Abstract
During propagation, yeast prions show a strict sequence preference that confers the specificity of prion assembly. Although propagations of [PSI+] and [RNQ+] are independent of each other, the appearance of [PSI+] is facilitated by the presence of [RNQ+]. To explain the [RNQ+] effect on the appearance of [PSI+], the cross-seeding model was suggested, in which Rnq1 aggregates act as imperfect templates for Sup35 aggregation. If cross-seeding events take place in the cytoplasm of yeast cells, the collision frequency between Rnq1 aggregates and Sup35 will affect the appearance of [PSI+]. In this study, to address whether cross-seeding occurs in vivo, a new [PSI+] induction method was developed that exploits a protein fusion between the prion domain of Sup35 (NM) and Rnq1. This fusion protein successfully joins preexisting Rnq1 aggregates, which should result in the localization of NM around the Rnq1 aggregates and hence in an increased collision frequency between NM and Rnq1 aggregates. The appearance of [PSI+] could be induced very efficiently, even with a low expression level of the fusion protein. This study supports the occurrence of in vivo cross-seeding between Sup35 and Rnq1 and provides a new tool that can be used to dissect the mechanism of the de novo appearance of prions.
Prions were originally defined as proteinaceous infectious particles responsible for transmissible spongiform encephalopathies in mammals (reviewed in reference 23). Since a non-Mendelian genetic element, [URE3], was identified as a yeast prion (37), however, this concept has been expanded to include protein-based genetic elements. In addition to [URE3], there are at least two more proteinaceous genetic elements in Saccharomyces cerevisiae, namely, [PSI+] and [RNQ+] (20, 22, 28). [Het-s] was also identified as a prion in the filamentous fungus Podospora anserina (2).
Despite the absence of any structural and functional homologies between various prion proteins from different organisms, they share a common feature, i.e., prion proteins can adopt two distinct conformational states. One of these, the aggregated prion state, can stimulate the soluble, nonprion conformation to convert into the prion form. Gaining intermolecular β-sheet structures, purified yeast prion proteins aggregate and form amyloid fibers in vitro (8, 12, 28, 32). Protein extract from yeast cells in the prion state can facilitate the in vitro polymerization of soluble prion protein from nonprion cells (21), and amyloid fibers of purified yeast prion proteins can convert the cells into the prion state when introduced into yeast cells, demonstrating the protein-only hypothesis (15, 31). Thus, intracellular prion aggregates are thought to have the same structural basis as amyloid fibers formed in vitro.
Yeast prion biology has provided invaluable insights into the prion concept at the molecular level. Because of its experimental convenience, [PSI+] has been investigated most intensively among various yeast prions. [PSI+] results from the aggregation of Sup35 protein, which is essential for terminating the translation at stop codons. When Sup35 is in the [PSI+] aggregated state, ribosomes often fail to release polypeptides at stop codons, causing a non-Mendelian trait which is easily detected by nonsense suppression. ade1 or ade2 nonsense mutants are used as marker genes to determine the [PSI+] state. These mutants cannot grow on adenine-deficient medium and form red colonies on medium supplemented with a limiting amount of adenine, such as yeast extract-peptone-dextrose (YPD). ade mutants in the [PSI+] state, however, can grow on adenine-deficient medium and form white colonies, as they produce functional Ade1 or Ade2 by virtue of a nonsense mutation readthrough. To sustain propagation, all yeast prions need the disaggregation activity of Hsp104, which can be inhibited by guanidine hydrochloride (GuHCl) (9). Since yeast prions are cured by growth on guanidine-containing medium, prion phenotypes can easily be distinguished from chromosomal suppressor mutants.
Sup35 (eRF3) of S. cerevisiae has a prion-determining N-terminal domain (N), a highly charged middle domain (M) that confers solubility on the molecule, and an essential C-terminal domain that binds guanine nucleotides and stimulates the polypeptide release reaction catalyzed by Sup45 (eRF1) (17, 29, 33). The de novo appearance of [PSI+] can be induced by overexpression of SUP35 or its prion domain-containing fragments (NM) (6). [PSI+] induction, however, can be achieved only in [RNQ+] cells that harbor the prion state of the Rnq1 protein (4, 19). Two hypotheses about how [RNQ+] can affect the appearance of [PSI+] have been suggested. One is an inhibitor titration model that postulates the molecules preventing the aggregation of Sup35 and the recruitment of these inhibitors to Rnq1 aggregates in [RNQ+] cells. The other is a cross-seeding model in which Rnq1 aggregates directly catalyze the polymerization of Sup35. In vitro cross-seeding between different amyloidogenic proteins was reported, and Rnq1 amyloid fiber can also act as a seed for Sup35 polymerization in vitro (7, 13). These in vitro data support the possibility of cross-seeding between Rnq1 and Sup35. However, because the milieu of cytoplasm is very different from that of a test tube, whether this cross-seeding really occurs in vivo is still obscure. For this study, we developed a new, robust [PSI+] induction method that confirms the cross-seeding events in the cytoplasmic environment.
