colony (∼36% of the WT colony area) that was covered with white aerial mycelium. Exogenous cAMP restored the phenotypes of all mutants to the above-mentioned WT phenotype (Fig. 3A).
Microscopic analyses.
We monitored germination, early colony formation, and conidiation in all mutants in the absence and presence of cAMP to further explore the aforementioned phenotype restoration by exogenous cAMP (Table 2; Fig. 4 and 5). On WA, the controls initially germinated from both apical cells of the spores, producing relatively long primary germ tubes within 12 h. Subsequently, secondary germ tubes from the same or other cells of the spore were formed. Within 48 h, tertiary hyphal filaments developed from the primary and secondary germ tubes. We did not observe significantly altered initial germination patterns for the MgGpa1, MgGpa2, and MgGpa3 mutants. However, at later stages, MgGpa1 mutants produced longer germ tubes than the WT or ectopic transformants did. MgGpb1 mutants grew significantly slower, with wavy germ tubes, and secondary and tertiary filaments as well as some hyphae showed anastomosis after growth for 80 h (data not shown).
FIG. 5.
Effects of disruption of the Gα- and Gβ-encoding genes MgGpa1, MgGpa3, and MgGpb1 on germination (24 h), early colony formation (48 h), and microconidiation (80 h) and restoration of the Mycosphaerella graminicola IPO323 control (WT) phenotype of the ΔMgGpa3 and ΔMgGpb1 mutants by adding exogenous cAMP (15 mM) to PDA at 20°C in the dark. Bar = 30 μm (also see the text for further details).
On PDA, the WT and ectopic controls formed shorter germ tubes that often showed differentiated budding cells on apical and intermediate cells of the spore as well as on secondary filaments within 24 h. These extended into filamentous colonies at 48 h, and these showed abundant microconidiation in the colony center after 80 h. Compared to the WT, the MgGpa1 mutants showed longer germ tubes and increased filamentation, with altered polarized growth, reduced branching (>24 h), and long uniform spores (>48 h) (Table 2; Fig. 4). MgGpa3 disruptant spores produced short germ tubes without secondary or tertiary filaments (>24 h). Colony formation was associated with extensive budding, resulting in dense colonies of typical uniform spores (>48 h) (Table 2; Fig. 5). The MgGpb1 mutant produced germ tubes that fused within 48 h and subsequently produced an extremely dense biomass between 48 and 80 h, which resulted from seemingly uncontrolled continuous anastomosis (Table 2; Fig. 5 and 6). This process greatly suppressed filamentous growth and significantly reduced microconidiation. The addition of cAMP to the medium reduced the colony size of the WT and MgGpa1 mutants but hardly affected the growth characteristics of the latter. For the MgGpa3 and MgGpb1 mutants, exogenous cAMP resulted in the formation of colonies that were very similar to the WT, suggesting that these genes may function in the cAMP pathway.
FIG. 6.
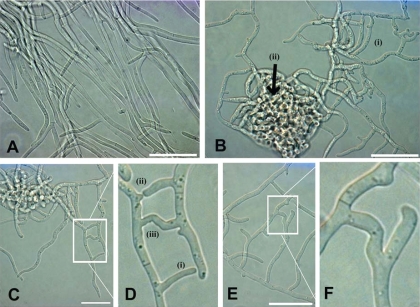
Disruption of MgGpb1 induces anastomosis. (A) Hyphae of the Mycosphaerella graminicola IPO323 WT strain grown on PDA for 80 h at 20°C in continuous darkness. (B) Extensive anastomosis in MgGpb1 mutants results in a network of interconnected hyphae (i) that show uncontrolled fusions producing dense local networks of mycelium (ii). (C and D) Three distinct phases are depicted during the anastomosis process, including (i) attraction phase, (ii) contact phase, and (iii) fusion phase. Panel D shows a higher magnification of the box in panel C. (E and F) Septum development after fusion. Panel F shows a higher magnification of the box in panel E. Bar = 30 μm.
Intracellular cAMP measurements.
In order to strengthen the macroscopic (Fig. 3) and microscopic (Fig. 5 and 6) observations in the presence and absence of exogenous cAMP, we performed intracellular cAMP measurements on all mutants. The MgGpa3 and MgGpb1 mutants showed significantly reduced levels of cAMP. In contrast, internal cAMP measurement data for the MgGpa1 mutant and the WT were not significantly different (Table 2).
