Abstract
Class III chitin synthases play important roles in tip growth and conidiation in many filamentous fungi. However, little is known about their functions in those processes. To address these issues, we characterized the deletion mutant of a class III chitin synthase-encoding gene of Aspergillus nidulans, chsB, and investigated ChsB localization in the hyphae and conidiophores. Multilayered cell walls and intrahyphal hyphae were observed in the hyphae of the chsB deletion mutant, and wavy septa were also occasionally observed. ChsB tagged with FLAG or enhanced green fluorescent protein (EGFP) localized mainly at the tips of germ tubes, hyphal tips, and forming septa during hyphal growth. EGFP-ChsB predominantly localized at polarized growth sites and between vesicles and metulae, between metulae and phialides, and between phalides and conidia in asexual development. These results strongly suggest that ChsB functions in the formation of normal cell walls of hyphae, as well as in conidiophore and conidia development in A. nidulans.
Chitin, a polymer of β-1,4-linked N-acetylglucosmine, is one of the major structural components of the fungal cell wall. Its metabolism, including synthesis, degradation, assembly, and cross-linking to other cell wall components, is thought to be very important for many fungi (5, 22, 24, 36, 45). Fungal chitin synthases have been classified into seven groups, classes I to VII, depending on the structures of their conserved regions (6). The genes encoding the synthases belonging to classes III, V, VI, and VII are only found in fungi with high chitin contents in their cell walls. We have identified six chitin synthase genes from Aspergillus nidulans and designated them chsA, chsB, chsC, chsD, csmA, and csmB; these gene products belong to classes II, III, I, IV, V, and VI, respectively (9, 13, 30, 31, 44, 52). The chsB deletion mutant grew very slowly and formed small colonies with highly branched hyphae, suggesting its important role in hyphal tip growth (3, 52). Repression of chsB expression in the deletion mutant of chsA, chsC, or chsD exaggerated the defects in the formation of aerial hyphae, the production of cell mass, or the growth under high-osmolarity conditions, respectively, compared to each single mutant. These results indicate that chsB functions at various stages of development (15, 16).
The deletion of class III chitin synthase-encoding genes leads to severe defects in most of the filamentous fungi thus far investigated. However, their detailed functions are currently unknown. In Neurospora crassa, inactivation of the gene encoding Chs-1, a class III chitin synthase with 63% identity to A. nidulans ChsB, leads to slow growth, aberrant hyphal morphology, and a decrease in chitin synthase activity. The mutant of chs-1 became sensitive to Nikkomycin Z, a chitin synthase inhibitor (53). In Aspergillus fumigatus, two genes encoding class III chitin synthases, chsC and chsG, have been identified. Their gene products showed 66 and 89% identity, respectively, to A. nidulans ChsB. The chsG deletion mutant showed slow growth and defects in conidiation, and its hyphae were highly branched. chsC deletion did not cause any phenotypic change. The chsC chsG double deletion mutant showed almost the same phenotype as the chsG single deletion mutant (28). Class III chitin synthases have been reported to be involved in the virulence of some pathogens. Deletion of Bcchs3a in the phytopathogenic fungus Botrytis cinerea and double deletion of WdCHS3 and class I chitin synthase WdCHS2 in the human pathogen Wangiella dermatitidis both caused a reduction of virulence (40, 48). On the other hand, the deletion mutant of a class III chitin synthase-encoding gene, CgChsIII, of the maize pathogen Colletotrichum graminicola did not exhibit the significant phenotypic difference from the wild-type strain (50). Deletion of a gene, chs1, encoding a class III chitin synthase of the maize pathogenic dimorphic fungi Ustilago maydis caused minor defects in the growth of haploid yeastlike cells and conjugation tube formation (49). These results indicate that the functions of class III chitin synthases has evolutionally diverged.
In the present study, we characterized the cytological defects of the A. nidulans chsB deletion mutant and investigated the localization of ChsB using FLAG- or enhanced green fluorescent protein (EGFP)-tagged ChsB. We reveal that the deletion mutant formed hyphae with aberrant cell wall structures and that ChsB tagged with EGFP primarily localized at polarized growth sites during germination, hyphal growth, septation, and conidiation. These findings suggest that ChsB functions at the polarized growth sites and forming septa during the hyphal growth and conidia development.
MATERIALS AND METHODS
Strains, media, and bacterial and fungal transformations.
The A. nidulans strains used in the present study are listed in Table 1. Complete medium (YG; 0.5% yeast extract, 1% glucose, 0.1% trace elements) and minimal medium (MMG) for A. nidulans were used (37). YG and MMG plates consisted of YG and MMG containing 1.5% agar. MMG was supplemented with arginine at 0.2 mg/ml, biotin at 0.02 μg/ml, pyridoxine at 0.5 μg/ml, and uridine at 2.44 mg/ml, when necessary. To support the growth of the mutants containing pyrG89, we supplemented the media with uridine at 2.44 mg/ml and uracil at 1.12 mg/ml. We used 2% glucose for Y2G medium. For YTF and MMTF media, 100 mM threonine and 0.1% fructose were used as carbon sources instead of glucose. All of the strains used in the present study were grown at 37°C. Bacterial and fungal transformations were performed as described previously (43).
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| FGSC A26 | biA1 | FGSCa |
| ABPU1 | biA1 pyrG89 argB2 pyroA4 wA3 | 30 |
| ABPU/A1 | biA1 pyrG89 argB2 pyroA4 wA3 [pSS1] | 10 |
| BM-3 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr-4-alcA(p)-chsB | 16 |
| BM-3/A1 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr-4-alcA(p)-chsB [pSS1] | 16 |
| FB-3, 4 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr-4-alcA(p)-chsBargB::chsB(p)-3XFLAG-chsB | This study |
| EB-5, 6 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr-4-alcA(p)-chsBargB::chsB(p)-egfp-chsB | This study |
| B-1, 3 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr-4-alcA(p)-chsB argB::chsB | This study |
| ΔB-18, 57 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::argB | This study |
FGSC, Fungal Genetics Stock Center, Kansas City, KS.
Plasmid constructions.
