Abstract
In trypanosomatid parasites, spliced leader (SL) trans splicing is an essential nuclear mRNA maturation step which caps mRNAs posttranscriptionally and, in conjunction with polyadenylation, resolves individual mRNAs from polycistronic precursors. While all trypanosomatid mRNAs are trans spliced, intron removal by cis splicing is extremely rare and predicted to occur in only four pre-mRNAs. trans- and cis-splicing reactions are carried out by the spliceosome, which consists of U-rich small nuclear ribonucleoprotein particles (U snRNPs) and of non-snRNP factors. Mammalian and yeast spliceosome complexes are well characterized and found to be associated with up to 170 proteins. Despite the central importance of trans splicing in trypanosomatid gene expression, only the core RNP proteins and a few snRNP-specific proteins are known. To characterize the trypanosome spliceosomal protein repertoire, we conducted a proteomic analysis by tagging and tandem affinity-purifying the canonical core RNP protein SmD1 in Trypanosoma brucei and by identifying copurified proteins by mass spectrometry. The set of 47 identified proteins harbored nearly all spliceosomal snRNP factors characterized in trypanosomes thus far and 21 proteins lacking a specific annotation. A bioinformatic analysis combined with protein pull-down assays and immunofluorescence microscopy identified 10 divergent orthologues of known splicing factors, including the missing U1-specific protein U1A. In addition, a novel U5-specific, and, as we show, an essential splicing factor was identified that shares a short, highly conserved N-terminal domain with the yeast protein Cwc21p and was thus tentatively named U5-Cwc21. Together, these data strongly indicate that most of the identified proteins are components of the spliceosome.
Trypanosomatid parasites utilize RNA splicing for the maturation of nuclear pre-mRNA in two distinct ways: first, as in other eukaryotes, cis splicing is used for intron removal. While this was unambiguously demonstrated for the Trypanosoma brucei poly(A) polymerase (PAP) mRNA (19), intron removal appears to be a rare event in trypanosomatids because a survey of the Tritryp genomes has identified only three additional putative intron-containing genes (10). Second, trypanosomatids process all of their nuclear pre-mRNAs by spliced leader (SL) trans splicing (reviewed in reference 15). In this splicing reaction, the capped, 39 nucleotide (nt)-long 5′ terminus of the SL RNA, the SL or miniexon, is fused to the 5′ end of each mRNA. This process is therefore a posttranscriptional mRNA capping mechanism and, since trypanosomatids transcribe their protein coding genes polycistronically, it resolves individual mRNAs from polycistronic precursors in conjunction with polyadenylation. SL trans splicing occurs in several organisms, including tunicate chordates, nematodes, and trematodes, but it is not found in insect and mammalian cells; thus, it is specific to the parasites and not present in the hosts of trypanosomatids. Hence, factors with specific functions in the trans-splicing process may be potential chemotherapeutic targets, but such factors have thus far only been found in the nematode Ascaris suum (6).
RNA splicing is carried out by the spliceosome which consists of the five small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4, U5, and U6, as well as of non-snRNP proteins. In the human system, there are ∼45 distinct spliceosomal snRNP proteins, and up to 170 proteins were found to be associated with spliceosomal complexes. The splicing machinery of Saccharomyces cerevisiae is of similar complexity because most of the human factors have orthologues in yeast (reviewed in references 11 and 38). The most prominent splicing factors are seven Sm proteins (SmB, SmD1 to SmD3, SmE, SmF, and SmG), also known as common proteins, which form a ring around a conserved binding site in single-stranded regions of the U1, U2, U4, and U5 snRNAs. The U6 snRNA binds a different protein ring composed of Sm-like proteins termed LSm2 to LSm8. In addition to these core RNP proteins, each U snRNA binds a set of specific RNP proteins. While cis and trans spliceosomes are very similar, data from the Ascaris system indicate that trans splicing does not require the U1 snRNP (8); instead the SL RNA splicing substrate itself binds the Sm proteins and is assembled into a spliceosomal RNP (26).
In trypanosomatids, all five spliceosomal U snRNAs have been identified (see reference 15 and references therein), and there are orthologues of all seven Sm proteins (28) and of LSm2 to LSm8 (36). In regard to Sm proteins, an interesting difference between T. brucei and the yeast and mammalian systems has recently been discovered. While in the latter the U snRNPs bind the same Sm complex, there is Sm variation in trypanosome snRNPs. The trypanosome U2 snRNA binds a specific Sm complex in which the canonical SmB and SmD3 subunits are replaced by the U2-specific paralogues Sm15K (also termed SSm1) and Sm16.5K (SSm2), respectively (37, 39). Similarly, the U4 snRNA binds an Sm complex in which SmD3 is substituted by a second paralogue termed SSm4 (37). Although the functional significance of Sm variation is not yet understood, the finding of a U2-specific Sm complex may explain an early observed difference in core RNP stability between the U2 and the other spliceosomal snRNPs (3).
Trypanosome U snRNPs have been investigated beyond their core structures. Recently, a study reported the detailed characterization of the tandem affinity-purified U1 snRNP (29). The human and S. cerevisiae U1 snRNPs harbor four U1-specific proteins, namely, U1-70K, U1A, U1C, and FBP11 (nomenclature of human proteins). In comparison, the trypanosome U1 snRNP was shown to be associated with divergent orthologues of U1-70K and U1C, as well as a trypanosome-specific protein termed U1-24K, which has no sequence similarity to either U1A or FBP11. The absence of U1A was plausible because trypanosome U1 snRNA is much smaller than its counterparts in other eukaryotes and apparently lacks stem-loop II, the U1A binding site in other systems (29, 34). A similar picture was obtained for the U2 and U5 snRNPs: the snRNAs are smaller than their human and yeast counterparts, they lack stem-loop structures and, despite the fact that both of these U snRNPs usually contain several specific proteins, only the orthologues of the U2-40K (4) and of the U5-specific PRP8 (18) have thus far been characterized in trypanosomes. No trypanosomal U4/U6 snRNP-specific protein has been described yet. Nevertheless, the few studies which have been conducted on trypanosomal U snRNPs thus far have uncovered unique RNP characteristics suggesting that RNA splicing in trypanosomatids deviates in several aspects from that in yeast and mammalian systems.
To comprehensively characterize trypanosome spliceosomal snRNP components, we have used a proteomics approach in T. brucei. We C-terminally fused the composite PTP (protein C epitope/tobacco etch virus protease site/protein A tandem domain) tag to the canonical SmD1 protein, tandem-affinity-purified SmD1-PTP and identified a set of 46 copurified proteins. With the exception of three LSm proteins, the set contained all characterized and putatively annotated T. brucei snRNP proteins, as well as 21 proteins annotated as “hypothetical.” A combination of tagging some of the unknown proteins, pull-down assays, and bioinformatics identified orthologues of the missing U1A protein, U5-40K, the U4/U6-specific PRP4, and a novel and essential U5-specific protein.
MATERIALS AND METHODS
Plasmids.
The plasmids for PTP tagging SmD1, LSm2, U1A, PRP4, U5-40K, and U5-Cwc21 were derived from the tagging vector pC-PTP-NEO (31). The following C-terminal sequences were cloned into these vectors by using the ApaI and NotI restrictions sites: SmD1 (nucleotide positions [relative to the initiation codon] +4 to +315), LSm2 (−61 to +375), U1A (+74 to +489), PRP4 (+1270 to +1827), U5-40K (+453 to +978), and U5-Cwc21 (+148 to +549). For transfection, the corresponding plasmids SmD1-PTP-NEO, LSm2-PTP-NEO, U1A-PTP-NEO, PRP4-PTP-NEO, U5-40K-PTP-NEO, and U5-Cwc21-PTP-NEO were linearized by using the restriction enzymes SnaBI, BsmI, BsmI, BsmI, BsiWI, and PshAI, respectively. For LSm2 and U5-Cwc21 RNA interference (RNAi), stem-loop constructs were generated according to the published cloning strategy (32) harboring, respectively, the regions from positions −61 to 375 and from positions 10 to 512.