In planta characterization of mutants.
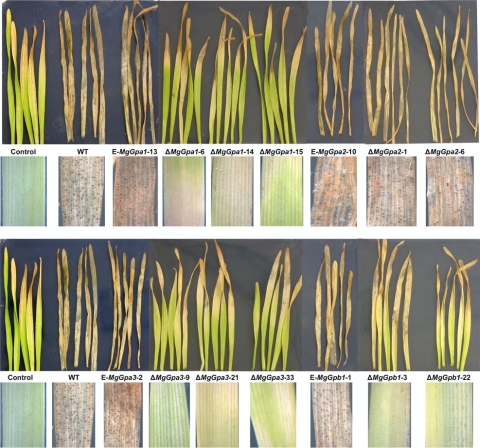
Ten-day-old seedlings of wheat cv. Obelisk inoculated with the WT or ectopic transformants showed chlorotic areas at 8 dpi that continued to expand starting from the leaf tips, which became necrotic at 12 dpi and eventually merged into large necrotic lesions containing numerous pycnidia between 16 and 18 dpi. Plants inoculated with MgGpa1, MgGpa3, or MgGpb1 mutants showed delayed symptom development, with only limited chlorotic areas, starting from the leaf tips, between 12 and 15 dpi. These chlorotic areas sometimes became necrotic but never contained pycnidia, the typical symptom of M. graminicola infection. On the leaf tips of water-treated controls, some chlorosis or necrosis was also observed, most likely as a result of senescence. An overview of disease symptoms caused by the WT and various disruptants and ectopic mutants and a summary of phenotypes of mutants of Gα- and Gβ-encoding genes in other filamentous plant-pathogenic fungi are shown in Fig. 7 and Table 3.
FIG. 7.
In planta phenotyping of MgGpa1, MgGpa2, MgGpa3, and MgGpb1 disruptants, the Mycosphaerella graminicola IPO323 WT strain, and ectopic transformants on 10-day-old seedlings of wheat cv. Obelisk 20 days after inoculation. Primary leaves were inoculated with either water (control) or the different M. graminicola strains. Note the strongly reduced pathogenicity for ΔMgGpa1, ΔMgGpa3, and ΔMgGpb1 mutants.
TABLE 3.
Effect of disruption of Gα- and Gβ-encoding genes on the development and pathogenicity of fungal plant pathogens
| Organism | Gene | Class | Growth | Conidiation | Germination | Virulence | Additional phenotypes | Reference |
|---|---|---|---|---|---|---|---|---|
| C. heterostrophus | Cga1 | Gα-I | Reduced | Reduced | Normal | Low appressorium formation, osmosensitive | 24 | |
| C. trifolii | Ctg1 | Gα-I | Reduced | Reduced | Reduced | Low appressorium formation, long germ tubes bearing appressoria | 65 | |
| C. parasitica | Cpg-1 | Gα-I | Reduced | Lost | Lost | Reduced pigmentation, increased cAMP level | 16 | |
| F. oxysporum | Fga1 | Gα-I | Normal | Reduced | Reduced | Decreased cAMP level, thermotolerant | 27 | |
| M. grisea | MagB | Gα-I | Reduced | Reduced | Normal | Reduced | Reduced penetration, long germ tubes bearing appressoria that are restored by exogenous cAMP | 41 |
| S. nodorum | Gna1 | Gα-I | Normal | Lost | Reduced | Defect in penetration, albino, osmosensitive | 61 | |
| U. maydis | Gpa1 | Gα-I | Normal | Normal | Normal | Normal | No phenotype in all aspects | 57 |
| B. cinerea | Bcg1 | Gα-I | Reduced | Normal | Normal | Reduced | Altered colony morphology, normal penetration, loss of protease secretion, | 22 |
| M. graminicola | Gpa1 | Gα-I | Normal | Reduced | Normal | Reduced | Altered colony morphology, hampered melanization, induced filamentation, altered polarized growth | This study |
| C. parasitica | Cpg-3 | Gα-II | Not functionally analyzed | 53 | ||||
| M. grisea | MagC | Gα-II | Normal | Reduced | Normal | Normal | Normal appressorium formation | 41 |
| U. maydis | Gpa2 | Gα-II | Normal | Normal | Normal | Normal | No phenotype in all aspects | 57 |
| B. cinerea | Bcg2 | Gα-II | Normal | Normal | Normal | Reduced | Normal colony morphology, virulence slightly reduced | 22 |
| M. graminicola | Gpa2 | Gα-II | Normal | Normal | Normal | Normal | No phenotype in all aspects | This study |
| C. parasitica | Cpg-2 | Gα-III | Reduced | Reduced | Normal | Decreased cAMP level | 16 | |
| F. oxysporum | Fga2 | Gα-III | Normal | Normal | Lost | Thermotolerant | 29 | |
| M. grisea | MagA | Gα-III | Normal | Normal | Normal | Normal | No phenotype in all aspects | 41 |
| U. maydis | Gpa3 | Gα-III | Lost | Elongated cells in liquid media, defective in mating | 57 | |||
| M. graminicola | Gpa3 | Gα-III | Reduced | Increased | Normal | Reduced | Altered colony morphology, hampered filamentation, reduced melanization | This study |
| U. maydis | Gpa4 | Gα-IV | Normal | Normal | Normal | Normal | No phenotype in all aspects | 57 |