To create strains in which the wild-type ChsB was expressed under the control of alcA promoter and ChsB tagged with three repeats of FLAG epitope or EGFP at its N terminus was expressed from its own promoter at the argB locus, we constructed the plasmids pFB-argB, pEB-argB, and pB-argB as follows: 4.9-kb HindIII-AflII fragment from pchsB that contains chsB(p)-chsB (16) was blunted and ligated with SphI-digested and blunted pSS1 (30) to yield pB-argB. A 1.5-kb PstI fragment from pB-argB containing the promoter region of chsB, 3XFLAG, and the coding region of the 5′ part of chsB was ligated with PstI-digested pUC118 to yield pUC118-pB. The 0.1-kb NcoI fragment from p6XFLAG containing 3XFLAG (18) was ligated with NcoI-digested pUC118-pB to yield pUC118-3XFLAGpB. A primer set of EGFP5′-forward (5′-CTAGAGGATCCCCGGGTACCGGTCG-3′) and EGFP3′-reverse (5′-CGGCCCATGGACTTGTACAGCTCGTCC-3′) was used to amplify a fragment containing entire coding region of EGFP from pEGFP (Clontech). We confirmed that no mutation was introduced into the sequence in the cloned PCR product. The fragment was digested by NcoI and ligated with NcoI-digested pUC118-pB to yield pUC118-EGFPpB. The 1.6-kb and 2.2-kb PstI fragments from pUC118-3XFLAG and pUC118-EGFPpB were ligated with PstI-digested pB-argB to yield pFB-argB and pEB-argB, respectively. To create a chsB deletion strain, we constructed pΔchsB as follows: a primer set of chsB-null fw (5′-GCTCTAGAGCCCAATGAGTGTGCACTCC-3′) and chsB-null rv (5′-GCTCTAGAGCCAGCTGAGCAGCTGTAAC-3′) (the underlined letters represent the XbaI recognition site) was used to amplify a 5.2-kb fragment from total DNA of FGSC A26. The 5.2-kb fragment was ligated with XbaI-digested pUC18 to yield pUC18-chsB. A 1.8-kb BamHI-SphI-digested fragment from pSS1 was blunted and ligated with BalI- and EcoRV-digested pUC18-chsB to yield pΔchsB.
Construction of A. nidulans strains.
We constructed chsB deletion mutants as follows. A 3.7-kb ApaLI fragment of pΔchsB was introduced into the ABPU1 strain. Deletion of chsB in some transformants was confirmed by Southern blot analysis. The total DNA of the transformants was extracted as described previously (33) and digested with XbaI. Southern blot analysis was performed as described previously (14). The 1.2-kb EcoRV- and XbaI-digested fragment from pUC18-chsB was used as a probe. A 5.5-kb band was detected in the wild-type strain, while a 4.0-kb band was detected in two transformants (data not shown). These were designated ΔB-18 and ΔB-57. Since no phenotypic difference was found between the two transformants under the various conditions tested, we used ΔB-18 for further analysis.
We constructed strains in which the native ChsB was expressed under the control of the alcA promoter at the chsB locus, and ChsB tagged with three copies of FLAG epitope or a copy of EGFP at its N terminus was expressed from the chsB promoter at the argB locus. A strain that expressed native ChsB instead of FLAG-ChsB or EGFP-ChsB at the argB locus was also constructed as a control strain. The BglII fragment of pFB-argB, pEB-argB, or pB-argB was introduced into the BM-3 strain (16). Integration at the argB locus was analyzed by Southern blot analysis. The transformants in which one copy of chsB(p)-3XFLAG-chsB, chsB(p)-egfp-chsB, or chsB was integrated at the argB locus were obtained and designated them FB-3 and FB-4 for FLAG-ChsB, EB-5 and EB-6 for EGFP-ChsB, and B-1 and B-3 for ChsB. Since no phenotypic difference was observed in the strains with the same genotype under the various conditions tested, we used FB-3, EB-5, and B-1 for further analysis.
Determination of conidiation efficiency.
Conidiation efficiency was determined by a previously described plating method (17). The conidia were spread on Y2G plates and incubated for 72 h.
TEM.
Fixation, embedding, and examination of samples with transmission electron microscopy (TEM) were carried out essentially as described previously (14, 18). Samples were prepared from the wild-type strain and the chsB deletion mutant grown on Y2G plates for 96 h. For ultrathin sections, mycelia obtained from plates were fixed for 5 h in 5% buffered glutaraldehyde (pH 7.0) with 0.1 M phosphate buffer and then fixed for 2 h in buffered 2% osmium tetroxide. After dehydration with a graded ethanol series, specimens were embedded in epoxy resin. The sections, cut on an ultramicrotome by using a glass knife, were stained with uranyl acetate and lead citrate and observed.
Fractionation of A. nidulans cell lysate and Western blot analysis.
Conidia of FB-3 and B-1 were inoculated into YG and cultured for 24 h. Preparation of cell lysates and fractionation by centrifugation were performed as described previously (43). The low-speed pellet (LSP), high-speed pellet (HSP), and high-speed supernatant (HSS) represent the 10,000 × g pellet, the 100,000 × g pellet, and the 100,000 × g supernatant, respectively. Western blot analysis was performed as described previously with slight modifications (18). Cell lysates were prepared using Multi-Beads Shocker (Yasui Kikai). To detect EGFP-ChsB, cell lysates were prepared as follows. Conidia of EB-5 and B-1 were inoculated into YG and cultured for 8 h. Their mycelia were frozen with liquid nitrogen and broken by grinding. FLAG-ChsB was detected by using a mouse anti-FLAG M2 monoclonal antibody (F3165, Sigma) at a 1:5,000 dilution and an anti-mouse immunoglobulin G (IgG), horseradish peroxidase-linked antibody (Cell Signaling Technology) at a 1:5,000 dilution. EGFP-ChsB was detected using a BD Living Colors monoclonal antibody (JL-8; Clontech) at a 1:1,000 dilution. A Can Get Signal immunoreaction enhancer solution (Toyobo) was used when necessary.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence microscopy was performed essentially as described previously (42). Cells were fixed and processed as described previously (8, 11). For cell wall digestion, the coverslips were overlaid for 1 h at room temperature with a 200-μl solution of phosphate-buffered saline (PBS; 13.7 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2HPO4) containing 6 mg of Yatalase (TaKaRa), 0.6 mg of lysing enzymes L2265 (Sigma), and 1% egg white (Sigma). The coverslips were washed in PBSP (i.e., PBS containing 0.1% Nonidet P-40) and immersed in ethanol at −20°C for 10 min. After being washed several times in PBSP, the coverslips were incubated for more than1 h at room temperature with a mouse anti-FLAG primary antibody (Sigma) at a 1:500 dilution or a rabbit anti-actin primary antibody (Sigma) at a 1:500 dilution in PBSB (PBS containing 0.1% bovine serum albumin). After being washed several times in PBSP, the coverslips were incubated for 1 h at room temperature with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibody (Sigma) at a 1:200 dilution or Cy3-conjugated anti-rabbit IgG secondary antibody (Sigma) at a 1:1,000 dilution in PBSB in the dark. The coverslips were washed several times in PBSP and stained with 0.01% calcofluor white (CFW; fluorescent brightener 28; Sigma) for 5 min. Finally, the coverslips were washed several times in PBSP, mounted on glass slides in 10 ml of glycerol-PBS (9:1 [vol/vol]; 0.01% p-phenylenediamine), and observed by using a confocal laser microscope.
Fluorescence microscopy.