Cells.
Procyclic cell culture, DNA transfections through electroporation, and the generation of cell lines by limiting dilution was carried out as described previously (7, 41). The cell line TbD1e was generated by replacing one SmD1 allele with a PCR product of the hygromycin phosphotransferase coding region fused to 120 bp of SmD1 5′ and 3′ gene flanks and by integrating pSmD1-PTP-NEO into the second allele. The selection conditions were 40 μg of G418/ml and 20 μg of hygromycin/ml. In the cell lines expressing PTP-tagged LSm2, U1a, PRP4, U5-40K, or U5-Cwc21, only the PTP construct was inserted, and one wild-type allele was retained. The RNAi cell lines were derived from the 29-13 strain and were selected as published previously (41). Correct integration of all constructs was determined by PCR assays, and the expression of PTP-tagged protein was confirmed by immunoblotting (see below).
For monitoring the cell growth of RNAi cells, double-stranded RNA (dsRNA) synthesis was induced by adding doxycycline to a final concentration of 2 μg/ml. Cells were counted and diluted to 2 × 106 cells/ml daily.
Protein analysis.
The PTP tag consists of the protein C epitope, a tobacco etch virus (TEV) protease cleavage site, and two tandemly repeated protein A domains. PTP-tagged proteins were detected by immunoblotting using the peroxidase-anti-peroxidase soluble complex (PAP reagent; Sigma), which recognizes the protein A domains of the PTP tag or a monoclonal anti-protein C epitope antibody HPC4 (Roche). For tandem affinity purification of SmD1-PTP, a crude extract of cytoplasmic and extracted nuclear components were prepared from 2.5 liters of TbD1e culture as detailed previously (13). The purification was exactly carried out as described in Schimanski et al. (31). The purified proteins of the final eluate were separated on an sodium dodecyl sulfate (SDS)-10 to 20% polyacrylamide gradient gel and stained with Coomassie blue (Gelcode Coomassie stain; Pierce). Alternatively, the final eluate was dialyzed against a low salt buffer (10 mM Tris-HCl [pH 7.7], 1 mM MgCl2, 10 mM KCl, 0.1 mM dithiothreitol, 0.1% Tween 20), concentrated ∼3-fold to 100 μl by using a vacuum concentrator, and directly subjected to trypsin digest and mass spectrometry.
Mass spectrometry.
Peptides derived from trypsin-digested proteins were separated by two-dimensional liquid chromatography, subjected to tandem mass spectrometry (LC-MS/MS), and identified by using the TurboSEQUEST program of the BioworksBrowser 3.1 software package (Thermo Electron) in a multiprocessor cluster platform as detailed elsewhere (30). Peptides were only considered when PeptideProphet probability values were ≥0.9.
RNA analysis.
PTP-tagged proteins were precipitated by 30 μl of settled immunoglobulin G (IgG) beads (GE Healthcare), which was mixed with 100 μl of crude extract (corresponding to an equivalent of ∼109 cells), incubated for 1 h at 4°C on a rotator, and pelleted by centrifugation. After the bead pellet was washed five times with 0.8 ml of PA-150 buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.7], 3 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Tween 20), total RNA was extracted from the beads by the guanidinium thiocyanate method (2) and resuspended in 40 μl of deionized water. Each RNA sample was analyzed by two primer extension reactions. In reaction A, the 5′-32P-end-labeled DNA oligonucleotides SL_PE (5′-CGACCCCACCTTCCAGATTC-3′), U2_PE (5′-ACAGGCAACAGTTTTGATCC-3′), U4_PE (5′-TACCGGATATAGTATTGCAC-3′), and U6_PE (5′-GGGAGAGTGCTAATCTTCTC-3′), which hybridize to the SL, U2, U4, and U6 snRNAs, respectively, were used, whereas reaction B contained the U1-specific oligonucleotide U1_PE (5′-AGCACGGCGCTTTCGTGATG-3′) and the U5-specific oligonucleotide U5_PE (5′-CCGCTCGAGGACACCCCAAAGTTT-3′). Primer extension reactions were carried out with 10 μl of total RNA and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Extension products were separated on 50% urea-8% polyacrylamide gels and visualized by autoradiography. The same RNA preparation and primer extension protocol was used to analyze the U snRNA abundance in doxycycline-induced RNAi cells.
To determine the relative abundance of RNAs in RNAi cells, total RNA preparations from whole-cell lysates were analyzed by semiquantitative reverse transcription-PCR (RT-PCR) assays. For pre-mRNA and 7SL RNA analysis RNA was reverse transcribed with random hexamer oligonucleotides and for mature mRNA analysis with an oligo(dT) primer. For each PCR, the cycle numbers of the linear amplification range was determined. The PAP sequences were amplified with the oligonucleotides Ex1-S (5′-GTGCAGCGGCACTCCCAAAAC-3′) and Ex2-AS (5′-CGTTAAAACAGATGGACAAATC-3′), which target the PAP coding sequences from positions 160 to 180 and from positions 267 to 288, respectively. The cDNAs of α-tubulin RNA were amplified with the antisense oligonucleotide 5′-GAGAGTTGCTCGTGGTAGGC-3′ (coding positions 841 to 860) and with the sense oligonucleotide 5′-GTGCATTGAACGTGGATCTG-3′ (positions 737 to 756) or with the pre-mRNA-specific sense oligonucleotide 5′-GTAAGTGGTGGTGGCGTAAG-3′, which anneals upstream of the SL addition site (positions −190 to −171 relative to the translation initiation codon). The 5′-terminal 123 bp of the 7SL cDNA were amplified with the oligonucleotides 5′-TTGCTCTGTAACCTTC-3′ and 5′-TCTACAGTGGCGACCTCAAC-3′.
Immunolocalization.
Procyclic T. brucei cells were harvested by centrifugation for 10 min at 600 × g and room temperature, washed, and resuspended in ice-cold phosphate-buffered saline (PBS; pH 7.4) (Calbiochem PBS tablet) at a final concentration of 2 × 107 cells/ml. A total of 2 × 105 parasites were allowed to settle on coverslips (13 mm) at room temperature for 15 min, fixed in 4% paraformaldehyde in PBS for 15 min at 4°C, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After four washes with PBS, fixed parasites were treated with 1% fish skin gelatin in PBS-0.1% Tween 20 for 1 h at room temperature to block nonspecific binding and then incubated for 1 h with rabbit anti-protein A serum (Sigma) diluted 1/40,000 in 1% fish skin gelatin solution. The coverslips were washed six times with PBS-0.05% Tween 20 and incubated for 45 min at room temperature with Alexa 594 conjugated to anti-rabbit secondary antibody (Molecular Probes) diluted 1/400 in 1% fish skin gelatin solution and DAPI (4′,6′-diamidino-2-phenylindole) at 1 μg/μl (diluted 1/200 in 1% fish skin gelatin solution). After six washes with PBS-0.05% Tween 20, the coverslips were mounted on glass slides with Vectashield mounting medium (Vector Laboratories). Images with identical exposure settings were taken with an Axiovert 200 microscope and prepared with AxioVision software (Zeiss).
RESULTS
Purification of spliceosomal snRNPs.