MATERIALS AND METHODS
Yeast strains and growth methods.
All strains used in this study are derivatives of strain 74-D694 (ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 [psi−] [RNQ+]). All [rnq−] strains were isolated by growth on YPD plus 3 mM GuHCl, and RNQ1 was deleted using the kanMX6 module. pRS304-TPSUP35-NM-RNQ1-GFP, pRS304-TPSUP35-NM-RNQ1(1-234)-GFP, pRS304-TPSUP35-NM-RNQ1(1-261)-GFP, pRS304-TPSUP35-NM-prnp-GFP, and pRS304-TPSUP35-NM-GFP were integrated at the nutritional marker locus trp1 after digestion with XbaI. In the case of the NM-GFP strain, pRS306-TPSUP35-NM-GFP was additionally integrated at the ura3 locus after StuI digestion. To replace the prion domain of chromosomal SUP35 with URE2 PrD, two-step replacement using pRS306-PSUP35-URE2 PrD-SUP35MC was exploited. The correct integrations of these plasmids were verified by PCR. The integrants cultured in YPD medium (1% yeast extract, 2% peptone, 2% glucose) were harvested by a brief centrifugation and suspended in SC medium for observation with a fluorescence microscope. All transformants were cultured in selective SC media to exponential phase (optical density at 600 nm, 0.8 to 1.0) to assay the appearance of [PSI+] and to determine the level of recombinant proteins. All selective SC media used in this study were supplemented with 0.54 mM adenine to prevent the overgrowth of Ade+ cells. The frequency of Ade+ cells was determined by spotting or spreading cells onto SC and SC−Ade plates in serial 10-fold dilutions. The average and standard deviation of Ade+ induction frequency were obtained for three independent transformants for each strain. To determine the [PSI+] state of Ade+ cells, colonies on an SC−Ade plate were spotted onto a 5-fluoroorotic acid (5-FOA) plate to select the cells that had lost the plasmid. 5-FOA-resistant cells were spotted again onto YPD and YPD-plus-3 mM GuHCl plates for a color assay. To induce the expression of recombinant genes from the CUP1 promoter, 50 μM CuSO4 was added to cultures at an optical density of 600 nm of 0.4, and cultures were incubated for 4 h. The polyethylene glycol-lithium acetate-single-stranded DNA method was used for the introduction of plasmids or PCR products into yeast cells (11).
Plasmid construction.
The sequences of oligonucleotide primers used for PCR in this study are available upon request. The PCR products of the truncated SUP35 promoter, full-length SUP35 promoter, and CUP1 promoter were cloned into p414GAL1 or p416GAL1, using SacI and BamHI. From the resulting plasmid, the truncated SUP35 promoter and the terminator of CYC1 were subcloned into pRS304 and pRS306.
A 3×FLAG construct was inserted as a primer sequence, and NM-3×FLAG was ligated as a BamHI-EcoRI fragment, followed by RNQ1 cloning using EcoRI and XhoI. The endogenous EcoRI site of the RNQ1 gene was removed without any change in the protein sequence. The NM-3×FLAG construct with a stop codon and 3×FLAG-RNQ1 were cloned as BamHI-XhoI fragments.
Green fluorescent protein (GFP) was cloned using EcoRI and XhoI. The EcoRI-EcoRI fragments of full-length RNQ1, RNQ1(1-234), RNQ1(1-261), and mouse prnp were cloned into pRS304-TPSUP35-NM-GFP.
To make pRS306-PSUP35-URE2 PrD-SUP35MC, the SUP35 promoter (EcoRI-BamHI), URE2 PrD (BamHI-SacII), and SUP35 MC (SacII-SacI) were cloned into pRS306. Using this plasmid as a template, URE2 PrD-M-3xFLAG was PCR amplified (with 3×FLAG in the primer sequence). All constructs derived from PCR products in this study were verified by DNA sequencing.
Antibodies and Western blot analysis.
The anti-FLAG antibody was purchased from Sigma, and anti-NM and anti-Rnq1 antisera were prepared in our laboratory. Total proteins of yeast were isolated by boiling in sodium dodecyl sulfate sample buffer as previously reported (16). Cells were treated with 0.1 M NaOH for 4 min and resuspended in sample buffer (60 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 4% β-mercaptoethanol, 5% glycerol, and 0.01% bromophenol blue), followed by boiling for 5 min. The resulting protein extracts were quantified with a 2-D Quant kit (GE Healthcare), using bovine serum albumin to prepare the standard curve. Thirty-five micrograms of lysate was loaded into each well of a 10% polyacrylamide gel, and after electrophoresis, proteins were transferred to an Immobilon-PSQ polyvinylidene difluoride membrane (Millipore). Proteins were detected with anti-FLAG antibody or antiserum against NM or Rnq1. The antibody-antigen interaction was revealed with ECL reagent (Millipore), and the chemiluminescence signal was detected with an LAS-3000 instrument (Fujifilm).