| F. oxysporum | Fgb1 | Gβ | Reduced | Reduced | Normal | Reduced | Altered colony morphology, faster conidial germination, decreased cAMP level, thermotolerant | 27 |
| M. grisea | Mgb1 | Gβ | Reduced | Reduced | Delayed | Lost | No appressorium formation, decreased cAMP level, more aerial hyphae, formation of fluffy colonies | 52 |
| C. parasitica | Cpgb-1 | Gβ | Increased | Reduced | Reduced | Reduced pigmentation | 31 | |
| C. heterostrophus | Cgb1 | Gβ | Normal | Lost | Lost | Impaired formation of appressoria, increased pigmentation, straight germination | 14 | |
| U. maydis | Bpp1 | Gβ | Normal | Increased filamentation and reduced budding growth, restored by adding exogenous cAMP | 48 | |||
| M. graminicola | Gpb1 | Gβ | Reduced | Reduced | Normal | Reduced | Altered colony morphology, hampered filamentation and melanization, increased anastomosis | This study |
DISCUSSION
The availability of a large EST library and verification with the genome sequence resulted in the identification and isolation of the Gα-encoding genes and the Gβ-encoding gene in M. graminicola. Gene replacement studies revealed that these G proteins are involved in the development and pathogenicity of this fungus. With the exception of MgGpa2, all genes are required for pathogenicity and play an important role in regulation of the dimorphic switch between filamentous and yeast-like growth, microconidiation, or melanization. Phylogenetic analyses indicated that the Gα-encoding genes belong to three different groups. The mammalian orthologs of MgGPA1 and MgGPA3 are Gαi and Gαs, respectively, whereas MgGPA2 has no close relative in mammals. Activation of the Gαi protein in mammals results mostly in inhibition of adenylyl cyclase and hence increases intracellular cAMP levels (5, 6). However, in fungi, the orthologous genes encoding Gαi proteins vary substantially in their regulation of intracellular cAMP levels. For instance, the Cpg-1 gene in C. parasitica negatively regulates the cAMP level, as disruption increased intracellular cAMP levels in this fungus (5). In contrast, the orthologs Gna1 in Neurospora crassa, Fga1 in Fusarium oxysporum, and MagB in M. grisea positively regulate the cAMP pathway, as disruption of these genes decreased intracellular cAMP levels (26, 28). In M. grisea, exogenous cAMP restored appressorium formation and pathogenicity of the MagB mutant (41). In the budding yeast, Gpa1 regulates the MAPK pathway and does not affect the cAMP pathway (3, 59).
Our data show that MgGpa1 is a negative regulator of filamentation, as mutants had significantly longer spores than the WT did. Moreover, phenotypes in YGB and on PDA were characterized by increased filamentous growth, and those on WA were characterized by longer germ tubes. However, the addition of exogenous cAMP hardly affected the phenotypes 80 h and 10 days after initiation of the experiments. Moreover, intracellular cAMP levels in MgGpa1 mutants and the WT were not significantly different. This suggests that MgGpa1 might be dispensable for cAMP regulation, similar to reports for yeast (3, 59). Alternatively, equal intracellular cAMP levels in both the MgGpa1 mutant and the WT strain could also be due to growth characteristics that may influence cAMP extraction efficiencies. For instance, all mutants and controls showed blastic conidiogenesis in YGB, except for MgGpa1 mutants, which showed highly increased filamentous growth. It is possible that such differences in growth pattern affected the intracellular cAMP measurements in the MgGpa1 mutants. In summary, our data suggest that MgGPA1 is functionally different from mammalian inhibitory Gαi proteins and that fungal Gαi-like proteins comprise three subgroups that can either negatively or positively regulate adenylyl cyclase activity or, similar to what has been observed in yeast, do not interfere with the cAMP pathway. This also shows that high protein sequence identities do not necessarily imply similar biological functions.
