Abstract
Aspergillus flavus, a mycotoxigenic filamentous fungus, colonizes several important agricultural crops, such as maize and peanuts. Two proteins, VeA and LaeA, known to form a nuclear complex in Aspergillus nidulans have been found to positively regulate developmental processes in several Aspergillus species. Here, an examination of near-isogenic A. flavus mutants differing in copy number of veA and laeA alleles (0, 1, or at least 2 each) revealed critical roles for VeA and LaeA in A. flavus development and seed colonization. In contrast to the wild type, both null mutants were unable to metabolize host cell lipid reserves and were inhibited by oleic acid in growth assays. The copy number of LaeA but not VeA appeared critical for a density-dependent sclerotial-to-conidial shift, since the multicopy laeA (MClaeA) strain produced relatively constant sclerotial numbers with increasing population size rather than showing the decrease in sclerotia seen in both the wild-type and MCveA strains. The MCveA-laeA strain yielded an intermediate phenotype. This study revealed unique roles of VeA and LaeA in seed pathogenesis and fungal biology, distinct from their cooperative regulatory functions in aflatoxin and sclerotial development.
Aspergillus flavus, an opportunistic pathogen of oil seeds, occurs as a saprophyte in soils worldwide and colonizes several important agricultural crops, such as maize, peanut, and cottonseed, before and after harvest (25, 29, 44). The pathogen generates asexual spores, conidia, as the source of inoculum and overwinters as sclerotia which germinate to produce conidia in the subsequent season (12). A. flavus and other aspergilli, such as Aspergillus parasiticus, can produce the polyketide-derived carcinogenic secondary metabolite aflatoxin (25, 33). In the United States, annual yield losses in the million-dollar range from aflatoxin contamination on peanut and maize crops are frequently reported (34). Aflatoxin-contaminated food and feed is also a major problem in developing countries, especially in Asia and Africa (45). Recently, an outbreak of aflatoxin poisoning from maize was reported to have killed a hundred people in Kenya (31). Therefore, measures to control Aspergillus infections and aflatoxin production are urgently needed to protect human and animal health.
The identification and characterization of molecules necessary for A. flavus conidial, sclerotial, and aflatoxin production are critical to develop rational control strategies. Recently, a heterotrimeric nuclear complex composed of three proteins, LaeA, VeA, and VelB, has been found to regulate sporulation and secondary metabolism in the related species Aspergillus nidulans (1). Although the role of VelB is not well defined, several studies have described the function of VeA and LaeA in the aspergilli. VeA is required for cleistothecial production in A. nidulans (24) and sclerotial production in both A. parasiticus (6) and A. flavus (13). In addition, the VeA gene regulates the expression of sterigmatocystin (a precursor of aflatoxin) and penicillin genes in A. nidulans (22) and aflatoxin genes in A. parasiticus (6) and A. flavus (13). The finding that VeA and LaeA are partnered in a transcriptional complex in A. nidulans (1) helps explain similarities in VeA and LaeA function. LaeA is a global regulator of secondary metabolite production in aspergilli, regulating the same set of metabolites as VeA in A. nidulans and A. flavus (2, 21), as well as dozens of putative toxins in the human pathogen Aspergillus fumigatus (32). LaeA is also necessary for sclerotial formation in A. flavus (21) and affects cleistothecial development in A. nidulans (J. W. Bok and N. P. Keller, unpublished data). Northern analysis shows that VeA and LaeA negatively regulate each other at the transcript level in A. nidulans (1) and LaeA negatively regulates veA in A. flavus (21), leading to the concept of a feedback mechanism maintaining morphological and secondary metabolite differentiation in the aspergilli.
Other factors have been reported which link morphological development with secondary metabolism. Of particular interest are a family of oxylipin-producing oxygenases (encoded by ppo and lox genes) which have been shown to balance ascospore and conidial production in A. nidulans (40, 41) and sclerotial and conidial production in A. flavus (19), as well as secondary metabolite production in both species (19, 38). Most recently, a density-dependent switch from sclerotial-to-conidial development in A. flavus was found to be affected by oxylipin production (18, 19). Both oxylipin production and the response to oxylipin signaling are dependent on an intact VeA protein (5, 7). VeA is also required for ppoA expression, and VeA-PpoA interactions affect both sexual and asexual development in A. nidulans (41).
The impact of the loss of these proteins on pathogenesis has been explored to some degree for LaeA and Ppo mutants but not yet reported for VeA. LaeA is a key determinant in aspergillosis caused by A. fumigatus and seed rot by A. flavus (3, 21), and Ppo loss impacts virulence attributes of A. fumigatus (11, 40), A. nidulans (39), and A. flavus (19). Considering the interdependence of oxylipin function with VeA coupled with the VeA-LaeA interaction, we postulated that VeA mutants would also be impaired in seed pathogenesis in a manner similar to that of LaeA mutants and, furthermore, that both mutants could be affected in density-dependent development. To explore these hypotheses, we created several A. flavus isogenic mutants differing only in copy number of veA and laeA genes, including ΔveA, ΔlaeA, multicopy laeA (MClaeA), and MCveA strains and a double MC strain (MCveA-laeA). The respective VeA and LaeA mutants exhibited critical differences in cell density responses and invasion of host tissues, despite gross similarities between sclerotial and aflatoxin production.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The Aspergillus flavus strains used and generated in this study are listed in Table 1. All strains were maintained as stocks in glycerol and grown at 29°C on glucose minimal medium (GMM) (36) amended with appropriate supplements for spore production.
TABLE 1.
Aspergillus flavus strains in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| NRRL 3357 | Wild type | 18 |
| NRRL 3357.5 | pyrG− | 18 |
| TSA 1.54 (ΔveA) | pyrG− ΔveA::AfpyrG | This study |
| TSA 2.46 (MCveA) | pyrG− AfpyrG veA | This study |
| TJW 71.1 (ΔlaeA) | pyrG− ΔlaeA::AfpyrG | 21 |
| TJW 79.13 (MClaeA) | pyrG− ΔlaeA::AfpyrG niaD−niaD laeA | 21 |
| TSA 2.8 (MCveA-laeA) | pyrG− AfpyrG veA niaD laeA | This study |
Af, A. fumigatus.
Fusion PCR and vector construction.