Fluorescence microscopy.
Fluorescence microscopy was carried out as described before (30). Briefly, yeast cells grown to exponential phase were observed microscopically in 96-well glass-bottomed microtiter plates (Whatman) pretreated with concanavalin A (Sigma) to ensure cell adhesion. Microscopy was performed on a Zeiss Axiovert 200 M inverted microscope with a Plan-Neofluar ×100/1.30 oil objective. Images were recorded on a Zeiss Axiocam MRm instrument with two-by-two binning.
RESULTS
NM fused to Rnq1 remarkably increases the appearance of [PSI+].
It is well known that the de novo appearance of [PSI+] can be induced by overexpression of SUP35 or its fragments containing the prion domain (NM) in [RNQ+] cells (6). From a cross-seeding viewpoint, this [PSI+] induction effect can be explained by the increase in the frequency of collisions between the NM domain and Rnq1 aggregates. We conjectured that if cross-seeding between Rnq1 and Sup35 is the cause of [PSI+] generation, raising the intermolecular collision frequency without increasing the amount of NM should be sufficient to induce the appearance of [PSI+]. To separate the effect of the intermolecular collision frequency from that of the molecular amount, a protein fusion method was exploited. We assumed that NM fused to Rnq1 may successfully join the preexisting Rnq1 aggregates in [RNQ+] cells (Fig. 1A) and that this will result in a high local concentration of NM around the Rnq1 aggregates irrespective of the total intracellular NM concentration. If this happens, there should be much more frequent intermolecular collision between NM and Rnq1 aggregates than when they are expressed separately, and repeated collisions will give rise to the appearance of [PSI+].
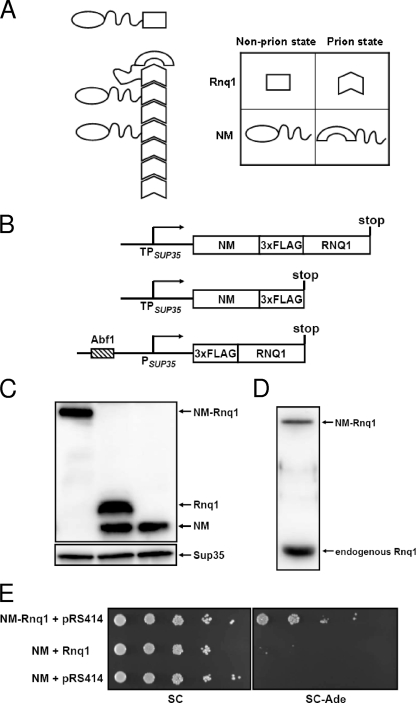
FIG. 1.
[PSI+] induction by the protein fusion method, which exploits an increased collision frequency. (A) Schematic representation of [PSI+] induction by the NM-Rnq1 fusion protein. (B) Constructs to test the protein fusion method. All constructs were cloned into ARS/CEN vectors. (C) Expression levels of the recombinant proteins were determined by Western blot analyses using anti-FLAG antibody. Sup35 was used as a loading control. (D) The protein level of NM-Rnq1 was compared to the level of the endogenous Rnq1 protein by Western blotting with anti-Rnq1 antiserum. (E) [RNQ+] cells were transformed with combinations of plasmids as indicated. Each transformant was grown in selective SC medium to exponential phase and spotted onto SC and SC−Ade plates in serial 10-fold dilutions.
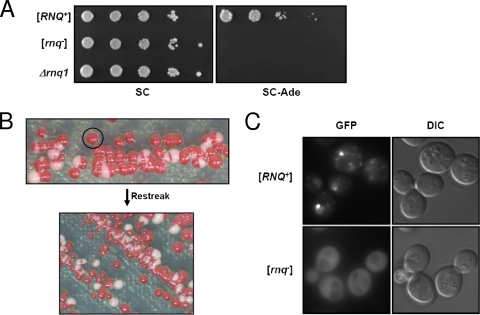
To verify this assumption, expression vectors including the NM-RNQ1 fusion construct were designed with two variants of the SUP35 promoter in ARS/CEN vectors to control the steady-state level of recombinant proteins (Fig. 1B). The two promoters used in this study differ only in length, and the shorter one (hereafter referred to as a truncated promoter or TPSUP35) does not comprise the putative Abf1 binding site. It was reported that the truncated promoter shows a lower expression level than the full-length SUP35 promoter (3). The expression levels of constructs were determined by Western blot analysis with an anti-FLAG antibody (Fig. 1C). Since the protein level of Rnq1 from the truncated SUP35 promoter was lower than those of other constructs (data not shown), RNQ1 was expressed from the full-length SUP35 promoter to adjust its expression level to the levels of NM-RNQ1 and NM expressed from the truncated SUP35 promoter (Fig. 1B and C). All constructs were introduced into [RNQ+] cells, and transformants grown to mid-exponential phase in selective medium