Conidia were spread on a thin MMG plate or were inoculated into MMG liquid medium and incubated at 37°C. Strains were stained with a solution containing 0.2 μg of 7-amino-4-chloromethylcoumarin (CMAC; Molecular Probes)/ml or 12 μg of CFW (F-6259; Sigma)/ml. Staining of N-(3-triethylammoniumpropyl)-4-[6-(4-diethylamino)phenylhexatrienyl] pyridinium dibromide (FM-4-64; Molecular Probes) was carried out as follows. Strains grown on MMG plate for 8 or 16 h at 37°C were stained with 10 μl of FM4-64 (1 μg/ml in dimethyl sulfoxide) for 10 min. Mycelia were visualized by using an Olympus BX52 fluorescence microscope. Images were taken and analyzed with ORCA-ER charge-coupled device camera (Hamamatsu) and Aquacosmos software (Hamamatsu). An Olympus FV500 fluorescence microscope and Fluoview (Olympus) were used for confocal laser microscopy. For CFW staining of the hyphae of ΔB-18, colonies were sliced with a razor before staining.
RESULTS
Cell wall formation of a chsB deletion mutant.
Although the characterization of the chsB deletion mutants of A. nidulans has already been reported (3, 52), cytological analysis of these mutants has not been performed. To avoid the possible interference by the production of N-terminal part of ChsB in the former chsB deletion mutant (52), we made a new chsB deletion mutant, ΔB-18, in which the expression of chsB was not expected (see Materials and Methods). ΔB-18 grew very slowly and formed very small colonies with highly branched hyphae. It did not form conidiophores and conidia (data not shown). The growth defects of ΔB-18 were similar to those of chsB deletion mutants obtained before by other group and were a little more severe than those of BM-3/A1 under the alcA-repressing conditions (3, 16, 52, data not shown), suggesting leaky expression of alcA(p)-chsB under the conditions used here.
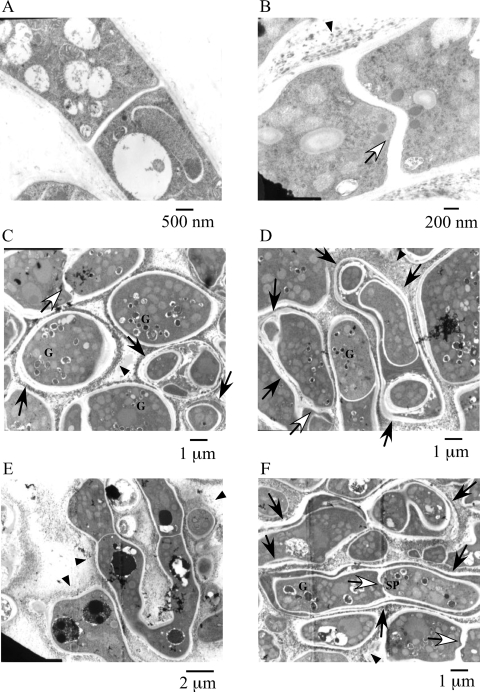
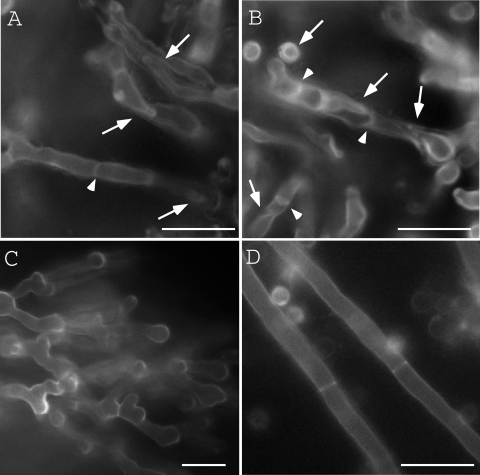
To investigate the cell wall and septum structures of ΔB-18 in detail, we observed them by TEM (Fig. 1). The hyphae of ΔB-18 were closely spaced. Its cell walls sometimes formed multilayer structures. Some of them seemed to be intrahyphal hyphae (Fig. 1C, D, and F, arrows). Amorphous structures were observed in the outside of the hyphae of ΔB-18 (Fig. 1B to F, arrowheads). These extracellular structures were similar to those observed in the class III chitin synthase mutant of Botrytis cinerea (40). Vesicles with electron dense contents were frequently seen in ΔB-18 (Fig. 1C, D, and F, indicated by a “G”). Since similar structures were observed in the hypA mutant of A. nidulans and they were thought to be aberrant Golgi equivalents (20, 39), we refer to these vesicles as Golgi equivalents. Some of the septa extended in a wavy manner (Fig. 1B, C, D, and F, open arrows). To investigate the overall structures of these multilayered cell walls and intrahyphal hyphae, the mutant colonies were stained with CFW and observed by fluorescence microscopy (Fig. 2). Multilayered cell walls and intrahyphal hyphae were seen (Fig. 2, arrows) and increased in an incubation time-dependent manner. “Wavy” septa were also observed (Fig. 2, arrowheads). Considering their morphologies, some of them would be the initial stage of the intrahyphal hyphae formation. These results suggest that ChsB is crucial for the formation of normal hyphal cell walls and conidia. Intrahyphal hyphae, multilayered cell walls, and wavy septa were hardly observed in the marginal regions of the colonies, indicating that they were predominantly formed in the old part of hyphae (Fig. 2C).
FIG. 1.
TEM of the chsB deletion mutant. Panels show ABPU/A1 (wild type) (A) and ΔΒ-18 (chsB deletion mutant) (B to F). Strains were grown on Y2G plates for 4 days. Arrows indicate double or multilayered cell walls. Open arrows indicate wavy septa. Arrowheads indicate amorphous structures. Septal pore and Golgi equivalents with electron dense contents are indicated by SP and G, respectively.
FIG. 2.
CFW staining of the hyphae of ΔB-18. ΔB-18 was grown on YG plates for 2 days (A) or 4 days (B and C), and ABPU/A1 was grown for 2 days (D). Internal regions (A, B, and D) and a marginal region (C) of the colonies are shown. Arrows indicate multilayered cell walls or intrahyphal hyphae. Arrowheads indicate wavy septa. Bars, 10 μm.
Construction of FLAG-ChsB, EGFP-ChsB-producing strains.
To determine the localization of ChsB, we constructed strains in which ChsB tagged with three copies of FLAG or a copy of EGFP at its N terminus was expressed from the promoter of chsB at the argB locus. The strains that expressed FLAG-ChsB and EGFP-ChsB were designated FB-3 and EB-5, respectively. The strain that expressed the native ChsB instead of FLAG-ChsB or EGFP-ChsB at the argB locus was also constructed as a control strain and was designated B-1. In these strains, the native ChsB was expressed under the control of the alcA promoter at the chsB locus. The alcA promoter was scarcely expressed in the presence of glucose as a carbon source (MMG, YG, or Y2G medium). FB-3 and EB-5 grew as well as the wild-type strain, their hyphal morphology was normal, and their conidiation efficiency was much higher than that of BM-3 under the alcA-repressing conditions (Table 2). These results demonstrated that FLAG-ChsB and EGFP-ChsB were nearly as functional as the native ChsB.