All primers used in this study are listed in Table 2. The veA replacement PCR products were constructed using fusion PCR following Szewczyk et al. (38). The 1.3-kb fragments upstream and downstream of the veA coding region were amplified by PCR with primers 5′F veA For and Rev for the upstream fragment and primers 3′F veA For and Rev for the downstream fragment, using NRRL 3357 (prototroph) genomic DNA as a template. Next, a 1.9-kb fragment of the pyrG auxotrophy marker gene was amplified from A. fumigatus AF293 genomic DNA using primers A. fumigatus pyrG For and Rev. These three amplified PCR products were cleaned with a QIAquick gel extraction kit (Qiagen), quantified, and fused using published procedures (38). The PCR product was amplified with primers Nested For and Rev (38). All PCR steps were performed using an Expand long template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. The final construct was confirmed with endonuclease digestion and PCR using primers Int veA For and Rev for internal veA and primers A. fumigatus pyrG For and Rev for pyrG. The veA complementation vector was constructed in two steps. First, the 1.9-kb A. fumigatus pyrG PCR fragment was amplified and ligated into the pCR2.1-TOPO vector (Invitrogen) to create pSA2.4. Next, a 4.4-kb SpeI fragment containing the A. flavus veA gene was amplified from A. flavus NRRL 3357 genomic DNA with primers MC veA For and Rev and ligated into the SpeI site of pSA2.4 to create the veA complementation vector, pSA3.13. The vector was confirmed by PCR with primers MC veA For and veA-pyrG Rev and endonuclease digestion.
TABLE 2.
Primer sequences in this study
| Primer | Sequencea (5′-3′) |
|---|---|
| 5′F veA For | ACAACCCTGGACTCTGGAAT |
| 5′F veA Rev | CGAAGAGGGTGAAGAGCATTGTTTGAGGCAGAGGACGCGTTGACTGTGATG |
| 3′F veA For | TGACGACAATACCTCCCGACGATACCTGGGTTGATTCCTGCTTTTCCTCC |
| 3′F veA Rev | TCTCGTTCTCCCATTTACCT |
| A. fumigatus pyrG For | TGCCTCAAACAATGCTCTTC |
| A. fumigatus pyrG Rev | CAAGGTATCGTCGGGAGGT |
| Nested For | AATCACGGACCTCGAAGCAG |
| Nested Rev | GGGGTCTTGATATGGCGAAT |
| Int veA For | CAACAAGACCGACATCACCTTC |
| Int veA Rev | CCATTCTTGGGATAGCTGCAAC |
| MC veA For | CAACGAACTAGTCCGCCTGCCCTTAACCTCCA |
| MC veA Rev | GCATACACTAGTCTCGCATGCCAGTGGATGGG |
| veA-pyrG Rev | CATCGGTTGACTACGCTCGCA |
| laeA-niaD For | GACCTGTGGTGAAACCTGAGG |
| veA Northern For | CTAGCTGGTCATTATTTGATCTCG |
| veA Northern Rev | GTTGTAGAGTGGACGATCATCATG |
| laeA Northern For | CCTTGTATGATGTATGTATGATGAGC |
| laeA Northern Rev | GACAGCGAAAGTGAAGAGGACATC |
| actin Northern For | GAAGCGGTCTGAATCTCCTG |
| actin Nothern Rev | ACAGTCCAAGCGTGGTATCC |
| aflR Northern For | AGAGTCTTCCTTCAGCCAGGTC |
| aflR Northern Rev | GTGGGGCTTTTCTTCATTCTCG |
Bold characters flag restriction enzyme (SpeI) site.
Fungal transformation procedure and mutant confirmation.
For fungal transformation, protoplasts were produced from freshly germinated conidia of NRRL 3357.5 (pyrG auxotroph) and transformed using a polyethylene glycol method (30). The final fusion PCR product (5 μg) was used for replacement of veA with pyrG after gel purification using a QIAquick gel extraction kit (Qiagen) to create strain TSA 1.54. The veA::pyrG vector, pSA3.13, was used alone or else cotransformed with pLRM11.1, a vector containing both laeA and niaD (21), to create MC strains with multiple copies of veA alone and MC strains with multiple copies of both veA and laeA (TSA 2.46 and TSA 2.8, respectively, were used for these studies).
Correct transformants were identified by analyzing genomic DNA using PCR screens followed by Southern analyses. Primers Int veA For and Rev, Nested For and Rev (4.3 kb for the wild type and 4.6 kb for transformants), and A. fumigatus pyrG For and Rev were used to identify pyrG replacement of veA. MC transformants were identified by PCR with primers MC veA For and veA-pyrG Rev and primers laeA-niaD For and laeA Northern Rev. Southern analysis was performed for each PCR-identified transformant to confirm single gene replacement of veA in TSA 1.54, at least 2 copies of veA in TSA 2.46, and at least 2 copies of veA and laeA in TSA 2.8. Probes were created with primers Nested For and Rev for the veA open reading frame (ORF) and primers laeA Northern For and Rev for the laeA ORF.
Northern analysis.
To examine the expression of veA and laeA transcripts, Northern analysis was performed. Fifty-milliliter amounts of liquid GMM were inoculated with 106 spores/ml of appropriate strains and incubated with shaking at 250 rpm at 29°C under dark conditions. After 48 h, the mycelium was collected and total RNA was extracted by using the Trizol method (Invitrogen). Blots were hybridized with a veA fragment amplified using the primers Northern For and Rev, an laeA fragment amplified using the primers Northern For and Rev, an actin fragment amplified using the primers actin Northern For and Rev, and an aflR fragment amplified using the primers aflR Northern For and Rev from NRRL3357 genomic DNA. Detection of signals was carried out with a Phosphorimager-SI (Molecular Dynamics).
Physiological experiments.
Conidial production, sclerotial formation, and colony diameter were measured for fungal strains following the methods of Horowitz Brown et al. (18, 19). Briefly, 8-ml amounts of 1.6% GMM plus 2% sorbitol agar were overlaid with 3-ml amounts of 0.7% agar GMM plus 2% sorbitol agar containing 102, 104, and 106 spores/plate of each A. flavus strain for culture. For conidial counts, three 1.5-cm plugs from each plate were homogenized in 5 ml of 0.01% Tween 80 (vol/vol) water, diluted to 1×, and counted with a hematocytometer. To visualize sclerotium formation, plates were sprayed with 70% ethanol to kill and wash away conidia. The exposed sclerotia were then collected, lyophilized, and weighed (dry weight per plate). Growth diameter was measured following a point inoculation of 5 μl of 106 spores/ml for each strain on 30 ml of 1.6% GMM. Cultures were grown at 29°C under continuous dark or light conditions for 3 days (conidia production), 7 days (sclerotia formation), and 3 and 6 days (colony diameter). Each treatment was replicated four times.