were 10-fold serially diluted and spotted onto a plate without adenine (−Ade) (Fig. 1E). NM-RNQ1 transformants generated Ade+ cells at a frequency of 3.0% ± 1.0%, while NM transformants gave rise to Ade+ cells at a frequency of (6.2 ± 0.9) × 10−5. To determine whether the Ade+ cells were in the [PSI+] state, cells that had lost the plasmid expressing NM-RNQ1 were selected by growth on a 5-FOA plate. All of the 5-FOA-resistant cells formed white or light pink colonies on a YPD plate, and >90% of cells (98 of 106 randomly selected Ade+ cells) formed red colonies on a plate containing GuHCl (data not shown). Moreover, when the prion domain (N domain) of chromosomal SUP35 was replaced with another Asn/Gln-rich domain, NM-Rnq1 no longer produced Ade+ cells, confirming that its effect is mediated through the N domain of Sup35 (see Fig. 5B, third row). From this result, we concluded that NM induces the appearance of [PSI+] more efficiently when fused to Rnq1. It should be emphasized that de novo [PSI+] generation could be induced at a high frequency even with low-level expression of the recombinant protein. The expression level of NM-RNQ1 from the truncated SUP35 promoter in the ARS/CEN vector was even lower than the level of the endogenous Rnq1 protein (Fig. 1D), which is less than 1/10 of the endogenous Sup35 level (10). To test whether the NM-Rnq1 fusion protein can form initial nuclei for the generation of [PSI+] per se, a plasmid expressing NM-RNQ1 was introduced into [RNQ+] and [rnq−] cells. The NM-Rnq1 fusion protein induced the appearance of [PSI+] in [RNQ+] cells but not in [rnq−] cells (Fig. 2A). The [rnq−] cells used in this study were isolated from isogenic [RNQ+] cells by growth on a GuHCl-containing plate. Because guanidine might affect other cellular phenotypes in addition to [RNQ+], we also made an RNQ1 deletion mutant to specifically cure the [RNQ+] phenotype. These Δrnq1 cells did not give rise to the de novo appearance of [PSI+], like [rnq−] cells (Fig. 2A). This result indicates that the preexisting Rnq1 aggregates and the NM-Rnq1 fusion protein cooperate to form the [PSI+] nucleus.
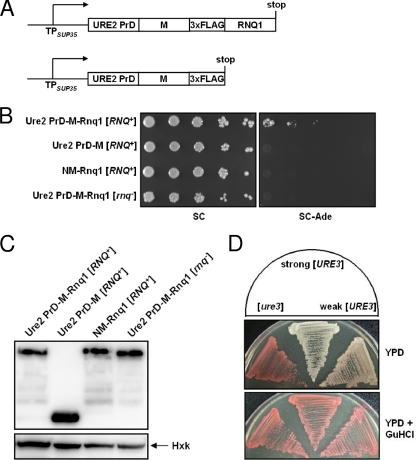
FIG. 5.
Induction of the prion state of the Ure2 prion domain by the protein fusion method. (A) Schematic representation of fusion constructs. (B) A strain in which the prion domain of chromosomal SUP35 is replaced with the Ure2 PrD was constructed. Ure2 PrD-M-Rnq1, Ure2 PrD-M, and NM-Rnq1 were produced, and the appearance of Ade+ cells was assayed by cell spotting. (C) Expression of recombinant proteins was determined by Western blotting with anti-FLAG antibody. Hexokinase (Hxk) was used as a loading control. (D) Strong and weak prion variants were isolated by using the protein fusion method.
FIG. 2.
NM-Rnq1 induces the appearance of [PSI+] only in [RNQ+] cells. (A) A plasmid carrying NM-RNQ1 was introduced into cells with different [RNQ+] states, and the [PSI+] inducibility of each transformant was determined using a spotting assay with 10-fold dilutions. (B) The NM-RNQ1-GFP integrant with the [RNQ+] state was streaked onto YPD medium. Cells from an isolated red colony continuously turned into a mixed population consisting of red, white, and some sectored colonies when transferred to a new YPD plate. (C) Fresh [RNQ+] cells from red colonies expressing NM-RNQ1-GFP and their isogenic [rnq−] cells were observed with a fluorescence microscope. In contrast to a dispersed state in [rnq−] cells, NM-Rnq1-GFP formed aggregates in [RNQ+] cells. DIC, differential interference contrast.
Incorporation of the fusion protein into preexisting [RNQ+] aggregates is required for [PSI+] induction.