Gα proteins that belong to group III show high protein sequence identity to stimulatory Gα proteins in N. crassa, C. parasitica, S. cerevisiae, and Schizosaccharomyces pombe. Unlike the Gαi-like proteins, they all positively regulate adenylyl cyclase and are functionally related to mammalian Gαs (16, 25, 33, 39, 42, 50). Significantly reduced intracellular cAMP levels in the MgGpa3 and MgGpb1 mutants confirmed our observation that the addition of exogenous cAMP to these mutants restored their phenotypes to that of the WT and hence suggest that MgGpa3 and MgGpb1 positively regulate adenylyl cyclase activity in M. graminicola. In many biological systems, cAMP is generated by adenylyl cyclase and acts as a second messenger that binds to the regulatory subunit of PKA. This leads to dissociation of the regulatory and catalytic subunits of this complex and consequently activates the latter. Hence, inhibition of adenylyl cyclase by disruption of Gαs genes or the catalytic subunit-encoding gene of PKA could lead to a similar phenotype. Indeed, the MgGpa3 and MgTpk2 (catalytic subunit-encoding gene of PKA) mutants of M. graminicola showed similar phenotypes, including reduced filamentation, increased microconidiation on PDA, and secretion of a dark brown pigment into YGB (43). Therefore, we suggest that MgGPA3 acts upstream of MgTPK2, positively regulates the cAMP pathway, and functionally belongs to the Gαs protein family.
Since MgGPA2 belongs to a distinct group of G proteins (group II) with no mammalian orthologs, we cannot predict its biochemical function yet. Interestingly, the phenotypes of all MgGpa2 mutants were identical to that of the WT or ectopic transformants under all in vitro and in planta conditions tested, which supports observations with U. maydis, Botrytis cinerea, and M. grisea where disruption of the MgGpa2 orthologs did not alter or only slightly affected in vitro phenotypes and pathogenicity (22, 41, 57). The phenotypes of all fungal Gα2-encoding gene mutants belonging to group II obtained so far indicate that this type of Gα protein does not play a major role in development and/or pathogenicity of plant-pathogenic fungi. Further studies are required to understand the role of the GPA2 subunits that are both specific to and highly conserved in filamentous fungi.
Spores of MgGpb1 mutants germinated normally, but the germ tubes had a wavy appearance and showed anastomosis on WA and PDA, which culminated on the latter medium into extensive fusions, resulting in dense areas in the colonies with little filamentous growth and strongly reduced conidiation. Although somatic cell fusion, known as homokaryon anastomosis, is common during vegetative growth of many filamentous fungi (18, 19, 23), it is very unusual for M. graminicola. Here we show that in M. graminicola, MgGpb1 negatively regulates vegetative cell fusion, resulting in its unique phenotype. The molecular mechanism of homokaryon cell fusion is poorly understood, but N. crassa ham-1 and ham-2 mutants were unable to undergo both self- and nonself-hyphal fusion during vegetative growth (69, 70). In S. cerevisiae and Cryptococcus neoformans, Gβ positively regulates the mating pheromone response pathway. Therefore, disruption of the encoding gene inhibits the response to sex pheromones (67, 68). In contrast, in S. pombe pheromone signaling is mediated by Gα (Gpa1) and the cAMP pathway is regulated by Gα (Gpa2) and Gβ (Gpb1). Interestingly, deletion of Gβ in S. pombe stimulates conjugation in nutrient-rich media, which is in accordance with our observations (37, 40). We showed that exogenous cAMP restored the phenotypes of the MgGpa3 and MgGpb1 mutants to the WT phenotype, indicating that these genes positively regulate the cAMP pathway. A possible mechanism for cAMP pathway regulation by MgGPB1 and MgGPA3 in M. graminicola could be that MgGPB1 is required for the efficient release of MgGPA3 from the heterotrimeric complex in order to stimulate adenylyl cyclase. Alternatively, or in addition, MgGPB1 could directly stimulate adenylyl cyclase, as observed for type II mammalian adenylyl cyclase, which is positively regulated by both Gα and Gβγ subunits (13, 15, 64).