To assay for growth on different fatty acids, the wild-type, ΔlaeA, and ΔveA strains were examined for growth on (i) 20 mM hexanoic acid (6 C), 6 mM oleic acid (18 C), and 4.9 mM erucic acid (22 C) as the sole carbon source, with the fatty acids substituting for the glucose in GMM, or (ii) GMM supplemented with these same molarities of fatty acids, following the method of Maggio-Hall and Keller (27). Growth diameter was measured following a point inoculation of 5 μl of 106 spores/ml for each strain on 30 ml of medium. Each treatment was replicated four times. The experiment was repeated twice.
Seed infections.
For seed/fungal studies, two cultivars (SunRunner and FloRunner) of peanut (Arachis hypogaea) and one (Northup King N33-P3) of non-fungicide treatment maize (Zea mays L.) were used. All the steps were aseptically performed as described by Kale et al. (21). Briefly, mature peanuts (20 peanut cotyledons) and maize (10 seeds) were surface sterilized and inoculated with suspensions of 105 spores/ml of each respective strain, as well as with a water control (mock inoculation). Seeds were placed in 50-ml Falcon tubes containing either sterile water or the spore suspensions and shaken for 30 min in a rotary shaker at 50 rpm, after which they were placed in a high-humidity chamber. Peanut cotyledons were incubated for 3 days for peanut cultivar SunRunner or 5 days for cultivar FloRunnner at 29°C under dark conditions, and maize kernels for 3 days. All seed experiments were repeated three times.
Histological study.
Infected and control peanut cotyledons of cultivar SunRunner were collected after 3 days of inoculation and sliced with a razor blade into 2-cm pieces which were immersed in ice-cold fixative FAA (3.7% formaldehyde, 5% acetic acid, 47.5% ethanol in water) in vials with vacuum pressure for 30 min. Tissues were then removed, incubated with fresh FAA overnight, dehydrated through a tert-butanol series following the method of Cseke et al. (10), and embedded in paraffin (Paraplast Plus). Paraffin blocks were sectioned in 10-μm slices, and serial sections were placed on glass slides and incubated at 37°C at least overnight, until tissues adhered to the slides. Dewaxing of tissues and staining with Gomori methenamine-silver (26) were performed in the University of Wisconsin—Madison School of Veterinary Medicine histology services laboratory. For lipid staining in peanut tissues, Nile red (16) was applied to tissues following the method of Tsitsigiannis et al. (41). A tetramethyl rhodamine 5-isothiocyanate filter in a fluorescent microscope (Olympus BX-60 with 546-nm excitation and 585-nm emission filters) was used to observe Nile red-stained tissues.
Aflatoxin analysis. (i) Extraction from medium.
Eight-milliliter amounts of 1.6% GMM-2% sorbitol agar were overlaid with 3-ml amounts of 0.7% GMM agar plus 2% sorbitol agar containing 102, 104, and 106 spores/plate of each fungal strain. Cultures were grown for 3 days at 29°C under dark or light conditions. Three 1.5-cm plugs from each plate were homogenized in 3 ml of 0.01% Tween 80 (vol/vol) water and vortexed vigorously for 1 min. One milliliter of chloroform was added, and the sample vortexed and incubated at room temperature for 30 min. The mixture was vortexed again and then centrifuged for 15 min. The lower layer was collected, allowed to dry for 3 days, and then resuspended in 100 μl of chloroform, and 40 μl of the suspension was spotted onto TLC plates (Whatman, Maidstone, England) using a chloroform/acetone (95:5, vol/vol) solvent system. Each treatment was repeated three times.
(ii) Extraction from seed.
Peanut cotyledons and maize kernels inoculated as described above were collected in 50-ml Falcon tubes with the addition of 5 ml of 0.01% Tween 80 and vortexed vigorously for 1 min. One milliliter was removed from each sample for conidium counting prior to aflatoxin extraction. Five milliliters of acetone was then added to the samples, followed by shaking for 10 min in a rotary shaker at 150 rpm. Samples were allowed to stand for 5 min at room temperature, and then 5 ml of chloroform was added to each sample, followed by shaking for 10 min at 150 rpm. Samples were allowed to stand for an additional 10 min at room temperature, vortexed briefly, and centrifuged for 15 min at 2,000 rpm to collect the organic lower phase. This phase was placed in a new tube and then dried completely for 3 days. Five milliliters of 0.1 M NaCl methanol/water (55:45) and 2.5 ml of hexane were added to each tube, and the mixture vortexed vigorously at high speed for 1 min. Samples were centrifuged at 2,000 rpm for 5 min. The hexane layer was collected, the remaining aqueous phase was washed with 2.5 ml of hexane, and then the collection process repeated as described above. The hexane extracts were combined, allowed to dry, and then resuspended in 500 μl of chloroform, and 10 μl of each extract was separated on a silica gel TLC plate using the chloroform/acetone (95:5 vol/vol) solvent system. Each treatment was repeated three times.
Statistical analysis.
Statistical differences were analyzed using the JMP software package, version 3.2.6 (SAS Institute, Inc., Cary, NC). Multiple comparisons of results for all strains were calculated for growth diameter, lipase activity, and sporulation on seed. To assess the density-dependent development of each strain, sclerotial and conidial numbers were compared at three population levels. Statistically significant mean values, indicated with different letters in the figures, are significant at P < 0.05.
RESULTS
Creation of veA and laeA mutant strains in A. flavus.
This study required creating near-isogenic strains varying in the number of laeA and veA alleles in the same A. flavus isolate. As ΔlaeA and MC strains of the genome-sequenced strain A. flavus 3357 already existed (21), the first goal was to obtain near-isogenic strains of A. flavus 3357 with loss of or overexpression of veA.