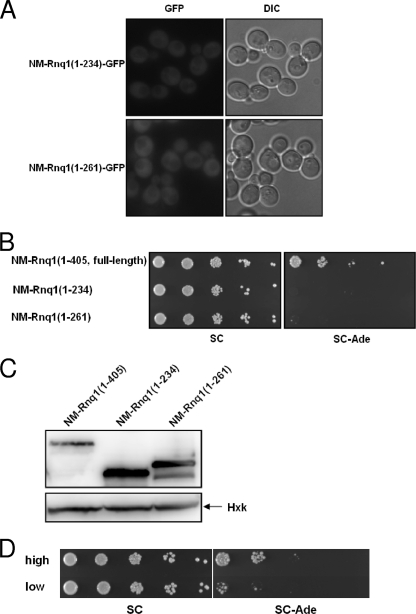
To investigate whether the Rnq1 prion domain in the fusion construct can still join preexisting Rnq1 aggregates, an NM-RNQ1-GFP integrant of the [RNQ+] state was created. This integrant gave rise to a mixed population consisting of red, white, and some sectored colonies on a YPD plate, and cells from an isolated red colony continuously turned into a mixed population when transferred to a new plate (Fig. 2B). Cells from isolated white colonies, however, always gave rise to white colonies unless guanidine was added to the medium (data not shown). This result means that most cells in red colonies are still [psi−], whereas those in white colonies are [PSI+]. To separate the effect of [RNQ+] from that of [PSI+] on NM-Rnq1-GFP aggregation, all of the experiments using this strain were carried out with freshly grown cells from red colonies (Fig. 2B). Isogenic [rnq−] cells were isolated by growth on a guanidine plate and showed red colony growth on a YPD plate. The protein level of NM-Rnq1-GFP, determined by Western blot analysis with anti-NM antiserum, was not affected by the presence of [RNQ+] (data not shown). The examination of NM-RNQ1-GFP integrants of [RNQ+] and [rnq−] cells with a fluorescence microscope showed clearly different distribution patterns of the fusion protein between the two strains. In contrast to a dispersed state in [rnq−] cells, NM-Rnq1-GFP formed aggregates in [RNQ+] cells (Fig. 2C). Owing to chromosomal integration and expression from a constitutive promoter, all cells showed even GFP signals, and cell-to-cell variation of the signal intensity was hardly detected. Although the frequency of Ade+ cell appearance was 1 to 10% (Fig. 2B; see Fig. 4C), nearly all [RNQ+] cells contained visible aggregates (Fig. 2C). This indicates that the observed aggregation of the NM-Rnq1-GFP protein in [RNQ+] cells was due to the prion domain of Rnq1 but not to NM, i.e., it was not the result of the appearance of [PSI+]. To demonstrate that incorporation of the fusion protein into the preexisting [RNQ+] aggregates is an important requisite for [PSI+] induction, we exploited Rnq1 deletion mutants which were reported to be unable to decorate Rnq1 aggregates (36). We confirmed that these truncated Rnq1s did not decorate the preexisting Rnq1 aggregates, using the NM-Rnq1(1-234)-GFP and NM-Rnq1(1-261)-GFP integrants (Fig. 3A). In contrast to the full-length Rnq1 fusion, NM-Rnq1(1-234) and NM-Rnq1(1-261) did not facilitate the appearance of [PSI+] compared to free NM (Fig. 3B) {the [PSI+] induction frequencies of NM-Rnq1(1-234), NM-Rnq1(1-261), and free NM were (7.9 ± 1.9) × 10−6, (3.9 ± 2.3) × 10−5, and (6.2 ± 0.9) × 10−5, respectively}.
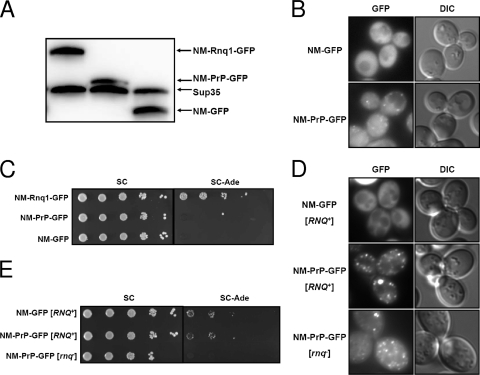
FIG. 4.
High local concentration of NM induced by aggregation of the NM-mouse PrP fusion construct does not enhance the appearance of [PSI+]. (A) Protein levels of NM-Rnq1-GFP, NM-PrP-GFP, and NM-GFP expressed from a truncated SUP35 promoter were determined by Western blotting with anti-NM antiserum. (B) Distribution patterns of NM-GFP and NM-PrP-GFP proteins expressed from the truncated SUP35 promoter were observed using fluorescence microscopy. (C) NM-RNQ1-GFP, NM-PrP-GFP, and NM-GFP integrants were spotted onto SC and SC−Ade plates in serial 10-fold dilutions to determine the frequency of Ade+ cells. (D) Distribution patterns of NM-GFP and NM-PrP-GFP proteins overproduced from the CUP1 promoter by treatment with 50 μM CuSO4 for 4 h. (E) Spotting assay to compare the Ade+ phenotype-inducing activities of strains overproducing the NM-GFP and NM-PrP-GFP proteins used for panel D.
FIG. 3.