TABLE 2.
Growth rate and conidiation efficiency of the strains B-1, FB-3, EB-5, and BM-3
| Straina | Production of ChsBb | Mean ± SD (%)c
|
|
|---|---|---|---|
| Colony diam (mm) | No. of conidia (106/mm2) | ||
| B-1 | ChsB | 38.6 ± 1.0 (100) | 2.9 ± 0.3 (100) |
| FB-3 | FLAG-ChsB | 36.6 ± 1.1 (95) | 2.8 ± 0.1 (97) |
| EB-5 | EGFP-ChsB | 32.4 ± 1.4 (92) | 1.8 ± 0.1 (62) |
| BM-3 | 5.8 ± 0.6 (15) | (>0.01) | |
Strains were grown on the Y2G plates for 72 h.
It is likely that a very small amount of ChsB was also produced from alcA(p)-chsB in each strain by the leaky expression of alcA promoter under the condition used in this study (see the text).
The ratio of growth rate and conidiation efficiency of each strain to that of the wild-type strain is shown in parentheses. The conidiation efficiency of BM-3/A1 was below the measurable level.
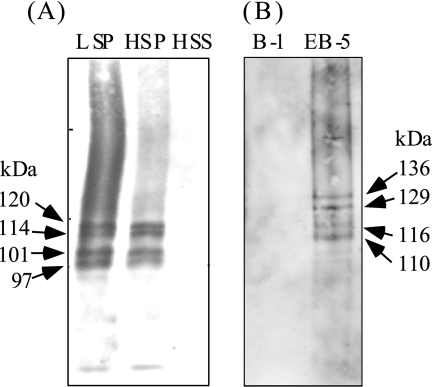
Western blot analysis of FLAG-ChsB in FB-3 and EGFP-ChsB in EB-5.
FLAG-ChsB in the cells was detected by Western blot analysis with anti-FLAG antibody. Four major bands were seen in the LSP and HSP fractions of FB-3 cell extract at approximate molecular masses of 120, 114, 101, and 97 kDa but not in the HSS fraction (Fig. 3A). No band was seen in the cell extract of the control strain B-1 (data not shown). In silico analysis revealed that ChsB has about four to seven transmembrane regions at its C terminus (four transmembrane regions were predicted by SOSUI [http://bp.nuap.nagoya-u.ac.jp/sosui/cgi-bin/adv_sosui.cgi] and seven transmembrane regions were predicted by TMHMM [http://www.cbs.dtu.dk/services/TMHMM-2.0/]). These findings suggest that ChsB is a membrane protein. Since the estimated molecular mass of FLAG-ChsB was 105.8 kDa, it is also suggested that some portion of FLAG-ChsB was modified posttranslationally. ChsB did not seem to be N glycosylated because the sizes of these four bands were not changed when they were treated with glycopeptidase F (data not shown). Four major bands at an approximate molecular masses of 136, 129, 116, and 110 kDa were also observed when the cell extract of EB-5 was subjected to Western blot analysis with anti-GFP antibody (Fig. 3B). The estimated molecular mass of EGFP-ChsB was 129 kDa.
FIG. 3.
Western blot analysis of FB-3 (A) and EB-5 (B) cell lysates. (A) FLAG-ChsB in the lysate of FB-3 was detected with anti-FLAG antibody. LSP, the 10,000 × g pellet; HSP, the 100,000 × g pellet; HSS, the 100,000 × g supernatant. Arrows indicate the positions of FLAG-ChsB and their approximate molecular masses. (B) The lysates of EB-5 and B-1 were subjected to Western blot analysis with anti-GFP antibody. Arrows indicate the positions of EGFP-ChsB and their approximate molecular masses.
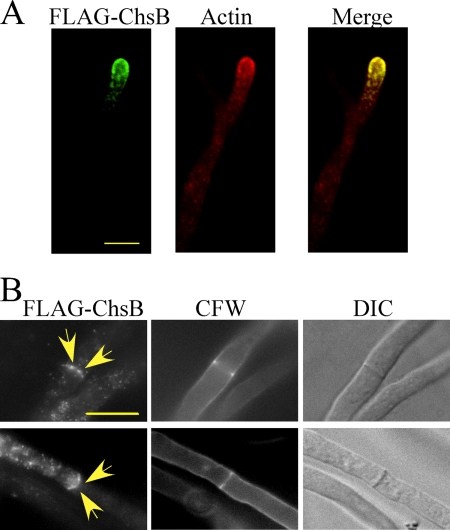
Localization of FLAG-ChsB.
Localization of FLAG-ChsB in the hyphae of FB-3 was determined by indirect immunofluorescence with anti-FLAG antibody. The fluorescence was primarily observed at the hyphal tips and septa (Fig. 4). Fluorescence at the hyphal tips was observed as apical crescent covering the hyphal tips. Punctuate loci near the cell surfaces and in the cytoplasm near the tips were also observed (Fig. 4A). No fluorescence was observed when the wild-type strain was subjected to indirect immunofluorescence (data not shown). Double staining of FLAG-ChsB and actin showed that most punctuate loci at the hyphal tips colocalized with actin, suggesting that these structures are associated with actin patches (Fig. 4A). FLAG-ChsB also localized at a small number of septa. This suggests that FLAG-ChsB localizes at forming septa (see below). In these septa, we observed a pair of spots with intense fluorescence (Fig. 4B, arrows).
FIG. 4.
Localization of FLAG-ChsB at hyphal tips and septa. Hyphae of FB-3 strain grown in MMG for 12 h were fixed, stained with anti-FLAG and anti-actin antibodies (A) or with anti-FLAG antibody (B), and visualized by indirect immunofluorescence microscopy. These hyphae were stained simultaneously with CFW (B). The images were obtained by confocal microscopy. Arrows indicate strong fluorescence of FLAG-ChsB. Bars, 5 μm.
Time-lapse of EGFP-ChsB at the hyphal tips and septa.