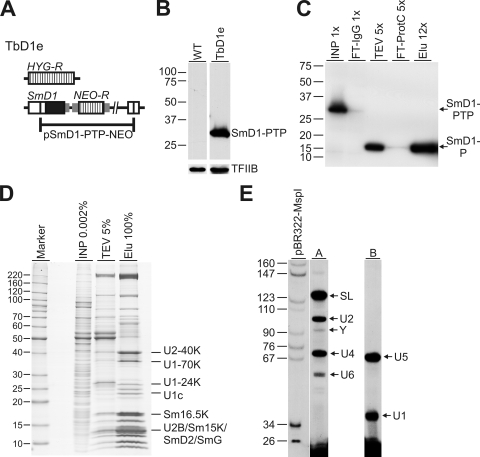
By integrating the plasmid pSmD1-PTP-NEO into one SmD1 allele and by knocking out the second SmD1 allele with a hygromycin phosphotransferase gene, we created the procyclic cell line TbD1e which exclusively expressed SmD1 as a C-terminal PTP fusion (Fig. 1A and B). Since SmD1 is almost certainly an essential protein and TbD1e cells did not exhibit a growth defect (data not shown), we concluded that PTP-tagged SmD1 was functional. Thus, we grew 2.5 liters of TbD1e cell culture, harvested the cells, prepared cell extract, and purified SmD1-PTP by tandem affinity purification. Immunoblot monitoring of the purification demonstrated that both chromatography steps were very efficient and that nearly 25% of the tagged protein in the input material was recovered in the final eluate (Fig. 1C). Separating the final eluate by denaturing polyacrylamide gel electrophoresis and staining the gel with Coomassie blue revealed a multitude of protein bands that were enriched through both purification steps (Fig. 1D). To verify that spliceosomal proteins were purified, we cut out six bands from the gel and identified its contents by trypsin digestion and LC-MS/MS. This approach identified the known snRNP proteins U2-40K, U1-70K, U1-24K, U1c, and Sm16.5K, as well as the four proteins Sm15K, SmD2, SmG, and the U2-specific U2B that comigrated at ∼13 kDa (Fig. 1D). Furthermore, we prepared total RNA from the final eluate and detected the spliceosomal U snRNAs by primer extension assays (Fig. 1E). Both the SL RNA and the five U snRNAs copurified with SmD1-P. In comparison, this primer extension profile closely resembled the profile obtained with total RNA prepared from cell lysates except that the U6 snRNA was underrepresented (data not shown); this finding is most likely due to the fact that U6 snRNA does not bind to SmD1 and was copurified only in the form of the U4/U6 snRNP. In sum, these data strongly indicated that the multitude of proteins detected in the final eluate of the SmD1-PTP purification represent components of the spliceosome.
FIG. 1.
PTP tandem affinity purification of SmD1. (A) Schematic outline (not to scale) of the SmD1 gene locus in the TbD1e cell line. Coding regions of the hygromycin (HYG-R) and neomycin (NEO-R) resistance genes and of SmD1 are represented by striped and open boxes, respectively. Introduced gene flanks providing RNA processing signals are indicated by small gray boxes. (B) Immunoblot of wild-type (WT) and TbD1e whole-cell lysates. SmD1-PTP was detected with a polyclonal anti-protein A antibody and, as a loading control, transcription factor IIB (TFIIB) with a polyclonal immune serum. (C) Immunoblot monitoring of the SmD1-PTP purification. Aliquots of the crude extract (INP), the IgG chromatography flowthrough (FT-IgG), the TEV protease eluate (TEV), the flowthrough of the anti-protein C affinity chromatography (FT-ProtC), and the final eluate (Elu) were separated on an SDS-10% acrylamide gel, blotted, and probed with a monoclonal anti-protein C epitope antibody. Relative amounts of aliquots are specified by x values. It should be noted that the size of the tagged SmD1 protein was reduced by ∼15 kDa due to the TEV protease digest (SmD1-PTP → SmD1-P). (D) The final eluate of a standard SmD1-PTP purification was separated on an SDS-10 to 20% polyacrylamide gel and stained with Coomassie blue. For comparison, small aliquots of the crude extract (INP) and the TEV protease eluate (TEV) were coanalyzed. Marker protein sizes in kilodaltons are specified on the left and proteins mass spectrometrically identified from excised gel bands are indicated on the right. (E) U snRNA profile in the final eluate. Total RNA prepared from the final eluate was subjected to two primer extension reactions. Extension products of the SL RNA (SL), U2, U4, and U6 snRNAs, as well as of the Y-structure intermediate were detected in reaction A and of the U1 and U5 snRNAs in reaction B as indicated on the right of each lane. DNA marker lengths in base pairs are listed on the left.
For the identification of these components we repeated the purification procedure, concentrated and dialyzed the final eluate, and subjected this bulk sample directly to trypsin digest and LC-MS/MS. Overall, this procedure identified 41 proteins by two or more significant peptide matches and six additional proteins by a single significant peptide match (Table 1) . Of note, none of these proteins was detected in previous PTP purifications of nuclear protein complexes such as RNA polymerase I (22, 23), the class I transcription factor A (1), or the general transcription factor TFIIH (14), suggesting that this set of proteins specifically copurified with SmD1. Accordingly, the protein set contained all Sm proteins, including the unique Sm paralogues, three U6-specific LSm proteins, and all of the specific snRNP proteins that had been characterized thus far (the references are listed in Table 1) or were putatively annotated as such in the T. brucei genome database at http://www.genedb.org. On the other hand, the known non-snRNP component of the spliceosome, PRP43 (16), and the U4/U6·U5 tri-snRNP-specific component PRP31 (16), were not detected, suggesting that the stringency of the purification procedure dissolved the spliceosome and that individual snRNPs were purified. This notion was verified by sedimenting an SmD1-P final eluate through a sucrose gradient: while this procedure separated individual snRNPs, cosedimentation of proteins at the bottom of the gradient was not detected (data not shown). Nevertheless, the presence of nearly all known spliceosomal snRNP proteins in the final eluate of the SmD1-PTP purification strongly suggested that the cadre of 21 unknown proteins (annotated as “conserved hypotheticals”) harbored trypanosome factors of RNA splicing. This was confirmed by a bioinformatic analysis which identified several domains of known splicing factors (see Table 1 and see below).
TABLE 1.