Incorporation of the fusion protein into preexisting [RNQ+] aggregates is required for [PSI+] induction. (A) Distribution patterns of NM-Rnq1(1-234)-GFP and NM-Rnq1(1-261)-GFP proteins expressed from the truncated SUP35 promoter were observed using fluorescence microscopy. (B) [PSI+] induction by truncated NM-Rnq1 fusion constructs. (C) Expression of NM-Rnq1(1-234) and NM-Rnq1(1-261) was determined by Western blotting with anti-FLAG antibody. Hexokinase (Hxk) was used as a loading control. (D) Inducibility of [PSI+] by NM-Rnq1 in [RNQ+] variants was determined. Each transformant was grown in selective SC−Ura medium to exponential phase and spotted onto SC and SC−Ade plates in serial 10-fold dilutions.
Heritable variants of the [RNQ+] prion were described previously (1). Since these [RNQ+] variants are distinguished by the efficiency with which they enhance the de novo appearance of [PSI+], we assumed that a direct interaction between NM-Rnq1 and the preexisting Rnq1 aggregates would result in different [PSI+] induction rates among these variants expressing NM-Rnq1. When the “low” and “high” variants of [RNQ+] were transformed with the NM-Rnq1 expression vector, the “low” variant generated [PSI+] at a significantly lower frequency (0.67% ± 0.57%) than that with the “high” variant (9.6% ± 5.3%) (Fig. 3D). Different [PSI+] induction frequencies between different [RNQ+] variants support, at least in part, a direct physical interaction between the NM domain and Rnq1 aggregates.
NM fused to mouse prion does not enhance the appearance of [PSI+].
Taken together, our data show that the NM-Rnq1 fusion protein can generate initial nuclei for [PSI+] by joining preexisting Rnq1 aggregates. It was surprising that a relatively small amount of the NM-Rnq1 protein efficiently generated a [PSI+] phenotype. Although the total NM level was low, the local concentration of NM around the Rnq1 aggregates may be very high in cells expressing NM-Rnq1 (Fig. 2C). Therefore, it is not certain whether crowding of NM in a restricted space is sufficient to generate a [PSI+] phenotype, irrespective of the cross-seeding activity of Rnq1 aggregates. To test whether a high local concentration of NM, and hence frequent collisions between NM molecules themselves, is sufficient to cause polymerization irrespective of proximity to Rnq1 aggregates, the Rnq1 domain of NM-Rnq1 was replaced with another amyloidogenic protein, mouse prion protein (PrP). Mouse PrP was reported to form aggregates in the cytoplasm of yeast (18). The inducibility of [PSI+] by diffused and localized NM was examined using NM-GFP and NM-PrP-GFP constructs cloned under the control of the truncated SUP35 promoter and integrated into a chromosome. Since the protein level of NM-GFP was slightly lower than that of NM-PrP-GFP, one more copy of the NM-GFP construct was integrated into a different locus of the chromosome to adjust the concentration of the NM domain (Fig. 4A). In contrast to the NM-GFP protein, which existed in a dispersed state throughout the cytoplasm, NM-PrP-GFP formed faint aggregates, indicating that the aggregation was due to the PrP domain (Fig. 4B). The frequencies of Ade+ cell appearance for the NM-GFP and NM-PrP-GFP strains were about 3.6 × 10−5 and 1 × 10−4, respectively. Although the frequency of Ade+ cell appearance for the NM-PrP-GFP strain was slightly higher than that for the NM-GFP strain, it was still much lower than the [PSI+] induction ratio of the NM-Rnq1 strain (Fig. 4C). To check whether the frequency of Ade+ cell appearance depends on the extent of aggregation of the NM fusion protein, a more intensive aggregation of the NM-PrP-GFP protein was induced by using the strong inducible CUP1 promoter. Although the cell-to-cell variation of the GFP signal intensity was increased, NM-PrP-GFP formed clearly visible aggregates (Fig. 4D). Aggregation of the PrP domain was independent of the presence of [RNQ+], as the NM-PrP-GFP protein aggregated in [rnq−] cells as well as in [RNQ+] cells (Fig. 4D). When protein expression was induced for 4 h, the frequency of Ade+ cell appearance was significantly increased (compare Fig. 4C and E). In spite of severe aggregation of overproduced NM-PrP-GFP, indicating a high local concentration of NM, the frequency of Ade+ cell appearance for the NM-PrP-GFP-overproducing cells was similar to that for cells overproducing NM-GFP in the [RNQ+] background (Fig. 4E). Moreover, overproduction of NM-PrP-GFP protein in [rnq−] cells did not give rise to Ade+ cells at all, despite the high local concentration of NM around the PrP aggregates, indicating the significance of [RNQ+] in the appearance of [PSI+]. Taken together, the results show that the high local concentration of NM alone does not seem to be sufficient to cause polymerization, but the Rnq1 aggregates in [RNQ+] cells seem to affect the conformational change of NM when these molecules collide with one another, resulting in the appearance of [PSI+].
The protein fusion method can be exploited to induce polymerization of the Ure2 prion domain.