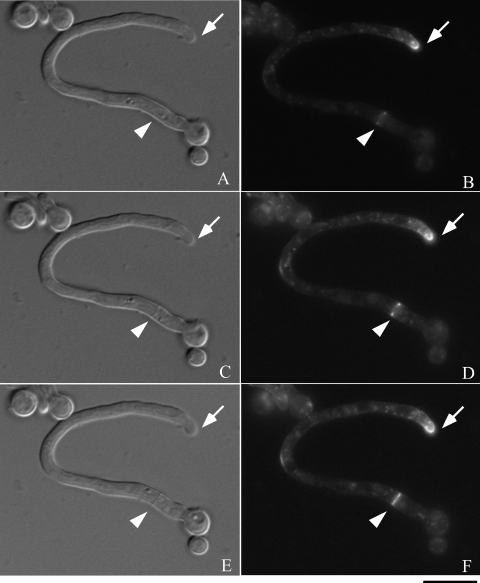
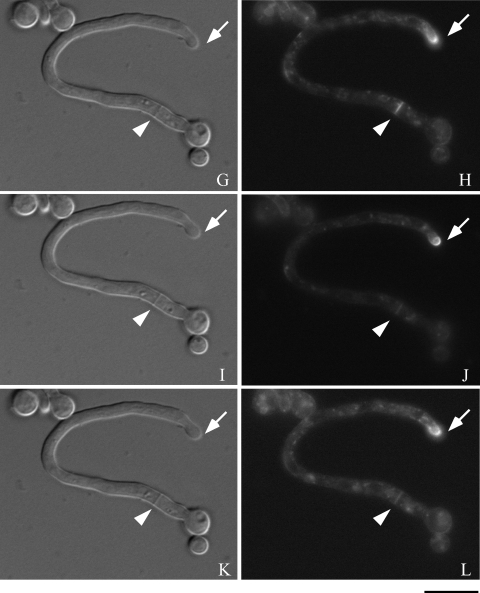
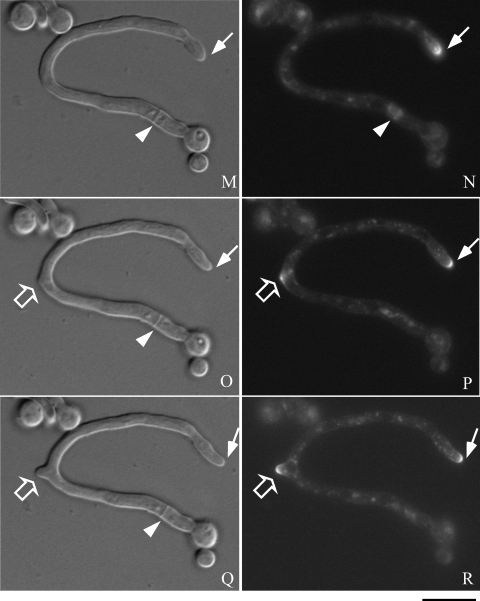
Next, to investigate the time-lapse movement of chsB in the hyphae, we observed the fluorescence of EGFP-ChsB in a living hypha of EB-5. Time-lapse images were taken after the conidia of EB-5 developed their germ tubes. During that period strong fluorescence was constantly observed at a hyphal tip as an apical crescent (Fig. 5B, D, F, H, J, L, N, P, and R). Relatively weak fluorescence of EGFP-ChsB was detected in the hypha as punctuate loci. EGFP-ChsB also localized to the newly formed branch site (Fig. 5P and R, open arrows). We observed temporary localization of EGFP-ChsB at the later branch site (Fig. 5D and F). Since we detected that EGFP-ChsB sometimes localized at the hyphal periphery and disappeared several minutes later, it is likely that the localization of EGFP-ChsB at the hyphal periphery is not necessarily related to the sites of hyphal branching (data not shown).
FIG. 5.
Time-lapse images of EGFP-ChsB localization in a germ tube. Hyphae of the EB-5 grown on an MMG plate for 8 h were observed by fluorescence microscopy. Panels A, C, E, G, I, K, M, O, and Q are DIC images. Panels B, D, F, H, J, L, N, P, and R are GFP images. (A and B) 0 min; (C and D) 3 min; (E and F) 6 min; (G and H) 9 min; (I and J) 12 min; (K and L) 15 min; (M and N) 30 min; (O and P) 45 min; (Q and R) 60 min. Solid arrows indicate a hyphal tip, arrowheads indicate a septation site, and open arrows indicate a branching site. Bars, 10 μm.
We also observed that EGFP-ChsB temporally localized at the forming septa. Strong fluorescence was originated from the periphery of the hypha and extended to the center for approximately 10 min (Fig. 5B, D, F, and H, arrowheads). Septum formation was simultaneously observed in differential interference contrast (DIC) images at the same location (Fig. 5A, C, E, and G, arrowheads). These results suggest that EGFP-ChsB moved following the leading edges of membrane at the forming septa. The fluorescence of EGFP-ChsB disappeared approximately 30 min after it reached the center of the septa (Fig. 5N and P, and data not shown).
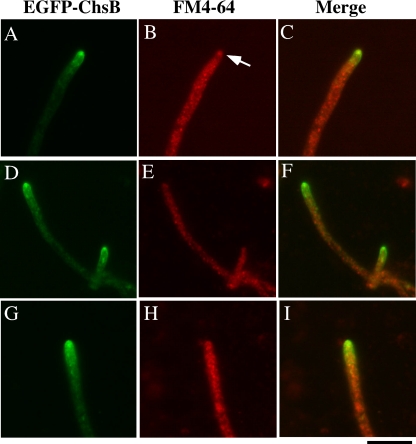
Recently, it was reported that Spitzenkörper was hardly seen in the germ tubes, suggesting that the mechanism of tip growth in germ tubes is not the same as that of mature hyphae (12, 41). Thus, we investigated the localization of EGFP-ChsB in the living mature hyphae grown for 16 h (Fig. 6). To clarify the presence or localization of Spitzenkörper, the hyphae were simultaneously stained with FM4-64. EGFP-ChsB was distributed as the apical crescent accompanied with a bright spot in most hyphal tips (Fig. 6A and D). Approximately half of these spots were colocalized with the spots stained with FM4-64, suggesting that EGFP-ChsB in the bright spots were associated with Spitzenkörper (Fig. 6B). However, another half was not stained with FM4-64 (Fig. 6E). Apical crescents without bright spots were also observed (Fig. 6G). These results suggest that ChsB changes its distribution in matured hyphal tips.
FIG. 6.
Localization of EGFP-ChsB in the hyphae grown on an MMG plate for 16 h. Hyphae were stained with FM-4-64 and observed by fluorescence microscopy. (A, D, and G) GFP images; (B, E, and H) FM4-64 fluorescent image; (C, F, and I) merged images. Each row of images (i.e., A to C, D to F, and G to I) shows the same field. An arrow indicates the Spitzenkörper. Bar, 10 μm.
In the elongated hyphae, some round-shaped fluorescence patterns were observed primarily in the cytoplasm of the old part of the hyphae. These fluorescent structures in the cytoplasm were not observed in the images of indirect immunofluorescence of FLAG-ChsB and were also stained with CMAC, which is known to stain vacuoles (data not shown). Since the degradation products of EGFP-ChsB were detected in the Western blot analysis when EB-5 was grown under this condition (data not shown), it is possible that partially degraded EGFP-ChsB was accumulated in vacuoles.
Localization of EGFP-ChsB in the conidia during conidial germination.