Mass spectrometric identification of spliceosomal proteins
| Protein category and accession no.a | Annotationb | Molecular mass (kDa) | Theoretical pI | No. of peptidesc | snRNA(s) | Source, reference(s), or characteristicsd |
|---|---|---|---|---|---|---|
| Characterized proteins | ||||||
| Tb927.2.4540 | SmB* | 12.3 | 9.9 | 8 | U1/U2/U4/U5/SL | 28 |
| Tb927.7.3120 | SmD1* | 11.7 | 7.9 | 7 | U1/U2/U4/U5/SL | 28 |
| Tb927.2.5850 | SmD2* | 12.5 | 10.0 | 8 | U1/U2/U4/U5/SL | 28 |
| Tb927.4.890 | SmD3* | 12.4 | 9.3 | 4 | U1/U2/U4/U5/SL | 28 |
| Tb927.6.2700 | SmE* | 9.6 | 7.9 | 7 | U1/U2/U4/U5/SL | 28 |
| Tb09.211.1695 | SmF* | 8.4 | 5.7 | 4 | U1/U2/U4/U5/SL | 28 |
| Tb11.01.5915 | SmG* | 8.9 | 5.8 | 5 | U1/U2/U4/U5/SL | 28 |
| Tb927.6.4340 | Sm15K/SSm2.1* | 12.8 | 10.3 | 4 | U2 | 37, 39 |
| Tb10.70.2250 | Sm16.5K/SSm2.2 | 14.7 | 9.7 | 2 | U2 | 37, 39 |
| Tb927.7.6380 | SSm4* | 23.2 | 10.2 | 8 | U4 | 37 |
| Tb927.8.5180 | LSm2 | 13.2 | 4.9 | 6 | U6 | 36; this study |
| Tb11.01.5535 | LSm4* | 14.2 | 10.9 | 3 | U6 | 17 |
| Tb927.5.4030 | LSm7* | 10.2 | 6.8 | 3 | U6 | 17 |
| Tb927.3.1780 | LSm8 | 14.0 | 7.8 | 3 | U6 | 17 |
| Tb927.8.4830 | U1-70K* | 31.7 | 10.0 | 5 | U1 | 29 |
| Tb10.6k15.2200/30 | U1A* | 18.0 | 8.5 | 3 | U1 | This study |
| Tb927.3.1090 | U1-24K | 24.2 | 4.9 | 3 | U1 | 29 |
| Tb10.70.5640 | U1C* | 21.7 | 9.4 | 4 | U1 | 29 |
| Tb10.61.2410 | U2-40K* | 36.5 | 6.3 | 16 | U2 | 4 |
| Tb927.3.3480 | U2B* | 13.6 | 10.1 | 4 | n.d.e | GeneDB |
| Tb10.70.7190 | PRP4 | 65.5 | 5.4 | 12 | U4/U6 | This study |
| Tb09.211.2420 | PRP8* | 277.0 | 8.5 | 47 | U5 | 18 |
| Tb11.01.7080 | U5-116K* | 105.5 | 5.0 | 22 | n.d. | GeneDB |
| Tb11.01.2940 | U5-40K* | 35.0 | 6.3 | 6 | U5 | This study |
| Tb927.8.2560 | U5-15K | 17.7 | 6.1 | 2 | n.d. | GeneDB |
| Tb09.211.2150 | PABP2† | 62.1 | 9.3 | 7 | n.d. | GeneDB |
| Tb11.01.3110 | HSP70† | 75.4 | 6.0 | 5 | n.d. | GeneDB |
| Tb09.160.4250/80 | TRYP1† | 22.4 | 6.1 | 2 | n.d. | GeneDB |
| New identifications | ||||||
| Tb927.5.2290 | RNA helicase* | 249.3 | 5.8 | 45 | n.d. | Putative (GeneDB) |
| Tb927.5.1340 | c.h.* | 92.2 | 7.0 | 10 | n.d. | Splicing factor SYF1, E = 6e−23 |
| Tb09.160.2900 | c.h.* | 63.2 | 9.2 | 9 | n.d. | PRP3 domain, E = e−37 |
| Tb10.6k15.0680 | c.h. | 87.7 | 8.7 | 8 | n.d. | Splicing factor CRN, E = 6e−41 |
| Tb11.01.7330 | c.h. | 111.0 | 7.3 | 7 | n.d. | U5-102kD (PRP6), E = e−5 |
| Tb09.160.2110 | U5-Cwc21* | 16.2 | 9.7 | 4 | U5 | Novel; this study |
| Tb10.70.1350 | c.h. | 55.4 | 6.2 | 4 | n.d. | Novel |
| Tb927.2.3400 | c.h. | 42.0 | 8.7 | 4 | n.d. | Novel |
| Tb927.8.1930 | c.h. | 31.7 | 6.7 | 4 | n.d. | Novel |
| Tb11.01.6640 | c.h.* | 17.0 | 4.7 | 3 | n.d. | Novel |
| Tb927.8.6280 | c.h. | 27.1 | 8.4 | 3 | n.d. | Novel |
| Tb11.02.0465 | c.h. | 12.1 | 5.0 | 2 | n.d. | Novel |
| Tb927.7.1890 | c.h. | 31.0 | 4.7 | 2 | n.d. | Novel |
| Single hits | ||||||
| Tb927.5.2910 | c.h. | 20.0 | 9.6 | 1 | n.d. | Novel |
| Tb927.8.4790 | c.h. | 26.0 | 7.0 | 1 | n.d. | Novel |
| Tb927.2.5240 | c.h. | 54.3 | 5.4 | 1 | n.d. | PRP19, E = 3e−42 |
| Tb927.7.6980 | c.h. | 195.0 | 7.1 | 1 | n.d. | Splicing Factor SF3b130, E = e−13 |
| Tb927.5.2140 | NORF1† | 93.3 | 7.8 | 1 | n.d. | Putative (GeneDB) |
| Tb10.70.5360 | La protein | 37.7 | 9.0 | 1 | n.d. | 21, 40 |
Proteins investigated in the present study are indicated in boldface. Accession numbers of the GeneDB database (http://www.genedb.org/) are given.
c.h., conserved hypothetical, i.e., a hypothetical protein that is conserved among trypanosomatids. *, Detected in two independent SmD1-PTP purifications. †, PABP2, poly(A) binding protein; HSP70, heat shock protein 70; TRYP1, tryparedoxin peroxidase; NORF1, nonsense mRNA reducing factor 1.
That is, the number of different trypsin-derived peptides identified by mass spectrometry.
GeneDB, the gene was putatively annotated in the genome database by sequence homology. E values were derived from standard BLASTP searches of the human genome database; only hits with E ≤ e−5 were recognized; nomenclature is given according to reference 11.
n.d., not determined.
Identification of LSm2 and U1A.
Among others, this analysis revealed an Sm-like protein (accession no. Tb927.8.5180) and a putative RNA-binding protein harboring an RNA recognition motif (RRM) that is encoded by two genes annotated as RPB14A (RNA-binding protein 14A; accession no. Tb10.6k15.2200) and RPB14B (Tb10.6k15.2230) (5). Both genes encode identical amino acid sequences, but RPB14A encodes 17 additional C-terminal residues. To assess the U snRNA specificity of these putative snRNP proteins, we generated cell lines in which the Sm-like protein or RBP14A were C-terminally PTP tagged. Both proteins were precipitated with IgG beads, which interact with the protein A domains of the PTP tag, and RNA preparations of the precipitates were analyzed by primer extension assays. Two reactions were carried out: reaction A harbored four primers which specifically hybridize to SL RNA and U2, U4, and U6 snRNAs, whereas in reaction B U1 and U5 snRNAs were detected (Fig. 2). In a positive control, SmD1-PTP was precipitated and, as expected, all six spliceosomal U snRNAs, as well as the SL intron, presumably in the form of the Y structure trans-splicing intermediate, coprecipitated with SmD1. All signals were specific because when the PTP-tagged version of the class I transcription factor A subunit 2 (1) was precipitated, none of the U snRNA-specific signals were detectable (lanes 3 and 4). This assay then clearly showed that the Sm-like protein preferentially brought down the U6 snRNA with some U4 snRNA, which most likely coprecipitated in the form of the U4/U6 snRNP (lanes 5 and 6). The presence of the Sm domain and the U6 interaction indicated that the protein was a U6-specific LSm protein. Although the original characterization of the trypanosome LSm2-8 proteins did not include the Sm-like protein (17), a recent revision of this work independently identified the Sm-like protein as the orthologue of LSm2 (36). Our results from a pulldown assay showed that, besides U6 and U4, a minor but specific U2 signal was detected (Fig. 2, compare lane 5 to lanes 1 and 3), suggesting that in trypanosomes, the U2-U6 interaction, which is characteristic of the catalytically active spliceosome (reviewed in reference 33), is conserved.
FIG. 2.