We observed that all M. graminicola strains were equally thermosensitive, whereas for N. crassa and F. oxysporum, thermotolerance was observed in Gα-encoding mutants and was positively correlated with decreased intracellular cAMP levels (28, 29, 71). Indeed, constitutive expression of Gna1 in N. crassa resulted in elevated cAMP levels and increased thermosensitivity (26, 71). The observed phenotypic differences could be due to the various lifestyles of these fungi.
Similar to MgGpa1, the orthologous group I Gα proteins in C. parasitica, M. grisea, Stagonospora nodorum, F. oxysporum, Colletotrichum trifolii, and B. cinerea affect pathogenicity (7, 16, 28, 41, 61, 65), whereas the orthologous proteins CGA1 and GPA1 in Cochliobolus heterostrophus and U. maydis, respectively, are dispensable for pathogenicity (24, 57). Such a functional dichotomy was also observed for group III Gα orthologs. M. graminicola MgGpa3 mutants were strongly reduced in pathogenicity, which was also observed in U. maydis and F. oxysporum (29, 57) but not in M. grisea and C. parasitica (7, 41). Similar observations were reported for Gβ-encoding genes. MgGpb1 mutants and orthologous mutants in F. oxysporum, M. grisea, C. parasitica, and C. heterostrophus were impaired in pathogenicity, whereas mutation of the Gβ-encoding gene in U. maydis did not disrupt pathogenicity (14, 27, 31, 48, 52).
Recently, we showed that impaired melanization in M. graminicola correlated with defects in pathogenicity for MgFus3, MgSlt2, and MgHog1 mutants (8, 44, 45), which is in agreement with our present results, where we observed that disruption of the MgGpa1, MgGpb1, and MgGpa3 genes affects melanization as well as pathogenicity. This is somewhat surprising, as M. graminicola penetrates its host by stomata, whereas melanization is reported to be very important for generating turgor pressure in the appressoria of appressorium-forming fungal plant pathogens like M. grisea and Colletotrichum lagenarium (32, 38, 46, 63). The melanin biosynthesis pathway in fungi is also well characterized (47, 66), and expression of three melanin biosynthesis genes was reduced only at the onset of germination in the Fus3 orthologous mutants of C. lagenarium, suggesting the involvement of this ortholog in melanization of appressoria (62). However, unlike the case in appressorium-forming pathogens, melanization in M. graminicola might affect other stages during infection of plants where melanization is important. Indeed, our previous studies revealed that nonmelanized mutants of MgSlt2 and the WT strain were equally effective in penetrating the host plant (44) but differed significantly in pathogenicity, which suggests that melanization is required at the time that the fungus starts to differentiate asexual fructifications in substomatal cavities (11, 36). However, to test this hypothesis, genes involved in melanin biosynthesis need to be studied in detail. In addition, expression profiling of the mutants generated in this study could possibly reveal potential cross talk between the melanin biosynthesis and cAMP pathways in M. graminicola.
Acknowledgments
Rahim Mehrabi was financially supported by the Agricultural Research and Education Organization (AREO) of Iran. The construction of EST libraries was funded by Syngenta, Jealott's Hill, United Kingdom. Gert H. J. Kema is a recipient of an OECD fellowship to USDA-ARS at Purdue University, West Lafayette, IN. Sarrah Ben M'Barek is cosponsored by a UNESCO-L'Oréal fellowship.