The sequence of the A. flavus 3357 veA ortholog was obtained by designing primers from the A. flavus ATCC MYA384 veA gene (GenBank DQ296645) (13). The sequences of the two genes were found to be 99% identical. All primers and probes in this study were designed from this sequence (Table 2). Figure 1A shows the strategy of replacement of veA with A. fumigatus pyrG. Transformants were first screened for loss of production of sclerotia on GMM plus 2% sorbitol medium, a phenotype associated with the A. flavus ATCC MYA384 ΔveA mutant (13). Several asclerotial A. flavus 3357 transformants were identified and their DNA extracted and analyzed by PCR and Southern analysis. Seventeen out of 100 transformants were found to contain the 4.6-kb and 4.3-kb fragments expected of KpnI (Fig. 1B) and SapI (data not shown) digests, respectively, as expected for a veA replacement with A. fumigatus pyrG. One of these strains, TSA 1.54, was chosen for further studies (Fig. 1B). A strain with at least two copies of veA was obtained by transforming NRRL 3357.5 with plasmid pSA3.13. Several strains were obtained, as determined by Southern analysis, and one, the MCveA strain TSA 2.46, was chosen for further studies (Fig. 1B). Next, a strain with at least two copies of both veA and laeA was obtained by transforming NRRL 3357.5 with plasmids pSA3.13 and pLRM11.1. One of these transformants, the MCveA-laeA strain TSA2.8, was chosen for further studies (Fig. 1B).
FIG. 1.
Deletion, MCveA, and MClaeA mutants of A. flavus. (A) Diagram of the strategy of replacement of A. flavus NRRL 3357.5 veA with A. fumigatus AF293 wild-type pyrG gene shows the restriction enzyme digestion sites of KpnI for Southern analysis with veA probe. To confirm gene replacement or MC transformants using Southern analysis, at least two restriction enzymes for each probe were utilized, KpnI (K) and SapI (data not shown) for veA and HindIII (H) and BamHI (data not shown) for laeA. A. fumi, A. fumigatus. (B) Southern analysis. The KpnI digest shows 6.8-kb and 1.1-kb veA fragments in the wild type and 4.7-kb, 2.2-kb, and 1.1-kb fragments in the ΔveA strain. The MCveA strain shows both wild-type 6.8-kb and 1.1-kb fragments, as well as 3.6-kb and 0.3-kb (not shown) fragments. The laeA probe presented a 5.6-kb fragment in the wild type; 4.5-kb, 3.2-kb, and 1.7-kb fragments in the ΔlaeA strain; and several extra bands in the MClaeA strain. The laeA mutants have been described before, in reference 21. WT, wild type.
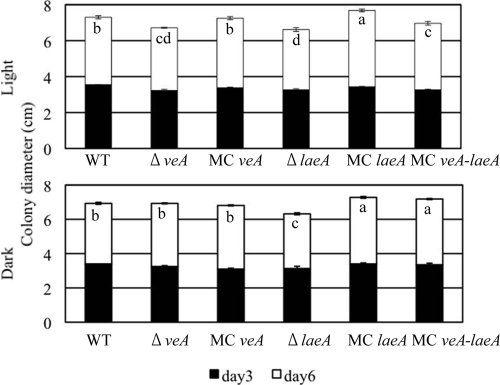
The strains with the six genotypes (the wild type and five mutants) exhibited clear differences in development and morphology, as described below, and additionally, the ΔlaeA strain showed a statistically significant inhibition in growth diameter compared to the growth of most other strains under both light and dark conditions. Conversely, the MClaeA strain's growth diameter was greater than the growth diameters of most other strains in both light and dark regimes (Fig. 2).
FIG. 2.
Colony diameters of veA and laeA mutants of A. flavus. A 5-μl amount of a suspension of 106 spores/ml of each strain was point inoculated on 30 ml of 1.6% GMM. Cultures were grown at 29°C under continuous dark or light conditions, and growth diameters measured at 3 and 6 days after inoculation. Letters indicate differences between strains that were statistically significant (P < 0.05) according to the Tukey-Kramer multiple comparison test. Error bars show the standard deviations of the results of four replications. Strains were grown in both light and dark conditions. WT, wild type.
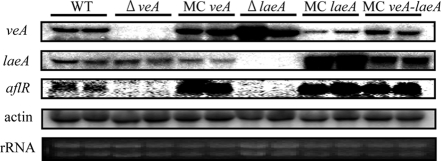
veA and laeA affect each other's transcription.
Kale et al. (21) recently found that laeA expression negatively affects transcription of veA in A. flavus; this result was replicated in our work (Fig. 3). We also found evidence for veA regulation of laeA expression. Although Northern analysis revealed that the ΔveA strain did not show an increase of laeA expression, the MCveA strain had decreased laeA expression compared to that of the wild type. The MCveA-laeA strain showed relatively high levels of expression of both veA and laeA but not as high as the individual MC strains.
FIG. 3.
Gene expression levels of veA and laeA in A. flavus mutants. Each strain was grown in liquid GMM culture with shaking (250 rpm at 29°C) under dark conditions. Total RNA was extracted from two replicates at 48 hrs after inoculation. Northern blots were probed with internal or ORF fragments of each gene (Table 2). rRNA and actin were the loading and expression controls. WT, wild type.
We also examined the expression of the aflatoxin-specific transcription factor aflR in all strains. As expected and as previously described (13, 15, 21), there was no aflR expression in ΔveA and ΔlaeA strains. Similarly to the MClaeA strain, both the MCveA and MCveA-laeA strain showed higher levels of aflR expression than the wild type with this treatment.
Conidial and sclerotial density-dependent production is affected by VeA and LaeA.
A recent study has shown that conidial and sclerotial production is density dependent in A. flavus, for which low cell densities resulted in high sclerotial formation and high cell densities in low sclerotial formation, with an inverse effect on conidial production (18). This quorum-like signaling system regulating the sclerotial-to-conidial shift was impaired in oxylipin-generating oxygenase mutants (18, 19). Because VeA has been shown to be important in oxylipin signaling responses (40, 41) and forms a complex with LaeA in the nucleus (1), we predicted that changes in veA and laeA expression could affect the density-dependent sclerotial-to-conidial shift.