To test whether the protein fusion method is applicable to other prion-prion interactions, we investigated [URE3] induction by [RNQ+] (1). To measure the appearance of the prion state of the prion domain of Ure2 (Ure2 PrD), the prion domain of chromosomal SUP35 (N) was replaced with Ure2 PrD (N-terminal 89 amino acids). When the fusion proteins Ure2 PrD-M-Rnq1 and Ure2 PrD-M (Fig. 5A) were expressed in an [RNQ+] strain, only Ure2 PrD-M-Rnq1 gave rise to Ade+ cells, while neither construct could form the nucleus for the polymerization of Ure2 PrD in the [rnq−] strain (Fig. 5B). These data indicate that Rnq1 aggregates can convert the conformation of Ure2 PrD and that this reaction is facilitated by localization of Ure2 PrD near the Rnq1 aggregates. The acquired Ade+ phenotype was stably maintained after the removal of the plasmid and could be cured by GuHCl, indicating that the phenotype manifests the prion state (Fig. 5D). Moreover, white and pink variants could be isolated, which is consistent with the case for previously reported [URE3] variants (27). This experiment supports the inducibility of [URE3] by [RNQ+] via cross-seeding and suggests that the protein fusion method can generally be used to generate prion states.
DISCUSSION
In vitro cross-seeding activity of Rnq1 aggregates on Sup35 was reported previously (7, 35). During the course of NM amyloid formation in the test tube, preformed Rnq1 aggregates shortened the lag phase in which initial polymerization of NM takes place (7). This in vitro cross-seeding activity was confirmed by an observation of the hybrid aggregates composed of NM and Rnq1 by electron microscopy (35). Besides yeast prion proteins, various amyloidogenic proteins, including β-amyloid and α-synuclein, which are responsible for Alzheimer's and Parkinson's diseases, respectively, show cross-seeding activity in vitro (13). Although cross-seeding between amyloidogenic proteins seems to be possible in the test tube, whether it occurs in the cytoplasmic environment is not certain. First of all, it is unclear whether in vitro amyloids adopt the higher-order structure of in vivo fibers faithfully. Moreover, because protein folding problems, including those of yeast prions, are associated with molecular chaperones and many other interacting partner proteins, intracellular events cannot be deduced simply from experiments using purified molecules (5, 34).
The colocalization of large aggregates formed by overproduced Rnq1 and Sup35 in [psi−] [RNQ+] cells was previously suggested as a manifestation of direct in vivo cross-seeding (7). However, it is not clear whether the large aggregates are propagating units or dead-end products (25). Moreover, microscopically observed colocalization does not necessarily represent a physiological physical interaction (24). The colocalization of Sup35 and Rnq1 aggregates might result from a sequestration of these aggregates in the perivacuolar compartment (14). In addition to the colocalization result, nonheritable Sup35 aggregates that do not generate stable [PSI+] have been shown to contain Rnq1 which does not separate even in the presence of detergent, supporting the cross-seeding model (26).
In this study, we presented data supporting the hypothesis that cross-seeding between Rnq1 and Sup35 occurs in vivo by showing that the increased collision frequency between Rnq1 and Sup35 results in the enhanced de novo appearance of [PSI+]. Even with a low level of expression, the NM-Rnq1 fusion protein constructed in this study dramatically increased the appearance of [PSI+]. It was previously reported that tandem repeats of the 27th immunoglobulin domain from human cardiac titin aggregate without a detectable lag phase, although the monomeric domain showed delayed polymerization in vitro (38). The report suggested that a high local concentration achieved by a tandem fusion can affect the polymerization of aggregation-prone proteins. The high local concentration of NM induced by the NM-PrP-GFP construct, however, did not enhance the appearance of [PSI+] in this study (Fig. 4C and E). This indicates that the frequency of collisions between NM and Rnq1 aggregates, but not a high local concentration of NM, is critical for the polymerization of NM. In addition to this result, the different frequencies of [PSI+] induction by the NM-Rnq1 fusion protein in different [RNQ+] strains (Fig. 3D) also support a physical interaction between NM and Rnq1 aggregates.
In this study, we developed a new, highly efficient method for [PSI+] induction that supports a cross-seeding activity of Rnq1 aggregates on Sup35. The convenience and high efficacy of the protein fusion method might make it a versatile tool for unraveling the process of prion formation.
Acknowledgments
We thank Susan W. Liebman (University of Illinois at Chicago) for generously providing the 74-D694 strain and its [RNQ+] variants and Won-Ki Huh for anti-Hxk antibody.