The Dutch Mycosphaerella group and Maarten A. De Waard are gratefully acknowledged for discussions and suggestions. Gert H. J. Kema gratefully acknowledges discussions with Steve Goodwin and Jin-Rong Xu on cell biological aspects of M. graminicola. We thank Jacques Withagen (Wageningen University and Research Center- Biometris) for statistical advice and Wouter Van Egmond and Peter Van Haastert at the University of Groningen for help with the cAMP assay. We thank two anonymous reviewers for useful suggestions that improved the manuscript.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Aimi, T., S. Kano, Q. Wang, T. Morinaga, and Q. A. Wang. 2001. Molecular cloning of three genes encoding G protein alpha subunits in the white root rot fungus, Rosellinia necatrix. Biosci. Biotechnol. Biochem. 65678-682. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L., G. Cook Jeanette, D. Voora, M. Baggott Daniel, R. Martinez Anthony, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 122887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boelker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25143-156. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B., S. Gao, G. H. Choi, and D. L. Nuss. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA 937996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childers, S. R., and S. A. Deadwyler. 1996. Role of cyclic AMP in the actions of cannabinoid receptors. Biochem. Pharmacol. 52819-827. [DOI] [PubMed] [Google Scholar]
- 7.Choi, G. H., B. Chen, and D. L. Nuss. 1995. Virus-mediated or transgenic suppression of a G-protein alpha subunit and attenuation of fungal virulence. Proc. Natl. Acad. Sci. USA 92305-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousin, A., R. Mehrabi, M. Guilleroux, M. Dufresne, T. Van der Lee, C. Waalwijk, T. Langin, and G. H. J. Kema. 2006. The MAP kinase-encoding gene MgFus3 of the non-appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7269-278. [DOI] [PubMed] [Google Scholar]
- 9.Delgado-Jarana, J., A.-L. Martinez-Rocha, R. Roldan-Rodriguez, M. I. G. Roncero, and A. Di-Pietro. 2005. Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signaling pathways. Fungal Genet. Biol. 4261-72. [DOI] [PubMed] [Google Scholar]
- 10.Dohlman, H. G., and J. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70703-754. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, K. E., and R. J. Howard. 2000. Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola. Mycol. Res. 1041074-1082. [Google Scholar]
- 12.Farr, D. F., G. F. Bills, G. P. Chamuris, and A. Y. Rossman. 1995. Fungi on plants and plant products in the United States. APS Press, St. Paul, MN.
- 13.Federman, A. D., B. R. Conklin, K. A. Schrader, R. R. Reed, and H. R. Bourne. 1992. Hormonal stimulation of adenylyl cyclase through Gi-protein beta gamma subunits. Nature 356159-161. [DOI] [PubMed] [Google Scholar]
- 14.Ganem, S., S. W. Lu, B. N. Lee, D. Y. T. Chou, R. Hadar, B. G. Turgeon, and B. A. Horwitz. 2004. G-protein beta subunit of Cochliobolus heterostrophus involved in virulence, asexual and sexual reproductive ability, and morphogenesis. Eukaryot. Cell 31653-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, B. N., and A. G. Gilman. 1991. Cloning and expression of a widely distributed (type IV) adenylyl cyclase. Proc. Natl. Acad. Sci. USA 8810178-10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein alpha subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 9314122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, S., and D. L. Nuss. 1998. Mutagenesis of putative acylation sites alters function, localization, and accumulation of a Gi alpha subunit of the chestnut blight fungus Cryphonectria parasitica. Mol. Plant-Microbe Interact. 111130-1135. [DOI] [PubMed] [Google Scholar]
- 18.Glass, N. L., D. J. Jacobson, and P. K. T. Shiu. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34165-186. [DOI] [PubMed] [Google Scholar]
- 19.Glass, N. L., C. Rasmussen, M. G. Roca, and N. D. Read. 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12135-141. [DOI] [PubMed] [Google Scholar]
- 20.Goedhart, P. W., and J. T. N. M. Thissen. 2001. Biometris Genstat procedure library manual. Biometris, Wageningen, The Netherlands.