Protein pulldown assays. Extracts were prepared from cells expressing the spliceosomal proteins SmD1, LSm2, U1A, PRP4, U5-40K, and U5-Cwc21 as C-terminals PTP fusions. The proteins were pulled down with IgG beads which bind to the protein A domains of the PTP tag. Coprecipitated U snRNAs were detected by two primer extension reactions as specified in the legend of Fig. 1E. As a negative control, the pulldown of the PTP-tagged class I transcription factor A subunit 2 (CITFA-2) was coanalyzed. On the left, pBR322-MspI marker sizes are indicated as well as the positions of the U snRNA-specific extension products.
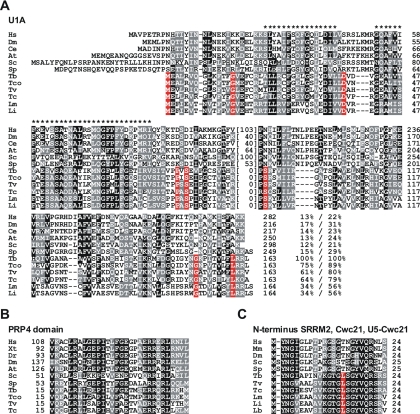
RPB14 specifically coprecipitated the U1 snRNA, which identified it as a U1 snRNP-specific protein and raised the possibility that it is the orthologue of the missing U1A protein. This was indeed confirmed by an alignment of U1A sequences from six model organisms and six trypanosomatids: sequence similarity was not restricted to the RRM domain but was observed throughout the alignment (Fig. 3A). Hence, we concluded that U1A is a component of the trypanosome U1 snRNP despite the fact that the U1 snRNA seems not to have a stem-loop II structure, the binding site of U1A in the human system.
FIG. 3.
Splicing factor sequence alignments. Sequences were aligned by using the CLUSTAL W program (http://www.ebi.ac.uk/Tools/clustalw2/) at default parameters. Black and gray-shaded letters identify identical or similar amino acid residues at positions with more than 50% sequence conservation. Positions with complete identity in trypanosomatid sequences and no similarity in the sequences of the other organisms are shaded in red. (A) Alignment of U1A sequences from Homo sapiens (Hs, accession number P09012), Drosophila melanogaster (Dm, P43332), Caenorhabditis elegans (Ce, NP_500505), Arabidopsis thaliana (At, NP_182280), S. cerevisiae (Sc, YBR119W), S. pombe (Sp, SPBC4B4.07c) and from the trypanosomatids T. brucei (Tb, Tb10.6k15.2200), Trypanosoma congolense (Tco, congo174c04.p1k_4), Trypanosoma vivax (Tv, tviv228a11.p1k_4), Trypanosoma cruzi (Tc, Tc00.1047053507723.120), Leishmania major (Lm, LmjF36.6985), and Leishmania infantum (Li, LinJ36.0680). Asterisks mark the RRM domain and numbers of amino acids in stretches of limited or no sequence conservation are provided in parentheses. The identity/similarity values specified at the end of each sequence were derived from pairwise comparisons with the T. brucei sequence using the EMBOSS program (http://www.ebi.ac.uk/Tools/emboss/align/) at default parameters. (B) Alignment of the PRP4 domain. Accession numbers: Hs (NP_004688), Xenopus tropicalis (Xt, AAH61324), Danio rerio (Dr, NP_956049), Dm (NP_648990), At (AAM14969), Sc (YPR178W), Sp (SPAC227.12), Tb (Tb10.70.7190), Tco (congo928f05.p1k_5), Tv (Tviv89a04.p1k_1), Tc (Tc00.1047053506855.340). (C) Alignment of the N termini of SRRM2/Cwc21 proteins. Accession numbers: Hs (AAH46104), Mus musculus (Mm, AAH19803), Dm (NP_647642), Sc (YDR482C), Sp (SPAC4A8.09c), Tb (Tb09.160.2110), Tv (Tviv1778d04.q1k_1), Tc (Tc00.1047053504797.130), Lm (LmjF01.0190), Li (LinJ01_V3.0190), Leishmania brasiliensis (Lb, LbrM01_V2.0220).
Identification of PRP4.
Our bioinformatic analysis suggested that the Tb10.70.7190 gene encodes the trypanosome orthologue of the U4/U6-specific protein PRP4 because of the presence of the conserved PRP4 domain (Fig. 3B; E = e−08) and seven C-terminal WD40 repeats (see Fig. S1 in the supplemental material; E = 6e−26). PRP4 is required for the first step in splicing, binds to the U4 snRNA and forms a complex with two other proteins, namely, PRP3 and a cyclophilin (9). To confirm that we had identified trypanosome PRP4 and the first trypanosomatid U4/U6 snRNP protein, we applied PTP-tagging and a IgG bead-mediated pulldown assay. As expected, both U4 and U6 snRNAs efficiently coprecipitated with the tagged protein (Fig. 2, lanes 9 and 10). Furthermore, it appears that the PRP4 complex is conserved in trypanosomes because another SmD1-copurified protein, encoded by the Tb09.160.2900 gene, was found to harbor a PRP3 domain (Table 1; E = e−37). While human PRP4 is also associated with the U4/U6·U5 tri-snRNP complex, we were unable to detect U5 snRNA in the T. brucei PRP4 pulldown assay. Possibly, the trypanosome U4/U6·U5 complex (42) is not as stable as its human counterpart and dissociated during precipitation. Alternative possibilities include a modified function of trypanosome PRP4 which does not include the tri-snRNP or a short, transient formation of the tri-snRNP which was not detected in the assay.
Identification of U5-specific snRNP components.
The human U5 snRNP occurs predominantly as a 20S particle with seven specific proteins ranging in size from 15 to 220 kDa. In trypanosomes, the largest protein, PRP8, was experimentally characterized (18), and genome annotation revealed putative orthologues of the U5-116K and U5-15K subunits. Our own analysis of the purified spliceosomal proteins revealed two further putative subunit orthologues of the U5 snRNP: the protein encoded by the Tb11.01.7330 gene is similar to the U5-102K subunit (Table 1; E = e−5), and the protein encoded in the Tb11.01.2940 gene has, as PRP4, seven WD repeats (data not shown) and, compared to the human genome, the highest similarity to the U5-40K subunit (E = 7e−14). Since several spliceosomal proteins possess WD domain repeats and since the latter protein is not conserved in S. cerevisiae, we analyzed whether this protein interacts with the U5 snRNA. A PTP tag-mediated pulldown of the U5-40K orthologue precipitated exclusively the U5 snRNA, verifying our bioinformatic approach (Fig. 2, lanes 11 and 12). Hence, it appears that the comparatively small U5 snRNA of trypanosomes is assembled in a U5 snRNP with a complexity similar to that of its human counterpart.