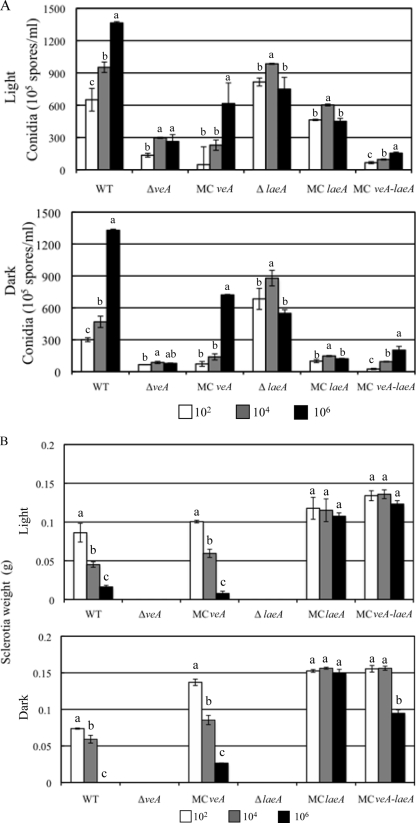
The relative abilities of the wild type and the veA and laeA mutants to form sclerotia and conidia were determined by inoculating 102, 104, and 106 conidia onto GMM plus 2% sorbitol plates which were placed in constant dark at 29°C for 3 (conidia) and 7 (sclerotia) days. Similar to prior results (18), sclerotial production diminished and conidial production increased in the wild type with increasing cell population levels (Fig. 4A and B). The veA and laeA null mutants were incapable of producing sclerotia at any population level and yielded relatively constant levels of conidial production regardless of population levels (Fig. 4A and B).
FIG. 4.
Effects of veA and laeA allele numbers on density-dependent conidial and sclerotial production in A. flavus. Each strain was grown from 102, 104, and 106 spores/plate as described in Materials and Methods. (A) Conidial counts. (B) Sclerotial weight. Letters indicate statistically significant differences (P < 0.05) for each strain at different population levels according to the Tukey-Kramer multiple comparison test. Error bars show standard deviations of the results of four replications. WT, wild type.
However, clear differences between effects of loss of or overexpression (MC) of veA compared to the results for cognate laeA mutants emerged in both conidial and sclerotial development. Previous studies have suggested a “balance” in sclerotial and conidial production, i.e., when sclerotial production is low, conidial is high and vice versa (18, 41). This appeared to hold true for the ΔlaeA strain (no sclerotial production at any cell density and high conidial counts at all densities) but not the ΔveA strain, for which conidial counts were very low at all population levels (Fig. 4A) despite the lack of sclerotial production (Fig. 4B). The MC mutants also showed clear differences in their density-dependent responses. The MCveA strain still exhibited a density-dependent response in sclerotial production with declining numbers in both light and dark regimes at high population levels (Fig. 4B). This was in contrast to the MClaeA strain, which maintained constant sclerotial numbers at all population levels (Fig. 4B). The MCveA-laeA double mutant exhibited an intermediate response. The trend to increased conidial numbers at high population levels was maintained in the MCveA and MCveA-laeA strains but not in the MClaeA strain (Fig. 4A). These results are summarized in Table 3.
TABLE 3.
Summary of density-dependent phenomena in A. flavus mutants
| Mutation | Morphological differentiation under indicated conditiona
|
|||
|---|---|---|---|---|
| Light
|
Dark
|
|||
| Conidia | Sclerotia | Conidia | Sclerotia | |
| None (WT) | + | + | + | + |
| ΔveA | ± | − | ± | − |
| ΔlaeA | − | − | − | − |
| MCveA | ± | + | ± | + |
| MClaeA | − | − | − | − |
| MCveA-laeA | + | ± | + | ± |
+, presence of density-dependent development; −, absence of density-dependent development; ±, intermediate response.
Density-dependent production of aflatoxin is controlled by LaeA.
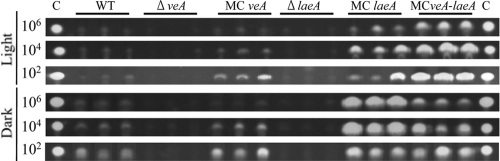
We also examined the strains for possible effects of laeA and veA expression on aflatoxin production at all cell densities, as aflatoxin production in the wild type is highest at low population levels (19). Regardless of cell densities, the ΔveA and ΔlaeA strains never produced observable aflatoxin under the growth conditions used here, whereas all the MC strains produced aflatoxin in all treatments (Fig. 5). The MCveA strain also showed a density-dependent decrease of aflatoxin with increasing cell population, similar to the wild type, whereas the MClaeA strain did not, and the double mutant showed an intermediate result. Aflatoxin production correlated with sclerotial production.
FIG. 5.
Aflatoxin production of veA and laeA mutants. Aflatoxin from each strain was assessed at three different spore inoculation levels. The experiment was replicated three times, as shown. C, aflatoxin B1 control; WT, wild type.
VeA and LaeA are important factors for seed colonization.
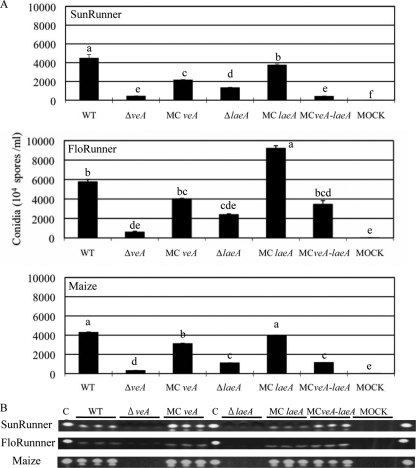
Recently, Kale et al. (21) reported that laeA mutants were aberrant in host colonization and aflatoxin production on both peanut and maize seed, but there are no reports for the role of VeA in A. flavus pathogenicity. Here, we examined and contrasted colonization attributes of the different veA and laeA mutants on two peanut cultivars and one maize hybrid.
Each fungal strain maintained similar growth patterns regardless of the host seed. Figure 6A shows that both null mutants produced fewer conidia than the wild type during growth on seeds, with the ΔveA strain developing significantly fewer conidia than the ΔlaeA strain. Visually, the ΔveA strain was most crippled in its ability to grow on any seed (data not shown). The MCveA and MCveA-laeA strains also produced fewer conidia than the wild type; however, the MClaeA strain was similar to the wild type in conidial production, depending on the host seed, as reported earlier (21). The MC strains also formed sclerotia on the seeds (data not shown).
FIG. 6.
Conidium production and aflatoxin production on peanut and maize seeds. Seeds of two peanut cultivars and one maize line were inoculated with 105 spores/ml of the wild type and the veA and laeA mutants and incubated for either 3 days (peanut cultivar SunRunnner and maize kernels) or 5 days (peanut cultivar FloRunner) after inoculation at 29°C under dark conditions. (A) For conidium counting, 1-ml amounts of homogenized suspensions of five peanut cotyledons or maize kernels of inoculated seeds were diluted to 1× and conidia counted. Letters indicate statistically significant differences (P < 0.05) of different strains, according to Tukey-Kramer multiple comparison test. Error bars show the standard deviations of the results of three replications. (B) Aflatoxin was extracted from inoculated peanut cotyledons and maize kernels and resuspended in 500 μl of chloroform, and 10 μl of each extract was spotted on a TLC plate and separated with chloroform/acetone (95:5, vol/vol). C, aflatoxin B1 control; WT, wild type; MOCK, control inoculated with water.