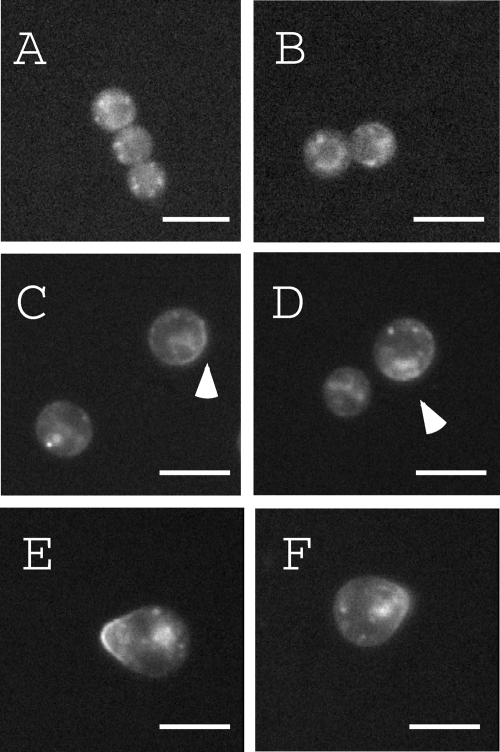
Localization of EGFP-ChsB in the conidia during conidial germination is shown in Fig. 7. A punctuate distribution of EGFP-ChsB was observed inside the conidia during isotropic growth of germinating conidia (Fig. 7A and B). After 4 h of incubation on an MMG plate, a portion of EGFP-ChsB localized at the surface region of conidia (Fig. 7C and C, arrowheads). After 6 h of incubation, EGFP-ChsB localized at the tips of germ tubes (Fig. 7E and F). These results suggest that EGFP-ChsB localized at the plasma membrane shortly before the emergence of germ tubes.
FIG. 7.
Localization of EGFP-ChsB during germination. Conidia of EB-5 were inoculated on MMG plates (A) and incubated for 2 h (B), 4 h (C and D), and 6 h (E and F). Arrowheads indicate the cell surface localization of EGFP-ChsB. Bars, 5 μm.
Localization of EGFP-ChsB during conidiophore development.
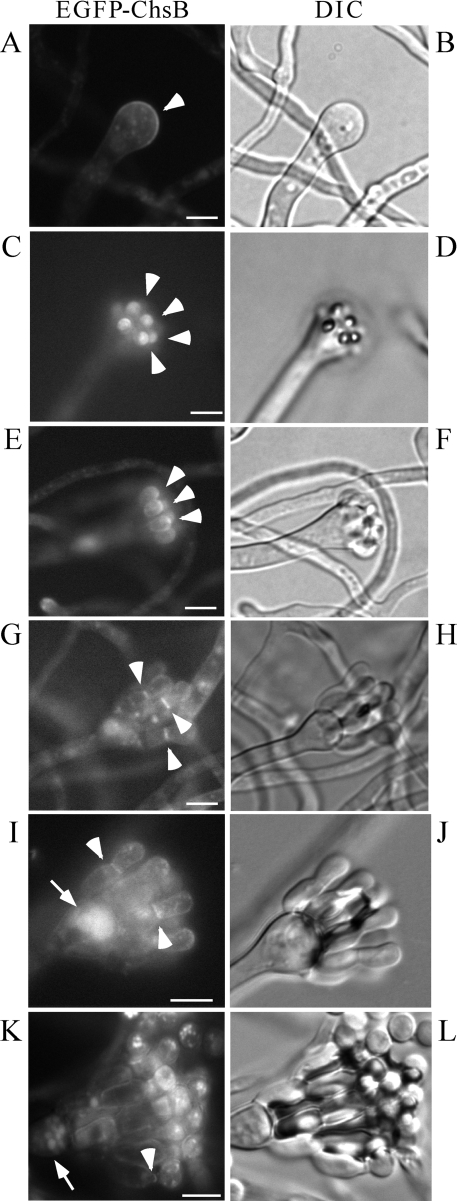
Next, we examined the localization of EGFP-ChsB during conidiophore development. EGFP-ChsB localized at the surface regions of the conidiophore vesicles before the metulae were developed. During the metula extension, EGFP-ChsB localized at their tips. Then, EGFP-ChsB localized at the septa between the metulae and phialides. EGFP-ChsB also localized between conidia and phialides (Fig. 8A, C, E, G, I, and K, arrowheads). These results suggest that ChsB also plays critical roles in the development of conidiophores and conidia. In the conidia, fluorescence was frequently observed as small dots and was occasionally observed at the rim of conidia (Fig. 8E, G, and I). Fluorescence was also observed in some round-shaped structures distributed primarily in the conidiophore vesicles (Fig. 8I and K, arrows). The shapes of these structures and the fact that they were stained with CMAC suggest that they were vacuoles (data not shown).
FIG. 8.
Localization of EGFP-ChsB in the conidiophore of EB-5. Conidia of EB-5 grown on MMG plate for 24 h and each step of conidiophore and conidia formation was observed by fluorescence microscopy. The formation of conidiophore vesicle (A and B), metulae (C to F), phialides (G to J), and conidia (K and L) is depicted. Panels A, C, E, G, I, and K are GFP images. Panels B, D, F, H, J, and L are DIC images. Arrowheads indicate the strong fluorescence of EGFP-ChsB, and arrows indicate vacuoles. Bars, 5 μm.
DISCUSSION
Although class III chitin synthases are only found in fungi with high chitin content and are thought to play a key role in their hyphal morphogenesis in many filamentous fungi, their functions at the molecular level remain poorly understood. In the present study, we showed the detailed cell wall structures of the deletion mutant of the class III chitin synthase-encoding gene, chsB, and its localizations in A. nidulans.
The deletion mutant of chsB exhibited severe growth defects and formed hyphae with multilayered cell walls and intrahyphal hyphae (Fig. 1 and 2). Formations of intrahyphal hyphae or double-layered cell walls were observed in some deletion mutants of the class V or VI chitin synthase-encoding genes and the genes involved in the biosynthesis of the glycosylphosphatidylinositol anchor that connects proteins working for cell wall structure maintenance (4, 14, 25, 26, 32, 44). In N. crassa and Aspergillus oryzae, it has been reported that new hyphae are generated from septa when Woronin body plugged a septal pore (19, 27, 45). Although the mechanisms of the formations of these abnormal structures are currently unknown, this implies that they are formed when septal pores are closed. The septal pores of the chsB deletion mutant are not necessarily closed (Fig. 1F, indicated by “SP”), but aberrant septal morphology of the chsB deletion mutant and temporal localization of EGFP-ChsB at forming septa of the wild-type cells suggest that the lack of ChsB might have damaged normal septum formation, which eventually generated intrahyphal hyphae and multilayered cell walls. The result that the intrahyphal hyphae and multilayered cell walls were predominantly observed in old part of hyphae supports this idea. It is also possible that intrahyphal hyphae and multilayered cell walls were formed by the invasions of other hyphae that were seen in the csmA deletion mutant (14). However, we did not observe the phenomenon in the chsB deletion mutant by CFW staining.