- 21.Goodwin, S. B. 2004. Minimum phylogenetic coverage: an additional criterion to guide the selection of microbial pathogens for initial genomic sequencing efforts. Phytopathology 94800-804. [DOI] [PubMed] [Google Scholar]
- 22.Gronover, C. S., D. Kasulke, P. Tudzynski, and B. Tudzynski. 2001. The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 141293-1302. [DOI] [PubMed] [Google Scholar]
- 23.Hickey, P. C., D. J. Jacobson, N. D. Read, and N. L. Glass. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37109-119. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz, B. A., A. Sharon, S. W. Lu, V. Ritter, T. M. Sandrock, O. C. Yoder, and B. G. Turgeon. 1999. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 2619-32. [DOI] [PubMed] [Google Scholar]
- 25.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 62455-2462. [DOI] [PubMed] [Google Scholar]
- 26.Ivey, F., Q. Yang, and K. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a Gai homolog in Neurospora crassa. Fungal Genet. Biol. 2648-61. [DOI] [PubMed] [Google Scholar]
- 27.Jain, S., K. Akiyama, T. Kan, T. Ohguchi, and R. Takata. 2003. The G protein beta subunit FGB1 regulates development and pathogenicity in Fusarium oxysporum. Curr. Genet. 4379-86. [DOI] [PubMed] [Google Scholar]
- 28.Jain, S., K. Akiyama, K. Mae, T. Ohguchi, and R. Takata. 2002. Targeted disruption of a G protein alpha subunit gene results in reduced pathogenicity in Fusarium oxysporum. Curr. Genet. 41407-413. [DOI] [PubMed] [Google Scholar]
- 29.Jain, S., K. Akiyama, R. Takata, and T. Ohguchi. 2005. Signaling via the G protein alpha subunit FGA2 is necessary for pathogenesis in Fusarium oxysporum. FEMS Microbiol. Lett. 243165-172. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen, L. N., B. M. Secher, and H. Hossy. 1999. Decision support systems featuring Septoria management, p. 251-262. In J. A. Lucas, P. Bowyer, and H. M. Anderson (ed.), Septoria on cereals: a study of pathosystems. CABI Publishing, Wallingford, United Kingdom.
- 31.Kasahara, S., and D. L. Nuss. 1997. Targeted disruption of a fungal G-protein beta subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10984-993. [DOI] [PubMed] [Google Scholar]
- 32.Kawamura, C., J. Moriwaki, N. Kimura, Y. Fujita, S. I. Fuji, T. Hirano, S. Koizumi, and T. Tsuge. 1997. The melanin biosynthesis genes of Alternaria alternata can restore pathogenicity of the melanin-deficient mutants of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10446-453. [DOI] [PubMed] [Google Scholar]
- 33.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Galpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 207693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kema, G. H. J., T. A. J. Van der Lee, O. Mendes, E. C. P. Verstappen, R. Klein Lankhorst, H. Sandbrink, A. Van der Burgt, L.-H. Zwiers, M. Csukai, and C. Waalwijk. 2008. Large scale gene discovery in the Septoria tritici blotch fungus Mycosphaerella graminicola with a focus on in planta expression. Mol. Plant-Microbe Interact. 211249-1260. [DOI] [PubMed] [Google Scholar]
- 35.Kema, G. H. J., and C. H. Van Silfhout. 1997. Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. III. Comparative seedling and adult plant experiments. Phytopathology 87266-272. [DOI] [PubMed] [Google Scholar]
- 36.Kema, G. H. J., D. Yu, F. H. J. Rijkenberg, M. W. Shaw, and R. P. Baayen. 1996. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology 86777-786. [Google Scholar]
- 37.Kim, D. U., S. K. Park, K. S. Chung, M. U. Choi, and H. S. Yoo. 1996. The G protein beta subunit Gpb1 of Schizosaccharomyces pombe is a negative regulator of sexual development. Mol. Gen. Genet. 25220-32. [DOI] [PubMed] [Google Scholar]
- 38.Kubo, Y., H. Nakamura, K. Kobayashi, T. Okuno, and I. Furusawa. 1991. Cloning of a melanin biosynthetic gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 4440-445. [DOI] [PubMed] [Google Scholar]
- 39.Kuebler, E., H. U. Moesch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 27220321-20323. [DOI] [PubMed] [Google Scholar]
- 40.Landry, S., M. T. Pettit, E. Apolinario, and C. S. Hoffman. 2000. The fission yeast git5 gene encodes a Gbeta subunit required for glucose-triggered adenylate cyclase activation. Genetics 1541463-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, S., and R. A. Dean. 1997. G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 101075-1086. [DOI] [PubMed] [Google Scholar]
- 42.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 167008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehrabi, R., and G. H. J. Kema. 2006. Protein kinase A subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Mol. Plant Pathol. 7656-677. [DOI] [PubMed] [Google Scholar]