Finally, we decided to investigate the SmD1-copurified protein of the Tb09.160.2110 gene, which had no obvious sequence homology to a known splicing factor, was annotated as a “hypothetical” and was found to be well conserved among trypanosomatid species (see Fig. S2 in the supplemental material). The pulldown of the PTP-tagged version of this protein coprecipitated efficiently and predominantly the U5 snRNA, identifying it as another U5 snRNP-specific protein (Fig. 2, lanes 13 and 14). Interestingly, the amino acid sequence of Tb09.160.2110 had no resemblance to the sequences of the missing 20S U5 subunits U5-200K and U5-100K (data not shown), suggesting it to be a novel U5 protein. However, a fraction of the human U5 snRNA is assembled into a larger complex called the 35S U5 snRNP (20). A hallmark of this large snRNP is the presence of the heteromeric PRP19 complex several subunits of which were shown to be essential splicing factors (see references 11 and 38 and references therein). Trypanosomes appear to possess the PRP19 complex because our bioinformatic analysis identified the SmD1-copurified protein Tb927.2.5240 as the trypanosome orthologue of PRP19 (Table 1, E = 3e−42). Direct sequence comparisons between trypanosomatid Tb09.160.2110 orthologues and human PRP19 components did not reveal convincing similarities (data not shown). However, in S. cerevisiae and Schizosaccharomyces pombe, the PRP19 complex was purified by tagging the subunit known as Cef1p in S. cerevisiae and Cdc5p in S. pombe (24). This revealed several new proteins and thus potentially new splicing factors termed Cwc/Cwf (for “complexed with Cef1p/Cdc5p”). Interestingly, the new U5-specific protein of T. brucei is strikingly similar in its 24 N-terminal amino acids to the yeast factor Cwc21/Cwf21 (Fig. 3C). Moreover, in the human system, this N-terminal domain is present in the serine/arginine repetitive matrix protein 2 (SRRM2), which may not only be expressed as the 300-kDa protein known as the splicing coactivator SRm300 but also as a small protein comprising only 194 amino acids (35). It is therefore possible that yeast Cwc21, as a component of the PRP19 complex, is an ancient splicing factor conserved in trypanosomatids and yeasts and that the small SRRM2 protein in humans is the orthologue of Cwc21 and an unrecognized component of the PRP19 complex and of the 35S U5 snRNP. We therefore and tentatively name the trypanosome protein U5-Cwc21.
Interestingly, the pulldown of U5-Cwc21 but not that of the U5-40K orthologue coprecipitated a minor but specific amount of U2 snRNA (Fig. 2, compare lanes 11 and 13). A possible explanation for this finding is that U5-Cwc21, as part of a putatively larger U5 snRNP, is more tightly associated with U2 snRNA in the U5·U2/U6 tri-snRNP complex of the activated spliceosome than the U5-40K orthologue. The puzzling observation is that the U6 snRNA did not coprecipitate because to our knowledge a U2/U5-specific interaction has not been reported. Although we cannot rule out that this result was obtained by a disruption of the U2/U6 interaction due to the ionic strength in the precipitation procedure, it is also possible that a unique interaction between the U2 and U5 snRNPs is important for RNA splicing in trypanosomes.
Immunolocalization of the new snRNP proteins.
Trypanosome snRNP proteins are almost exclusively found in the nucleus (25). To verify that the newly characterized snRNP proteins exhibit the same cellular distribution, we detected the PTP-tagged proteins in procyclic trypanosomes by a polyclonal antibody which recognizes the protein A domains of the PTP tag. As shown in Fig. 4, LSm2, U1A, PRP4, and U5-40K exhibit this expected localization pattern and are detected exclusively in the nucleus. U5-Cwc21 is also predominantly localized in the nucleus; however, we consistently found some of the protein in the cytoplasm, indicating that this protein may have functions in addition to its role as a component of the U5 snRNP. The nuclear staining pattern was typically not uniform but, as best seen with U1A, of particulate appearance, indicating that trypanosome splicing factors are concentrated in nuclear speckles as described in other systems (reviewed in reference 12).
FIG. 4.
Localization of splicing factors by immunofluorescence microscopy. Cells were fixed and stained with DAPI. PTP-tagged splicing factors were detected by using a rabbit polyclonal anti-protein A antibody (α-ProtA) and Alexa 594-conjugated anti-rabbit IgG secondary antibody. Bar, 10 μm. As a control, wild-type cells (WT) were analyzed that do not express the PTP tag.
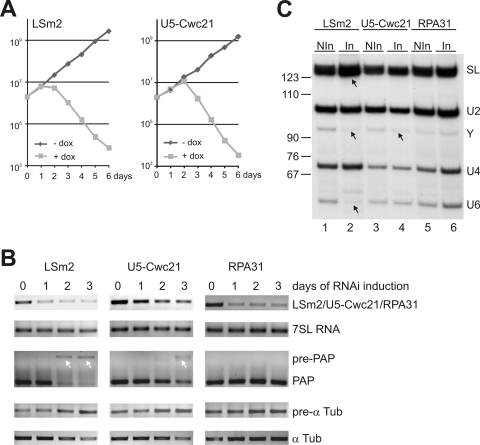
Functional analysis of LSm2 and U5-Cwc21.
In a final step, we verified the discovery of new trypanosomal RNA splicing factors by a functional analysis of LSm2 and U5-Cwc21 in vivo. For each gene, we created three clonal procyclic cell lines for inducible expression silencing by RNAi. Consistently, we found that knocking down LSm2 expression was lethal and effectively halted cell growth on the second day (Fig. 5A). A similar phenotype was observed with the U5-Cwc21 knockdown, although in this case the cells grew normally until day 2 (Fig. 5A). The finding that U5-Cwc21 was an essential gene was surprising because a knockout of Cwc21 in S. cerevisiae was not lethal (24). Next, a semiquantitative RT-PCR analysis showed that silencing of both genes resulted in RNA splicing defects (Fig. 5B). To exclude the possibility that dying trypanosomes nonspecifically exhibit such defects, we coinvestigated a knockdown of the essential RNA polymerase I subunit RPA31 which affected trypanosome growth even faster than silencing of LSm2 (23). The induction of dsRNA synthesis specifically reduced the target mRNAs, whereas the level of 7SL RNA remained constant during the course of the experiment (Fig. 5B, compare top two panel rows). We then designed a competitive RT-PCR assay for the intron-containing PAP pre-mRNA and its mature product. In uninduced cells, this assay did not detect PAP pre-mRNA, indicating that intron removal in this pre-mRNA occurs very rapidly. However, in cells in which LSm2 was silenced, PAP pre-mRNA was clearly detected from day 2 concomitant with a signal reduction of mature PAP mRNA (Fig. 5B). The same cis-splicing defect, albeit less pronounced, was observed when U5-Cwc21 was silenced, whereas it was not detected in the control knockdown of RPA31 (third panel row). LSm2 and U5-Cwc21 silencing also affected trans splicing because it reproducibly caused an increase of α tubulin pre-mRNA (fourth panel row) and a decrease of the corresponding mature mRNA (bottom panel row) as detected in noncompetitive RT-PCR assays. When we analyzed RNA of the single copy RBP6z gene, which encodes an RNA polymerase I subunit (22), the same results were obtained, suggesting that the trans-splicing defect was global (data not shown).
FIG. 5.
Silencing of LSm2 and U5-Cwc21 expression. (A) Growth of representative cell lines for LSm2 and U5-Cwc21 knockdowns in the presence (+dox) and absence (−dox) of dsRNA synthesis-inducing doxycycline. (B) Semiquantitative RT-PCR analysis of RNA abundance in total RNA preparations from cells in which dsRNA synthesis was induced for 0, 1, 2, or 3 days. As a control, the knockdown of the RNA polymerase I subunit RPA31 was coanalyzed. The first panels show the knockdown of the respective mRNAs and the second panels the analysis of 7SL RNA which is not sensitive to splicing defects. The third panels show a competitive PCR of the PAP pre-mRNA/mRNA. The white arrows point to the pre-mRNA amplification products. Pre-mRNA (pre-α Tub) and mature mRNA of α tubulin (α Tub) was amplified in the fourth and fifth panels, respectively. (C) Total RNA preparations of noninduced LSm2, U5-Cwc21, and RPA31 RNAi cells (NIn) and cells which were doxycycline induced for 3 days (In) were analyzed by primer extension assays to detect the SL RNA (SL) and its Y intermediate as well as the U2, U4, and U6 snRNAs. Arrows in lanes 2 and 4 point to the increased SL RNA signal, the decreased Y signal and the lost U6 signal.