Putative dilated Golgi equivalents were frequently seen in the chsB deletion mutant. Similar structures were also observed in the hypA mutant of A. nidulans. hypA is an ortholog of TRS120 of S. cerevisiae, which encodes a regulatory component of the TRAPP II complex (39). This complex is involved in vesicle transport through the Golgi in S. cerevisiae. The tip growth was severely inhibited in the chsB deletion mutant. Thus, it is possible that this inhibition would disturb the vesicle transport in the cells and lead to form these abnormal structures.
Since FLAG-ChsB and EGFP-ChsB localized at the tips of germ tubes and hyphal tips, the function of ChsB is likely to be required for normal tip growth. During the growth of hyphal tips, it is conceivable that the enzymes involved in cell wall synthesis are transported to the tips. A strong fluorescence signal was constantly detected at the tip of the hypha in the time-lapse analysis of EGFP-ChsB (Fig. 5), suggesting that ChsB is continuously synthesized and is transported to hyphal tips during hyphal growth. In S. cerevisiae, the class II chitin synthase Chs2p is synthesized and transported to the bud neck. It is then internalized by endocytosis and transported to the vacuole for degradation (24). Since endocytosis occurs at the subapical regions of hyphae in A. nidulans (2, 34, 41, 46), ChsB is likely endocytosed at these regions of the hyphae and transported to the vacuole for degradation. We could not exclude the possibility that ChsB is recycled at the apical region of hyphae in A. nidulans like the class IV chitin synthase Chs3p of S. cerevisiae that is recycled to the plasma membrane at the bud neck after it is retrieved from the plasma membrane by endocytosis and transported through the early endosome and trans-Golgi network (54, 55). The punctuate loci observed in the cytoplasm are likely vesicles that transport EGFP-ChsB to hyphal tips.
Riquelme et al. reported that chitin synthases belonging to classes I and VII colocalized with Spitzenkörper at hyphal tips in N. crassa (35). We found that EGFP-ChsB was distributed as apical crescent at the tips of germ tubes and mature hyphae. In the latter case, approximately half of apical crescents were accompanied with single bright spots that were coincident with Spitzenkörper, and we could not detect this particular organization at the tips of germ tubes (data not shown). Recently, Köhli et al. reported that in Ashbya gossypii, Spitzenkörper was formed at the tips of fast-growing hyphae but not at the tips of slowly growing hyphae (21). Thus, EGFP-ChsB is also likely to change its localization at the hyphal tips depending on the hyphal elongation speed, because the elongation speeds of germ tubes are slower than those of mature hyphae (12, 41). The difference in the distribution of A. nidulans class III chitin synthase from those of N. crassa class I and VII enzymes might be also reflecting the difference in their classes or reflecting species specificity, since colocalization of a class VI chitin synthase of C. graminicola with Spitzenkörper was not detected (1).
FLAG-ChsB and EGFP-ChsB also localized at the forming septa. Chitin synthases of A. nidulans can be separated into two groups depending on their localization pattern at forming septa. The movement of strong fluorescence of EGFP-ChsB from the periphery to the center at the growing septum was similar to that of the class II chitin synthase, ChsA, of A. nidulans. On the other hand, ChsC, CsmA, and CsmB distributed as X-like shapes similar to the formin SepA and the actin cytoskeleton and separated into two rings during the septum formation (18, 29, 38, 42, 44). Based on their localization pattern, it is possible that ChsB and ChsA have redundant function in the formation of septa. However, the strain BDA-3 in which chsA was deleted and in which the expression of chsB was under the control of the alcA promoter (15) did not show aberrant septum formation under the alcA-repressing condition (data not shown). Localization of multiple chitin synthases at the septa has been also reported in other fungi (23, 49). In addition, it is suggested that classes I and IV chitin synthases, Chs3p and Chs8p, synthesize short-chitin rodlets and long-chitin microfibrils, respectively, at the same location in Candida albicans (23). Similar functional differences of the chitin synthases may also exist in A. nidulans.
EGFP-ChsB localized at the surface regions of conidiophore vesicles, the tips of metulae, the septa between the metulae and phialides, and between phialides and conidia (Fig. 8). These localization patterns are similar to those of the septin AspB of A. nidulans (51). In S. cerevisiae, the localization of class IV chitin synthase Chs3p at the bud neck depends on the interaction with septin through Chs4p and Bni4p (7). Thus, it is possible that these localization patterns of EGFP-ChsB are dependent on AspB. Since the chsB deletion mutant did not form conidiophores and conidia, our results suggest that the function of ChsB is crucial for the formation of conidiophore and conidial cell walls.
In the Western blot analysis of FB-3 and EB-5, four major bands of different molecular mass were observed (Fig. 3). This result suggests that ChsB was modified posttranslationally in A. nidulans. Posttranslational modification of class III chitin synthase (WdChs3p) was also suggested in W. dermatitidis (47). In that case, a smear band of high molecular weight was detected by Western blot analysis. Since the major bands of FLAG-ChsB and EGFP-ChsB (Fig. 3 and data not shown) were not smeared, it is likely that the modification(s) on ChsB was different from those of WdChs3p. We suggested previously that A. nidulans ChsA was also modified posttranslationally (18). The activities and/or localization of these chitin synthases may be regulated by these modifications.