This work was supported by the 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Science & Technology (grant MG08-0201-1-0), and by a Korea Science and Engineering Foundation grant funded by MOST (R01-2007-000-20590-0), Republic of Korea. H.-J. Kim and Y. Ryu were supported by second-stage BK21 research fellowships from the Korean government.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Bradley, M. E., H. K. Edskes, J. Y. Hong, R. B. Wickner, and S. W. Liebman. 2002. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4)16392-16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coustou, V., C. Deleu, S. Saupe, and J. Begueret. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 949773-9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagkessamanskaya, A., M. Ter-Avanesyan, and W. H. Mager. 1997. Transcriptional regulation of SUP35 and SUP45 in Saccharomyces cerevisiae. Yeast 131265-1274. [DOI] [PubMed] [Google Scholar]
- 4.Derkatch, I. L., M. E. Bradley, J. Y. Hong, and S. W. Liebman. 2001. Prions affect the appearance of other prions: the story of [PIN+]. Cell 106171-182. [DOI] [PubMed] [Google Scholar]
- 5.Derkatch, I. L., M. E. Bradley, and S. W. Liebman. 1998. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl. Acad. Sci. USA 952400-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkatch, I. L., Y. O. Chernoff, V. V. Kushnirov, S. G. Inge-Vechtomov, and S. W. Liebman. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1441375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derkatch, I. L., S. M. Uptain, T. F. Outeiro, R. Krishnan, S. L. Lindquist, and S. W. Liebman. 2004. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 10112934-12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dos Reis, S., B. Coulary-Salin, V. Forge, I. Lascu, J. Begueret, and S. J. Saupe. 2002. The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J. Biol. Chem. 2775703-5706. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira, P. C., F. Ness, S. R. Edwards, B. S. Cox, and M. F. Tuite. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 401357-1369. [DOI] [PubMed] [Google Scholar]
- 10.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425737-741. [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and R. H. Schiestl. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 231-34. [DOI] [PubMed] [Google Scholar]
- 12.Glover, J. R., A. S. Kowal, E. C. Schirmer, M. M. Patino, J. J. Liu, and S. Lindquist. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89811-819. [DOI] [PubMed] [Google Scholar]
- 13.Han, H., P. H. Weinreb, and P. T. Lansbury, Jr. 1995. The core Alzheimer's peptide NAC forms amyloid fibrils which seed and are seeded by β-amyloid: is NAC a common trigger or target in neurodegenerative disease? Chem. Biol. 2163-169. [DOI] [PubMed] [Google Scholar]
- 14.Kaganovich, D., R. Kopito, and J. Frydman. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 4541088-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King, C. Y., and R. Diaz-Avalos. 2004. Protein-only transmission of three yeast prion strains. Nature 428319-323. [DOI] [PubMed] [Google Scholar]
- 16.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16857-860. [DOI] [PubMed] [Google Scholar]
- 17.Liu, J. J., N. Sondheimer, and S. L. Lindquist. 2002. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc. Natl. Acad. Sci. USA 99(Suppl. 4)16446-16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma, J., and S. Lindquist. 1999. De novo generation of a PrPSc-like conformation in living cells. Nat. Cell Biol. 1358-361. [DOI] [PubMed] [Google Scholar]
- 19.Osherovich, L. Z., and J. S. Weissman. 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106183-194. [DOI] [PubMed] [Google Scholar]
- 20.Patino, M. M., J. J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273622-626. [DOI] [PubMed] [Google Scholar]
- 21.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1997. In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277381-383. [DOI] [PubMed] [Google Scholar]
- 22.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 153127-3134. [PMC free article] [PubMed] [Google Scholar]
- 23.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 9513363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajan, R. S., M. E. Illing, N. F. Bence, and R. R. Kopito. 2001. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA 9813060-13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripaud, L., L. Maillet, and C. Cullin. 2003. The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. EMBO J. 225251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salnikova, A. B., D. S. Kryndushkin, V. N. Smirnov, V. V. Kushnirov, and M. D. Ter-Avanesyan. 2005. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 2808808-8812. [DOI] [PubMed] [Google Scholar]
- 27.Schlumpberger, M., S. B. Prusiner, and I. Herskowitz. 2001. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 217035-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sondheimer, N., and S. Lindquist. 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5163-172. [DOI] [PubMed] [Google Scholar]
- 29.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 144365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung, M. K., and W. K. Huh. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24767-775. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, M., P. Chien, N. Naber, R. Cooke, and J. S. Weissman. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428323-328. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, K. L., N. Cheng, R. W. Williams, A. C. Steven, and R. B. Wickner. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 2831339-1343. [DOI] [PubMed] [Google Scholar]
- 33.Ter-Avanesyan, M. D., A. R. Dagkesamanskaya, V. V. Kushnirov, and V. N. Smirnov. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uptain, S. M., and S. Lindquist. 2002. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56703-741. [DOI] [PubMed] [Google Scholar]
- 35.Vitrenko, Y. A., E. O. Gracheva, J. E. Richmond, and S. W. Liebman. 2007. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J. Biol. Chem. 2821779-1787. [DOI] [PubMed] [Google Scholar]
- 36.Vitrenko, Y. A., M. E. Pavon, S. I. Stone, and S. W. Liebman. 2007. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr. Genet. 51309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickner, R. B. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264566-569. [DOI] [PubMed] [Google Scholar]
- 38.Wright, C. F., S. A. Teichmann, J. Clarke, and C. M. Dobson. 2005. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438878-881. [DOI] [PubMed] [Google Scholar]