The colonized seeds were next examined for aflatoxin contamination. All MC strains and the wild type produced aflatoxin in all hosts, in contrast to the lack of aflatoxin production by both the ΔveA and ΔlaeA strain (Fig. 6B). The considerably higher aflatoxin production by some MC mutants in vitro (Fig. 5), however, was not replicated in growth on seed under the conditions in this study.
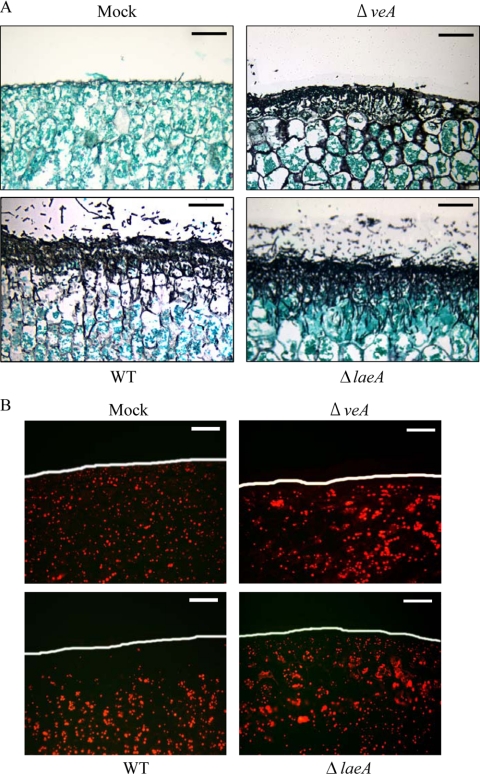
To further investigate the ability of the strains to colonize seed, histological studies were performed. We were specifically interested in assaying for maceration effects and reasoned that this could be partially measured by host cell lipid utilization. The staining techniques did not show any obvious difference in host penetration by MC strains compared to that of the wild type (data not shown). However, the two null mutants exhibited different host invasion patterns. The results in Fig. 7A and B show that wild-type hyphae penetrated several layers of host epidermal and mesophyll cells, with accompanying dissolution of host lipid reserves. Although the ΔlaeA strain also penetrated the host cells intracellularly, host lipid reserves were largely intact and the cell integrity appeared less damaged (Fig. 7A and B). In contrast, hyphae of the ΔveA strain grew intercellularly in epidermal cells and did not appear to penetrate peanut cells as well as hyphae of other strains (Fig. 7A). This mutant, like the ΔlaeA mutant, was also less able to degrade host cell lipid reserves than the wild type (Fig. 7B). However, an in vitro assay for general lipase activity revealed no significant difference between these strains (data not shown).
FIG. 7.
Histological examination reveals differences in seed ingress and lipid utilization of ΔveA and ΔlaeA strains compared to these functions in the wild type. (A) Tissues were stained with Gomori methenamine-silver for detection of fungal hyphae. (B) Tissues were stained with Nile red for lipid body detection in seeds. To observe tissues, a bright-field microscope was used for Gomori stain and a tetramethyl rhodamine 5-isothiocyanate filter in a fluorescent microscope was used for Nile red. Seeds infected with the wild-type fungus show diminishment of lipid bodies near the surface (white line) of the seed. Scale bars = 100 μm. WT, wild type; Mock, control inoculated with water.
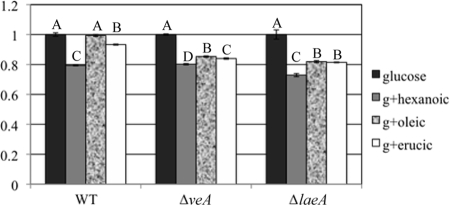
The wild type and the ΔlaeA and ΔveA strains were then grown on media amended with different fatty acids either as sole carbon source or supplemented with glucose to determine if there might be any gross difference in the ability to utilize or be inhibited by short-, medium-, or long-chain fatty acids. The results did not support any critical difference between the wild type and the two mutants when grown on a fatty acid as the sole carbon source, but the two mutants showed significant inhibition of growth compared to that of the wild type when cultured on GMM amended with oleic acid (Fig. 8). This experiment was repeated twice with similar results (data not shown).
FIG. 8.
Loss of veA and laeA sensitizes the fungus to oleic acid. Inhibition of colony diameters of ΔveA and ΔlaeA mutants but not the wild type is observed when GMM is supplemented with 6 mM oleic acid at 3 (data not shown) and 6 days after inoculation. Letters indicate statistically significant differences (P < 0.05) at 6 days after inoculation with different strains, according to Tukey-Kramer multiple comparison test. Error bars show the standard deviations of the results of four replications. WT, wild type; g, glucose; hexanoic, hexanoic acid; oleic, oleic acid; erucic, erucic acid.
DISCUSSION
In this study, we characterized the function and cross-regulation of VeA and LaeA in A. flavus development and pathogenesis. The results, while confirming that VeA and LaeA share functions in regulating aflatoxin and sclerotial production, also demonstrate distinct roles of VeA and LaeA in terms of vegetative growth, conidiation, density-dependent responses, and pattern of colonization of host tissues.
A requirement for LaeA in density-dependent sensing.