Finally, we analyzed the RNAi effect on the abundance of U snRNAs by primer extension assays (Fig. 5C). As was demonstrated before (36), LSm2 silencing caused a loss of U6 snRNA (compare lanes 1 and 2). Conversely, the knockdown of U5-Cwc21 did not affect the abundance of U5 snRNA (data not shown). This was not unexpected because the formation of stable core RNPs requires the binding of the Sm/LSm complexes but not of snRNP-specific proteins. Moreover, LSm2 silencing resulted in a decrease of the Y-structure intermediate and an increase of SL RNA which strongly indicates that the LSm complex is essential prior to the first step of splicing (compare lanes 1 and 2). This defect, but again less pronounced, was also observed when U5-Cwc21 was silenced (compare lanes 3 and 4) but not detectable when RPA31 was silenced (compare lanes 5 and 6).
Together, these results show that LSm2 and U5-Cwc21 are essential cis- and trans-splicing factors required for parasite growth. The defects seen in both knockdowns strongly indicate that both factors are required before or at the first splicing step. As the RT-PCR analysis (Fig. 5B) suggests, the less-severe splicing defects observed with the U5-Cwc21-silencing compared to Lsm2 silencing may be due to a lower knockdown efficiency of U5-Cwc21. On the other hand, the coanalysis of RPA31-silenced cells clearly showed that the observed defects were specific to the knockdowns of the splicing factors.
DISCUSSION
Tandem affinity purification of SmD1 using the PTP system revealed a set of 47 proteins which contained all known snRNP proteins except three LSm proteins, as well as 21 proteins whose function was unknown. By a bioinformatic approach, seven of the latter proteins, including PRP4 and U5-40K, were annotated as orthologues of known snRNP proteins. Of those which could not be annotated in this way, two proteins exhibited domains reminiscent of snRNP proteins. One protein contained an Sm-like domain identifying it as a potential core snRNP protein, and the other one had an RRM motif. Generating cell lines which expressed a PTP-tagged version of these proteins enabled specific pulldown assays which revealed that the Sm-like and RRM-containing proteins were specifically bound to U6 and U1 snRNA, respectively. Although the result of the Sm-like protein was in accordance with a recent study that identified this protein independently as the U6 core protein LSm2 (36), sequence alignments confirmed that the RRM-containing protein was the missing U1 snRNP component U1A. One question emanating from this latter finding is how does the trypanosome U1A interact with the U1 snRNA in the absence of stem-loop II (27)? Due to the presence of the RRM domain it can be anticipated that trypanosome U1A will directly interact with its cognate snRNA. Thus, the determination of the exact binding site may reveal whether assembly and RNP structure of the trypanosome U1 snRNP and of its human counterpart are different.
Another interesting aspect of this work in relation to the U1 snRNP is the observed expression level of the U1 snRNP components. The bands of U1-specific proteins are among the strongest in the SmD1 purification (see Fig. 1D). As we know from the nematode system, U1 is important for cis splicing but not for SL trans splicing (8). In comparison to SL trans splicing, which is required for the maturation of every mRNA, cis splicing is a rare event in T. brucei; there is experimental evidence for only a single intron in the trypanosome genome disrupting the PAP gene (19), and annotation of the completed genome predicted the presence of only up to three additional introns (10). Hence, it is unclear why trypanosomes strongly express four U1-specific proteins (U1A from two different genes) only to accommodate the removal of so few introns. Possibly, in trypanosomes the U1 snRNP is important for the integrity of the spliceosome in general, or it has a yet-unrecognized, trypanosome-specific role in SL trans splicing.
The last protein we investigated did not exhibit a clear sequence homology to known splicing factors but shared a short, highly conserved domain with the putative yeast splicing factor Cwc21p and the human splicing coactivator SRm300/SRRM2. Yeast Cwc21p is a dispensable protein and to our knowledge SRRM2 has not been shown to be a snRNP protein. In contrast, trypanosome U5-Cwc21 is an essential protein and appears to be required for the first splicing step. Hence, at the current standing, U5-Cwc21 is a novel splicing factor which has either a trypanosome-specific function or a function which has not yet been characterized for its putative orthologues in other systems. Purification of U5-Cwc21 will show whether it is part of a trypanosome PRP19 complex.
Ten polypeptides of our SmD1 copurified protein set remain at the status “conserved hypothetical.” The fact that this set contained nearly all known trypanosome snRNP proteins and that the experimental analysis of five such proteins revealed new spliceosomal snRNP components strongly indicates that these uncharacterized proteins represent trypanosome factors of RNA splicing; they may be extremely divergent orthologues of known splicing factors or are candidates for trypanosome-specific RNA splicing factors. The latter may be specifically required for the SL trans-splicing process which does not occur in the insect and mammalian hosts of trypanosomatid parasites. While we know that SL trans-splicing is carried out by the same spliceosomal U snRNPs which remove introns, two SL RNA-associated proteins, which are specifically required for the trans-splicing process, have been characterized in the nematode Ascaris suum (6). The trypanosome set of spliceosomal proteins may harbor similar factors, and therefore this study sets the stage for a thorough analysis of the unusual way trypanosomes have adapted the RNA splicing process.
Supplementary Material
Acknowledgments
Mass spectrometric analyses were carried out at the Seattle Biomedical Research Institute Proteomics Core.
This study was supported by a grant from the National Institutes of Health to A.G. (AI059377) and by an FAPESP grant to R.M.B.C. (2007/07476-0). D.L.A. was supported by a CAPES/PDEE grant (BEX 1338/06-4), and J.H.L. was supported by a grant of the Korea Science and Engineering Foundation (C00093).