Acknowledgments
This study was performed using the facilities of the Biotechnology Research Center, The University of Tokyo. This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Amnuaykanjanasin, A., and L. Epstein. 2006. A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma 227155-164. [DOI] [PubMed] [Google Scholar]
- 2.Araujo-Bazán, L., M. A. Peñalva, and E. A. Espeso. 2008. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol. Microbiol. 67891-905. [DOI] [PubMed] [Google Scholar]
- 3.Borgia, P. T., N. Iartchouk, P. J. Riggle, K. R. Winter, Y. Koltin, and C. E. Bulawa. 1996. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20193-203. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, S. M., A. Piwowar, M. Al Dabbous, J. Vierula, and S. J. Free. 2006. Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa. Eukaryot. Cell 5587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulawa, C. E. 1993. Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 47505-534. [DOI] [PubMed] [Google Scholar]
- 6.Choquer, M., M. Boccara, I. R. Goncalves, M. C. Soulie, and A. Vidal-Cros. 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2712153-2164. [DOI] [PubMed] [Google Scholar]
- 7.DeMarini, D. J., A. E. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 13975-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnault, K., B. el Moudni, J. P. Bouchara, D. Chabasse, and G. Tronchin. 1999. Association of a myosin immunoanalogue with cell envelopes of Aspergillus fumigatus conidia and its participation in swelling and germination. Infect. Immun. 671238-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 23675-78. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara, M., M. Ichinomiya, T. Motoyama, H. Horiuchi, A. Ohta, and M. Takagi. 2000. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. 127359-366. [DOI] [PubMed] [Google Scholar]
- 11.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horio, T. 2007. Role of microtubules in tip growth of fungi. J. Plant Res. 12053-60. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi, H. 2009. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development, and septum formation. Med. Mycol. S1S47-S52. [DOI] [PubMed] [Google Scholar]
- 14.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 1813721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichinomiya, M., H. Horiuchi, and A. Ohta. 2002. Different functions of the class I and class II chitin synthase genes, chsC and chsA, are revealed by repression of chsB expression in Aspergillus nidulans. Curr. Genet. 4251-58. [DOI] [PubMed] [Google Scholar]
- 16.Ichinomiya, M., T. Motoyama, M. Fujiwara, M. Takagi, H. Horiuchi, and A. Ohta. 2002. Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology 1481335-1347. [DOI] [PubMed] [Google Scholar]
- 17.Ichinomiya, M., A. Ohta, and H. Horiuchi. 2005. Expression of asexual developmental regulator gene abaA is affected in the double mutants of classes I and II chitin synthase genes, chsC and chsA, of Aspergillus nidulans. Curr. Genet. 48171-183. [DOI] [PubMed] [Google Scholar]
- 18.Ichinomiya, M., E. Yamada, S. Yamashita, A. Ohta, and H. Horiuchi. 2005. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 41125-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jedd, G., and N. H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2226-231. [DOI] [PubMed] [Google Scholar]
- 20.Kaminskyj, S. G., and M. R. Boire. 2004. Ultrastructure of the Aspergillus nidulans hypA1 restrictive phenotype shows defects in endomembrane arrays and polarized wall deposition. Can. J. Bot. 82807-814. [Google Scholar]
- 21.Köhli, M., V. Galati, K. Boudier, R. W. Roberson, and P. Philippsen. 2008. Growth-speed-correlated localization of exocyst and polarisome components in growth zones of Ashbya gossypii hyphal tips. J. Cell Sci. 1213878-3889. [DOI] [PubMed] [Google Scholar]
- 22.Latgé, J. P., I. Mouyna, F. Tekaia, A. Beauvais, J. P. Debeaupuis, and W. Nierman. 2005. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med. Mycol. 43(Suppl. 1)S15-S22. [DOI] [PubMed] [Google Scholar]
- 23.Lenardon, M. D., R. K. Whitton, C. A. Munro, D. Marshall, and N. A. Gow. 2007. Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol. Microbiol. 661164-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesage, G., and H. Bussey. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70317-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H., H. Zhou, Y. Luo, H. Ouyang, H. Hu, and C. Jin. 2007. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 641014-1027. [DOI] [PubMed] [Google Scholar]
- 26.Martín-Urdíroz, M., M. I. Roncero, J. A. González-Reyes, and C. Ruiz-Roldán. 2008. ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot. Cell 7112-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maruyama, J., S. Kikuchi, and K. Kitamoto. 2006. Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 43642-654. [DOI] [PubMed] [Google Scholar]
- 28.Mellado, E., A. Aufauvre-Brown, N. A. Gow, and D. W. Holden. 1996. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 20667-679. [DOI] [PubMed] [Google Scholar]
- 29.Momany, M., and J. E. Hamer. 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskel. 38373-384. [DOI] [PubMed] [Google Scholar]
- 30.Motoyama, T., M. Fujiwara, N. Kojima, H. Horiuchi, A. Ohta, and M. Takagi. 1996. The Aspergillus nidulans genes chsA and chsD encode chitin synthases which have redundant functions in conidia formation. Mol. Gen. Genet. 251442-450. (Corrected and republished, 253:520-528, 1997.) [DOI] [PubMed] [Google Scholar]
- 31.Motoyama, T., N. Kojima, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation of a chitin synthase gene (chsC) of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 582254-2257. [DOI] [PubMed] [Google Scholar]
- 32.Müller, C., C. M. Hjort, K. Hansen, and J. Nielsen. 2002. Altering the expression of two chitin synthase genes differentially affects the growth and morphology of Aspergillus oryzae. Microbiology 1484025-4033. [DOI] [PubMed] [Google Scholar]
- 33.Oakley, C. E., C. F. Weil, P. L. Kretz, and B. R. Oakley. 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53293-298. [DOI] [PubMed] [Google Scholar]
- 34.Peñalva, M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42963-975. [DOI] [PubMed] [Google Scholar]
- 35.Riquelme, M., S. Bartnicki-García, J. M. González-Prieto, E. Sánchez-León, J. A. Verdín-Ramos, A. Beltrán-Aguilar, and M. Freitag. 2007. Spitzenkörper localization and intracellular traffic of green fluorescent protein-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell 61853-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41367-378. [DOI] [PubMed] [Google Scholar]
- 37.Rowlands, R. T., and G. Turner. 1973. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans. Mol. Gen. Genet. 126201-216. [DOI] [PubMed] [Google Scholar]
- 38.Sharpless, K. E., and S. D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, X., Y. Sha, and S. Kaminskyj. 2004. Aspergillus nidulans hypA regulates morphogenesis through the secretion pathway. Fungal Genet. Biol. 4175-88. [DOI] [PubMed] [Google Scholar]
- 40.Soulié, M. C., C. Perino, A. Piffeteau, M. Choquer, P. Malfatti, A. Cimerman, C. Kunz, M. Boccara, and A. Vidal-Cros. 2006. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell. Microbiol. 81310-1321. [DOI] [PubMed] [Google Scholar]
- 41.Taheri-Talesh, N., T. Horio, L. Araujo-Bazán, X. Dou, E. A. Espeso, M. A. Peñalva, S. A. Osmani, and B. R. Oakley. 2008. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 191439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 161961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeshita, N., A. Ohta, and H. Horiuchi. 2002. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 298103-109. [DOI] [PubMed] [Google Scholar]
- 44.Takeshita, N., S. Yamashita, A. Ohta, and H. Horiuchi. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 591380-1394. [DOI] [PubMed] [Google Scholar]
- 45.Trinci, A. P., and A. J. Collinge. 1974. Occlusion of the septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma 8057-67. [DOI] [PubMed] [Google Scholar]
- 46.Upadhyay, S., and B. D. Shaw. 2008. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol. Microbiol. 68690-705. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Z., and P. J. Szaniszlo. 2000. WdCHS3, a gene that encodes a class III chitin synthase in Wangiella (Exophiala) dermatitidis, is expressed differentially under stress conditions. J. Bacteriol. 182874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Z., L. Zheng, H. Liu, Q. Wang, M. Hauser, S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2001. WdChs2p, a class I chitin synthase, together with WdChs3p (class III) contributes to virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 697517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, I., D. Assmann, E. Thines, and G. Steinberg. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18225-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werner, S., J. A. Sugui, G. Steinberg, and H. B. Deising. 2007. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant-Microbe Interact. 201555-1567. [DOI] [PubMed] [Google Scholar]
- 51.Westfall, P. J., and M. Momany. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanai, K., N. Kojima, N. Takaya, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 581828-1835. [DOI] [PubMed] [Google Scholar]
- 53.Yarden, O., and C. Yanofsky. 1991. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 52420-2430. [DOI] [PubMed] [Google Scholar]
- 54.Ziman, M., J. S. Chuang, and R. W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 71909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 91565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]