Quorum-sensing systems in bacteria contribute to the production of virulence factors and biofilm formation in interactions between bacteria and host (14, 42). In fungi, a quorum-sensing system governing morphological shifts and virulence has been uncovered in the human pathogen Candida albicans (17, 35). Recently, oxylipin-deficient lipoxygenase and dioxygenase mutants have been found to affect a newly discovered quorum-sensing-like, density-dependent sclerotial-to-conidial morphology shift in A. flavus (18, 19). Because oxylipin signaling is dependent on VeA function (5, 7, 40) and VeA is part of a nuclear complex with LaeA (1), we asked if VeA or LaeA mutants could be affected in this quorum-like morphology shift in A. flavus. Both null mutants were blocked in sclerotial formation regardless of cell population, and perhaps due to an inability to produce sclerotia, conidial production was relatively stable for each mutant at all three population levels, although it was much higher in the ΔlaeA strain. The MC strains showed clear differences in density-dependent development in that an extra copy of LaeA but not VeA abolished this quorum-like phenomenon (Fig. 4A and B). To date, there are no chemical data identifying molecules regulating the sclerotial-to-conidial switch in A. flavus, although oxylipins are hypothesized to fulfill this function at least in part (18, 19). Quorum-sensing molecules for Candida albicans (farnesol and tyrosol) and Saccharomyces cerevisiae (phenylethanol and tryptophol) are aromatic alcohols and control the morphological switch from the yeast to filamentous growth in these fungi (8, 9). Interestingly, the yeast-to-filamentous growth switch in the fungus Ceratocystis ulmi is attenuated by lipoxygenase inhibitors and may implicate oxylipins in quorum sensing in this tree pathogen (20). We speculate that A. flavus MClaeA mutants are aberrant in oxylipin production and/or sensing but that this can be remediated to some degree when VeA levels also increase, as demonstrated by the intermediate density-dependent phenotype of the MCveA-laeA strain. The effects of gene loss and gain on density-dependent development are summarized in Table 3.
VeA and LaeA feedback regulation.
Both veA and laeA have been reported to be global regulators of secondary metabolites in A. flavus (13, 21), as well as in other aspergilli (2, 6). Here, the results indicate that the MCveA and MClaeA strains—particularly the MClaeA strain—produce more aflatoxin and sclerotia than the wild type. The MCveA-laeA double mutant did not show increased toxin production compared to that of the single mutants or an additive effect on sclerotial production. Prior work indicated that LaeA negatively regulated veA expression (21), and here, we show evidence for VeA regulation of LaeA (Fig. 3), as was described for A. nidulans (1). These results support a mechanism of mutual repression of veA and laeA expression and may explain, in part, a dampening of the expression of both genes in the MCveA-laeA strain compared to the expression of the single genes in the MCveA and MClaeA strains which, in turn, may affect aflatoxin and sclerotial output in the double mutant.
Requirement for VeA and LaeA in host cell penetration and degradation.
Host lipid reserves are depleted during seed colonization by Aspergillus (23, 37), with lipase and esterase activities implicated in seed pathogenesis (37, 39). Both null mutants were impaired in seed colonization, where neither strain could degrade lipid reserves despite hyphal penetration of at least some layers of the host seeds (Fig. 7A and B).
The crippled ability of both null mutants to utilize lipid reserves brings to mind several lipid biosynthesis mutants also impaired in Aspergillus colonization of seed, including β-oxidation mutants (28), odeA mutants [(delta)12-desaturase] (43), and the oxylipin oxygenase mutants in A. nidulans (4, 39) and A. flavus (19). The inhibition of both null mutants by oleic acid (not seen in the wild type) (Fig. 8) suggests a possible toxic effect of this fatty acid on these strains which may relate to their impairment in growth on seed. It is less likely that the inhibition is associated with defects in β-oxidation, since the mutants grew equally as well as the wild type on oleic acid as a sole carbon source (data not shown), although we cannot rule out this possibility. Regardless of mechanism, the results of all of these studies together may support lipid utilization and/or signaling as an important factor in Aspergillus seed pathogenesis.
Interestingly, the hyphal penetration patterns of the two null mutants as revealed by Gomori staining were quite diverse, whereas hyphae of the ΔveA strain remained largely intercellular (Fig. 7A). This inability to penetrate intracellularly may indicate loss of degradative enzymes in this strain and may explain its poor production of conidia on host seed. However, we note that the strain is crippled in conidial production on medium also. The relative decrease of conidial production by the ΔlaeA strain on seed (compared to its vigorous conidial production in medium) might be attributable to a loss in lipid assimilation or the possible toxicity effects mentioned above. Histology of the MC strains presented an invasion and lipid degradation pattern similar to that of the wild type. The relatively decreased conidial production on seed from these strains is possibly a function of their skewed sclerotial development rather than an inability to obtain nutrients from the seed.
In conclusion, this study provides evidence for distinct roles of LaeA and VeA in the development and pathogenesis of A. flavus despite the considerable overlapping of functions previously reported (13, 21). The loss of both genes blocks the production of sclerotia and aflatoxin, but under our conditions, only laeA overexpression abolishes density-dependent phenomena, including a sclerotial-to-conidial shift and decreased aflatoxin production with cell population increase. The null mutants, while both were reduced in host lipid utilization, displayed distinct cell ingress abilities as reflected in patterns of hyphal penetration of host cells.
ADDENDUM
A study with some similar conclusions has been published by Duran and colleagues (13a).
Acknowledgments
Funding has been provided for this research from the USDA Cooperative State Research, Education, and Extension Service (CSREES) project (WIS01200) and NSF IOB-0544428, subagreement S060039, to N.P.K.
We thank Jin Woo Bok, Russell N. Spear, and Sarah J. Swanson for their technical assistance.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Bayram, O., S. Krappmann, M. Ni, J. W. Bok, K. Helmstaedt, O. Valerius, S. Braus-Stromeyer, N. J. Kwon, N. P. Keller, J. H. Yu, and G. H. Braus. 2008. The VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 3201504-1506. [DOI] [PubMed] [Google Scholar]
- 2.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 41574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodhagen, M., D. I. Tsitsigiannis, E. Hornung, C. Goebel, I. Feussner, and N. P. Keller. 2008. Reciprocal oxylipin-mediated cross-talk in the Aspergillus-seed pathosystem. Mol. Microbiol. 67378-391. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, A. M., L. L. Hinze, H. W. Garner, and N. P. Keller. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 653668-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo, A. M., J. W. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 704722-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champe, S. P., P. Rao, and A. Chang. 1987. An endogenous inducer of sexual development in Aspergillus nidulans. J. Gen. Microbiol. 1331383-1387. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., M. Fujita, Q. Feng, J. Clardy, and G. R. Fink. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 1015048-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., and G. R. Fink. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 201150-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cseke, L. J., P. B. Kaufman, G. K. Podila, and C. J. Tsai. 2004. Handbook of molecular/cellular methods in biology and medicine, 2nd ed. CRC Press, Boca Raton, FL.