- 44.Mehrabi, R., T. van der Lee, C. Waalwijk, and G. H. J. Kema. 2006. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant-Microbe Interact. 19389-398. [DOI] [PubMed] [Google Scholar]
- 45.Mehrabi, R., L.-H. Zwiers, M. A. de Waard, and G. H. J. Kema. 2006. MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant-Microbe Interact. 191262-1269. [DOI] [PubMed] [Google Scholar]
- 46.Money, N. P., and R. J. Howard. 1996. Confirmation of a link between fungal pigmentation, turgor pressure, and pathogenicity using a new method of turgor measurement. Fungal Genet. Biol. 20217-227. [Google Scholar]
- 47.Moriwaki, A., J. Kihara, T. Kobayashi, T. Tokunaga, S. Arase, and Y. Honda. 2004. Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 2381-8. [DOI] [PubMed] [Google Scholar]
- 48.Muller, P., A. Leibbrandt, H. Teunissen, S. Cubasch, C. Aichinger, and R. Kahmann. 2004. The Gb subunit-encoding gene bpp1 controls cyclic AMP signaling in Ustilago maydis. Eukaryot. Cell 3806-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mumby, S., C. Kleuss, and A. Gilman. 1994. Receptor regulation of G-protein palmitoylation. Proc. Natl. Acad. Sci. USA 912800-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakafuku, M., T. Obara, K. Kaibuchi, I. Miyajima, A. Miyajima, H. Itoh, S. Nakamura, K. Arai, K. Matsumoto, and Y. Kaziro. 1988. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. USA 851374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neer, E. J., C. J. Schmidt, R. Nambudripad, and T. F. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371297-300. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura, M., G. Park, and J. R. Xu. 2003. The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol. Microbiol. 50231-243. [DOI] [PubMed] [Google Scholar]
- 53.Parsley, T. B., G. C. Segers, D. L. Nuss, and A. L. Dawe. 2003. Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third G alpha homologue. Curr. Genet. 4324-33. [DOI] [PubMed] [Google Scholar]
- 54.Payne, R. W., and P. W. Lane. 1993. Genstat 5 release 3 reference manual. Clarendon Press, Oxford, United Kingdom.
- 55.Pennington, S. R. 1994. GTP-binding proteins. 1. Heterotrimeric G proteins. Protein Profiles 1169-233. [PubMed] [Google Scholar]
- 56.Reference deleted.
- 57.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Boelker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 161934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Schwartz, M. A., and H. D. Madhani. 2004. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38725-748. [DOI] [PubMed] [Google Scholar]
- 60.Simon, M. I., M. P. Strathmann, and N. Gautam. 1991. Diversity of G proteins in signal transduction. Science 252802-808. [DOI] [PubMed] [Google Scholar]
- 61.Solomon, P. S., K. C. Tan, P. Sanchez, R. M. Cooper, and R. P. Oliver. 2004. The disruption of a G alpha subunit sheds new light on the pathogenicity of Stagonospora nodorum on wheat. Mol. Plant-Microbe Interact. 17456-466. [DOI] [PubMed] [Google Scholar]
- 62.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13374-383. [DOI] [PubMed] [Google Scholar]
- 63.Talbot, N. J. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57177-202. [DOI] [PubMed] [Google Scholar]
- 64.Tang, W. J., and A. G. Gilman. 1991. Type-specific regulation of adenylyl cyclase by G protein beta gamma subunits. Science 2541500-1503. [DOI] [PubMed] [Google Scholar]
- 65.Truesdell, G. M., Z. Yang, M. B. Dickman, and Z. H. Yang. 2000. A Galpha subunit gene from the phytopathogenic fungus Colletotrichum trifolii is required for conidial germination. Physiol. Mol. Plant Pathol. 56131-140. [Google Scholar]
- 66.Tsuji, G., S. Tsuge, T. Shiraishi, and Y. Kubo. 2003. Expression pattern of melanin biosynthesis enzymes during infectious morphogenesis of Colletotrichum lagenarium. J. Gen. Plant Pathol. 69169-175. [Google Scholar]
- 67.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. L. MacKay. 1989. The Ste4 and Ste18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell 56467-477. [DOI] [PubMed] [Google Scholar]
- 69.Wilson, J. F., and J. A. Dempsey. 1999. A hyphal fusion mutant in Neurospora crassa. Fungal Genet. Newsl. 4631. [Google Scholar]
- 70.Xiang, Q., C. Rasmussen, and N. L. Glass. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Gai causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zwiers, L. H., and M. A. de Waard. 2001. Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 39388-393. [DOI] [PubMed] [Google Scholar]