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Brandenburg, J., B. Schimanski, E. Nogoceke, T. N. Nguyen, J. C. Padovan, B. T. Chait, G. A. Cross, and A. Günzl. 2007. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 264856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 3.Cross, M., A. Günzl, Z. Palfi, and A. Bindereif. 1991. Analysis of small nuclear ribonucleoproteins (RNPs) in Trypanosoma brucei: structural organization and protein components of the spliced leader RNP. Mol. Cell. Biol. 115516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross, M., B. Wieland, Z. Palfi, A. Günzl, U. Röthlisberger, H. W. Lahm, and A. Bindereif. 1993. The trans-spliceosomal U2 snRNP protein 40K of Trypanosoma brucei: cloning and analysis of functional domains reveals homology to a mammalian snRNP protein. EMBO J. 121239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Gaudenzi, J., A. C. Frasch, and C. Clayton. 2005. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot. Cell 42106-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denker, J. A., D. M. Zuckerman, P. A. Maroney, and T. W. Nilsen. 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417667-670. [DOI] [PubMed] [Google Scholar]
- 7.Günzl, A., A. Bindereif, E. Ullu, and C. Tschudi. 2000. Determinants for cap trimethylation of the U2 small nuclear RNA are not conserved between Trypanosoma brucei and higher eukaryotic organisms. Nucleic Acids Res. 283702-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannon, G. J., P. A. Maroney, and T. W. Nilsen. 1991. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J. Biol. Chem. 26622792-22795. [PubMed] [Google Scholar]
- 9.Horowitz, D. S., R. Kobayashi, and A. R. Krainer. 1997. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA 31374-1387. [PMC free article] [PubMed] [Google Scholar]
- 10.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, M. Berriman, E. Sisk, M. A. Rajandream, E. Adlem, R. Aert, A. Anupama, Z. Apostolou, P. Attipoe, N. Bason, C. Bauser, A. Beck, S. M. Beverley, G. Bianchettin, K. Borzym, G. Bothe, C. V. Bruschi, M. Collins, E. Cadag, L. Ciarloni, C. Clayton, R. M. Coulson, A. Cronin, A. K. Cruz, R. M. Davies, J. De Gaudenzi, D. E. Dobson, A. Duesterhoeft, G. Fazelina, N. Fosker, A. C. Frasch, A. Fraser, M. Fuchs, C. Gabel, A. Goble, A. Goffeau, D. Harris, C. Hertz-Fowler, H. Hilbert, D. Horn, Y. Huang, S. Klages, A. Knights, M. Kube, N. Larke, L. Litvin, A. Lord, T. Louie, M. Marra, D. Masuy, K. Matthews, S. Michaeli, J. C. Mottram, S. Muller-Auer, H. Munden, S. Nelson, H. Norbertczak, K. Oliver, S. O'neil, M. Pentony, T. M. Pohl, C. Price, B. Purnelle, M. A. Quail, E. Rabbinowitsch, R. Reinhardt, M. Rieger, J. Rinta, J. Robben, L. Robertson, J. C. Ruiz, S. Rutter, D. Saunders, M. Schafer, J. Schein, D. C. Schwartz, K. Seeger, A. Seyler, S. Sharp, H. Shin, D. Sivam, R. Squares, S. Squares, V. Tosato, C. Vogt, G. Volckaert, R. Wambutt, T. Warren, H. Wedler, J. Woodward, S. Zhou, W. Zimmermann, D. F. Smith, J. M. Blackwell, K. D. Stuart, B. Barrell, and P. J. Myler. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 12.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4605-612. [DOI] [PubMed] [Google Scholar]
- 13.Laufer, G., G. Schaaf, S. Bollgönn, and A. Günzl. 1999. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 195466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J. H., H. S. Jung, and A. Günzl. 2009. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. doi: 10.1093/nar/gkp236. [DOI] [PMC free article] [PubMed]
- 15.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, X. H., Q. Liu, L. Liu, C. Tschudi, and S. Michaeli. 2006. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 14529-39. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Q., X. H. Liang, S. Uliel, M. Belahcen, R. Unger, and S. Michaeli. 2004. Identification and functional characterization of Lsm proteins in Trypanosoma brucei. J. Biol. Chem. 27918210-18219. [DOI] [PubMed] [Google Scholar]
- 18.Lücke, S., T. Klockner, Z. Palfi, M. Boshart, and A. Bindereif. 1997. Trans mRNA splicing in trypanosomes: cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 164433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mair, G., H. Shi, H. Li, A. Djikeng, H. O. Aviles, J. R. Bishop, F. H. Falcone, C. Gavrilescu, J. L. Montgomery, M. I. Santori, L. S. Stern, Z. Wang, E. Ullu, and C. Tschudi. 2000. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 6163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 2982205-2208. [DOI] [PubMed] [Google Scholar]
- 21.Marchetti, M. A., C. Tschudi, H. Kwon, S. L. Wolin, and E. Ullu. 2000., Import of proteins into the trypanosome nucleus and their distribution at karyokinesis, J. Cell Sci. 113899-906. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, T. N., B. Schimanski, A. Zahn, B. Klumpp, and A. Günzl. 2006. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 14927-37. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, T. N., B. Schimanski, and A. Günzl. 2007. Active RNA polymerase I of Trypanosoma brucei harbors a novel subunit essential for transcription. Mol. Cell. Biol. 276254-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 222011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palfi, Z., and A. Bindereif. 1992. Immunological characterization and intracellular localization of trans-spliceosomal small nuclear ribonucleoproteins in Trypanosoma brucei. J. Biol. Chem. 26720159-20163. [PubMed] [Google Scholar]
- 26.Palfi, Z., A. Günzl, M. Cross, and A. Bindereif. 1991. Affinity purification of Trypanosoma brucei small nuclear ribonucleoproteins reveals common and specific protein components. Proc. Natl. Acad. Sci. USA 889097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palfi, Z., W. S. Lane, and A. Bindereif. 2002. Biochemical and functional characterization of the cis-spliceosomal U1 small nuclear RNP from Trypanosoma brucei. Mol. Biochem. Parasitol. 121233-243. [DOI] [PubMed] [Google Scholar]
- 28.Palfi, Z., S. Lücke, H. W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Seraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 978967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palfi, Z., B. Schimanski, A. Günzl, S. Lücke, and A. Bindereif. 2005. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 332493-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panigrahi, A. K., Y. Ogata, A. Zikova, A. Anupama, R. A. Dalley, N. Acestor, P. J. Myler, and K. D. Stuart. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 41942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 61069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92315-326. [DOI] [PubMed] [Google Scholar]
- 34.Stark, H., P. Dube, R. Lührmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409539-542. [DOI] [PubMed] [Google Scholar]
- 35.Strausberg, R. L., E. A. Feingold, L. H. Grouse, J. G. Derge, R. D. Klausner, F. S. Collins, L. Wagner, C. M. Shenmen, G. D. Schuler, S. F. Altschul, B. Zeeberg, K. H. Buetow, C. F. Schaefer, N. K. Bhat, R. F. Hopkins, H. Jordan, T. Moore, S. I. Max, J. Wang, F. Hsieh, L. Diatchenko, K. Marusina, A. A. Farmer, G. M. Rubin, L. Hong, M. Stapleton, M. B. Soares, M. F. Bonaldo, T. L. Casavant, T. E. Scheetz, M. J. Brownstein, T. B. Usdin, S. Toshiyuki, P. Carninci, C. Prange, S. S. Raha, N. A. Loquellano, G. J. Peters, R. D. Abramson, S. J. Mullahy, S. A. Bosak, P. J. McEwan, K. J. McKernan, J. A. Malek, P. H. Gunaratne, S. Richards, K. C. Worley, S. Hale, A. M. Garcia, L. J. Gay, S. W. Hulyk, D. K. Villalon, D. M. Muzny, E. J. Sodergren, X. Lu, R. A. Gibbs, J. Fahey, E. Helton, M. Ketteman, A. Madan, S. Rodrigues, A. Sanchez, M. Whiting, A. Madan, A. C. Young, Y. Shevchenko, G. G. Bouffard, R. W. Blakesley, J. W. Touchman, E. D. Green, M. C. Dickson, A. C. Rodriguez, J. Grimwood, J. Schmutz, R. M. Myers, Y. S. Butterfield, M. I. Krzywinski, U. Skalska, D. E. Smailus, A. Schnerch, J. E. Schein, S. J. Jones, and M. A. Marra. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 9916899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tkacz, I. D., S. Cohen, M. Salmon-Divon, and S. Michaeli. 2008. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 16022-31. [DOI] [PubMed] [Google Scholar]
- 37.Tkacz, I. D., Y. Lustig, M. Z. Stern, M. Biton, M. Salmon-Divon, A. Das, V. Bellofatto, and S. Michaeli. 2007. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA 1330-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahl, M. C., C. L. Will, and R. Lührmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136701-718. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P., Z. Palfi, C. Preusser, S. Lücke, W. S. Lane, C. Kambach, and A. Bindereif. 2006. Sm core variation in spliceosomal small nuclear ribonucleoproteins from Trypanosoma brucei. EMBO J. 254513-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westermann, S., and K. Weber. 2000. Cloning and recombinant expression of the La RNA-binding protein from Trypanosoma brucei. Biochim. Biophys. Acta 1492483-487. [DOI] [PubMed] [Google Scholar]
- 41.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for conditional gene knockouts and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 9989-101. [DOI] [PubMed] [Google Scholar]
- 42.Xu, Y., H. Ben Shlomo, and S. Michaeli. 1997. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: the Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl. Acad. Sci. USA 948473-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.