- 11.Dagenais, T. R., D. Chung, S. S. Giles, C. M. Hull, D. Andes, and N. P. Keller. 2008. Defects in conidiophore development and conidium-macrophage interactions in a dioxygenase mutant of Aspergillus fumigatus. Infect. Immun. 763214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diener, U. L., R. J. Cole, T. H. Sanders, G. A. Payne, L. S. Lee, and M. A. Klich. 1987. Epidemiology of aflatoxin formation by Aspergillus flavus. Annu. Rev. Phytopathol. 25249-270. [Google Scholar]
- 13.Duran, R. M., J. W. Cary, and A. M. Calvo. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotia formation. Appl. Microbiol. Biotechnol. 731158-1168. [DOI] [PubMed] [Google Scholar]
- 13a.Duran, R. M., J. W. Cary, and A. M. Calvo. 2009. The role of veA in Aspergillus flavus infection of peanut, corn and cotton. Open Mycol. J. 327-36. [Google Scholar]
- 14.Fray, R. G. 2002. Alternating plant-microbe interaction through artificially manipulating bacterial quorum-sensing. Ann. Bot. 89245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgianna, D. R., and G. A. Payne. 2009. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet. Biol. 46113-125. [DOI] [PubMed] [Google Scholar]
- 16.Greenspan, P., E. P. Mayer, and S. D. Fowler. 1985. Nile Red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 672982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horowitz Brown, S., R. Zarnowski, W. C. Sharpee, and N. P. Keller. 2008. Morphological transition governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 745674-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horowitz Brown, S., J. B. Scott, J. Bhaheetharan, W. C. Sharpee, L. Milde, R. A. Wilson, and N. P. Keller. Oxygenase coordination is required for morphological transition and the host-fungal interaction of Aspergillus flavus. Mol. Plant Microbe Interact., in press. [DOI] [PubMed]
- 20.Jensen, E. C., C. Ogg, and K. W. Nickerson. 1992. Lipoxygenase inhibitors shift the yeast/mycelium dimorphism in Ceratocystis ulmi. Appl. Environ. Microbiol. 582505-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kale, S. P., L. Milde, M. K. Trapp, J. C. Frisvad, N. P. Keller, and J. W. Bok. 2008. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 451422-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, N., W. Brooks, and A. M. Calvo. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 21178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, N. P., N. J. Kantz, and T. H. Adams. 1994. Aspergillus nidulans veA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 601444-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H., K. Hank, K. Kim, D. Han, K. Jahng, and K. Chae. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 3772-80. [DOI] [PubMed] [Google Scholar]
- 25.Klich, M. A. 2007. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8713-722. [DOI] [PubMed] [Google Scholar]
- 26.Lillie, R. D. 1965. Histopathologic technique and practical histochemistry, 3rd ed. McGraw-Hill Book Company, New York, NY.
- 27.Maggio-Hall, L. A., and N. P. Keller. 2004. Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 541173-1185. [DOI] [PubMed] [Google Scholar]
- 28.Maggio-Hall, L. A., R. Wilson, and N. P. Keller. 2005. Fundamental contribution of β-oxidation to polyketide mycotoxin production in planta. Mol. Plant Microbe Interact. 18783-793. [DOI] [PubMed] [Google Scholar]
- 29.Michailides, T. J., and T. Thomidis. 2007. First report of Aspergillus flavus causing fruit rots of peaches in Greece. Plant Pathol. 56352. [Google Scholar]
- 30.Miller, B. L., K. Y. Miller, and W. E. Timberlake. 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 51714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muture, B. N., and G. Ogana. 2005. Aflatoxin levels in maize and maize products during the 2004 food poisoning outbreak in Eastern Province of Kenya. East Afr. Med. J. 82275-279. [DOI] [PubMed] [Google Scholar]
- 32.Perrin, R. M., N. D. Fedorova, J. W. Bok, R. A. Cramer, J. R. Wortman, H. S. Kim, W. C. Nierman, and N. P. Keller. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettit, R. E. 1984. Yellow mold and aflatoxin, p. 35-36. In D. M. Porter, D. H. Smith, and R. Rodeiguez-Kabana (ed.), Compendium of peanut diseases. The American Phytopathological Society, St. Paul, MN.
- 34.Rubens, J., and K. F. Cardwell. 2005. The cost of mycotoxin management in the United States, p. 1-13. In H. K. Abbas (ed.), Aflatoxin and food safety. CRC Press, Boca Raton, FL.
- 35.Shchepin, R., J. M. Hornby, E. Burger, T. Niessen, P. Dussault, and K. W. Nickerson. 2003. Quorum sensing in Candida albicans: probing farnesol's mode of action with 40 natural and synthetic farnesol analogs. Chem. Biol. 10743-750. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smart, M. G., D. T. Wicklow, and R. W. Caldwell. 1990. Pathogenesis in aspergillus ear rot of maize: light microscopy of fungal spread from wounds. Phytopathology 801287-1294. [Google Scholar]
- 38.Szewczyk, E., T. Nayak, C. E. Oakley, H. Edgerton, Y. Xiong, N. Taheri-Talesh, S. A. Osmani, and B. R. Oakley. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 13111-3120. [DOI] [PubMed] [Google Scholar]
- 39.Tsitsigiannis, D. I., and N. P. Keller. 2006. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol. Microbiol. 59882-892. [DOI] [PubMed] [Google Scholar]
- 40.Tsitsigiannis, D. I., T. M. Koweiski, R. Zarnowski, and N. P. Keller. 2005. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology 1511809-1821. [DOI] [PubMed] [Google Scholar]
- 41.Tsitsigiannis, D. I., R. Zarnowski, and N. P. Keller. 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 27911344-11353. [DOI] [PubMed] [Google Scholar]
- 42.Williams, P. 2007. Quorum sensing, communication and cross-kingdom signaling in the bacterial world. Microbiology 1533923-3938. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, R. A., A. M. Calvo, P. K. Chang, and N. P. Keller. 2004. Characterization of the Aspergillus parasiticus delta12-desaturase gene: a role for lipid metabolism in the Aspergillus-seed interaction. Microbiology 1502881-2888. [DOI] [PubMed] [Google Scholar]
- 44.Yu, J., T. E. Cleveland, W. C. Nierman, and J. W. Bennett. 2005. Aspergillus flavus genomics: gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 22194-202. [DOI] [PubMed] [Google Scholar]
- 45.Yu, J., G. Payne, B. C. Campbell, B. Guo, T. E. Cleveland, J. F. Robens, N. P. Keller, J. W. Bennett, and W. C. Nierman. 2008. Mycotoxin production and prevention of aflatoxin contamination in food and feed. In G. H. Goldman and S. A. Osmani (ed.), The aspergilli. CRC Press, Boca Raton, FL.