Abstract
An epidemic of infections after video-assisted surgery (1,051 possible cases) caused by rapidly growing mycobacteria (RGM) and involving 63 hospitals in the state of Rio de Janeiro, Brazil, occurred between August 2006 and July 2007. One hundred ninety-seven cases were confirmed by positive acid-fast staining and/or culture techniques. Thirty-eight hospitals had cases confirmed by mycobacterial culture, with a total of 148 available isolates recovered from 146 patients. Most (n = 144; 97.2%) isolates presented a PRA-hsp65 restriction pattern suggestive of Mycobacterium bolletii or Mycobacterium massiliense. Seventy-four of these isolates were further identified by hsp65 or rpoB partial sequencing, confirming the species identification as M. massiliense. Epidemic isolates showed susceptibility to amikacin (MIC at which 90% of the tested isolates are inhibited [MIC90], 8 μg/ml) and clarithromycin (MIC90, 0.25 μg/ml) but resistance to ciprofloxacin (MIC90, ≥32 μg/ml), cefoxitin (MIC90, 128 μg/ml), and doxycycline (MIC90, ≥64 μg/ml). Representative epidemic M. massiliense isolates that were randomly selected, including at least one isolate from each hospital where confirmed cases were detected, belonged to a single clone, as indicated by the analysis of pulsed-field gel electrophoresis (PFGE) patterns. They also had the same PFGE pattern as that previously observed in two outbreaks that occurred in other Brazilian cities; we designated this clone BRA100. All five BRA100 M. massiliense isolates tested presented consistent tolerance to 2% glutaraldehyde. This is the largest epidemic of postsurgical infections caused by RGM reported in the literature to date in Brazil.
Outbreaks, pseudooutbreaks, and cases of health-care-associated infections caused by rapidly growing mycobacteria (RGM) have been reported since the first case was described in 1938 (13). In virtually all nosocomial infections caused by this group of microorganisms, there were failings in the sterilization processes of solutions, surgical instruments, or medical devices (13, 14, 45). Recent publications indicate an increasing number of infections secondary to breast augmentation and video-assisted surgeries (7, 9, 19, 23, 25, 40-43). The growing number of cases and reports may be due, at least in part, to the well-known tolerance to alkaline glutaraldehyde among Mycobacterium chelonae-Mycobacterium abscessus group isolates and to the low susceptibility to high-level disinfectants (20, 22, 39).
Outbreaks of RGM infections unrelated to medical procedures also can occur and usually are associated with exposure to recreational water containing a large number of bacteria and inadequate chlorination (15, 44), highlighting the ubiquity of these organisms in the environment. In fact, RGM have been recovered from many different environmental sources, including soil and water distribution systems (8, 45). RGM are considered opportunistic pathogens and can cause chronic lung disease, particularly the species included in the M. chelonae-M. abscessus group (8, 46).
M. chelonae-M. abscessus group taxonomy has undergone several updates due to the discrimination of new species by sequencing multiple housekeeping genes and, to a lesser extent, by the evaluation of phenotypic characteristics. Mycobacterium bolletii and Mycobacterium massiliense are newly proposed species that are closely related to M. abscessus and M. chelonae, and consequently they should be considered part of the same group (1, 4). The species M. massiliense was proposed in 2004 based on nonconventional phenotypic characterization and genotypic studies of two isolates recovered from sputum and bronchoalveolar fluid from a patient in Marseille, France (4). Following the original description, reports from different areas of the world have described the occurrence of this species both in culture collections previously classified as M. abscessus and as the cause of invasive opportunistic infections and outbreaks (9, 18, 34, 38, 40).
In Brazil, outbreaks caused by RGM have been detected since 1998, mainly in the cities of Rio de Janeiro and São Paulo. Eight outbreaks following laser in situ keratomileusis (surgery for myopia correction), mesotherapy sessions (intradermal injections), or breast implants have been described, most of them associated with species belonging to the M. chelonae-M. abscessus group. (16, 30-32). Recently, two outbreaks of surgical-site infection following video-assisted surgeries were reported in Brazil (9, 40).
Since August 2006, a large number of surgical wound infections caused by RGM following video-assisted surgery have been diagnosed in the state of Rio de Janeiro, in the southeast region of Brazil. From August 2006 to July 2007, more than 1,000 suspected cases were reported to the Secretary of Health of the Rio de Janeiro state.
The aim of this work was to define species identification, antimicrobial and glutaraldehyde susceptibility patterns, and the clonality of representative mycobacterial isolates recovered from postsurgery wound infections during the epidemic.
MATERIALS AND METHODS
Case definition.
A possible case was defined as a patient who submitted to video-assisted surgery from August 2006 to July 2007 in the state of Rio de Janeiro, Brazil, presenting persistent inflammation signs at the surgical incision site and who had not responded to standard antimicrobial therapy. A probable case was defined as a patient that fulfilled the possible case definition and had granulomas in tissue obtained from the surgical wound or surrounding structures or a positive acid-fast staining but negative culture for mycobacteria. A confirmed case was defined as a patient that fulfilled the possible case definition and had a positive culture for mycobacteria in a clinical sample collected from the surgical wound or surrounding tissues.
Primary isolation of mycobacteria from clinical samples.
Tissue fragments and secretions were obtained after biopsy or needle aspiration procedures, respectively, and processed for baciloscopy and culture at local clinical laboratories (24). These were inoculated onto 5% sheep blood agar and chocolate agar, Lowenstein-Jensen (LJ) slants, and in some laboratories, also in mycobacteria growth indicator tube (MGIT) broth (Becton-Dickinson, Franklin Lakes, NJ).
Phenotypic tests.
All isolates were tested for growth rate, pigment production, and acid fastness as described elsewhere (24).
Outbreak strains, unrelated controls, and environmental isolate.
RGM strains from previously reported outbreaks of surgical wound infections that occurred in Brazil in Belém city (F1832) and Goiânia city (F1334 and F1345), one epidemiologically unrelated isolate (CRM-0270), and the M. massiliense type strain CIP108297 were used as unrelated controls. The isolate CRM-0270 was recovered in 2007 from a sputum sample collected from a patient who lived in the city of Rio de Janeiro who had not previously submitted to bronchoscopy. One isolate (CRM-0029), recovered from a trocar used in a cholecystectomy performed in hospital H18 during the epidemic period, also was included for comparison purposes.
PRA-hsp65.
All 146 isolates were analyzed initially using the PCR-restriction enzyme analysis of the hsp65 gene (PRA-hsp65), as previously described by Telenti et al. (36) and Ringuet et al. (28), with minor modifications. In brief, a loopfull of recent growth on LJ was suspended in 70 μl of 1× Tris-EDTA (TE), boiled for 10 min, and frozen at −20°C. Amplification was carried out with the primers Tb11 (5′-ACC AAC GAT GGT GTG TCC AT-3′) and Tb12 (5′-CTT GTC GAA CCG CAT ACC CT-3′). The PCR mixture consisted of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 1% formamide, 200 μM each deoxynucleoside triphosphate (dNTP), 0.5 μM primers, 1.25 U Platinum Taq polymerase (Invitrogen Life Technologies Brazil, São Paulo, Brazil), and 5 μl of bacterial lysate. Amplification was achieved with a GeneAmp PCR system 2400 (Perkin-Elmer, Branchburg, NJ) under the following conditions: 45 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. Amplicons were digested with either BstEII (Promega Corporation, Madison, WI) or HaeIII (Promega); restriction fragments were analyzed after 3% FingerPrinting agarose gel (BioAmerica Inc., Miami, FL) electrophoresis and staining with ethidium bromide. Fifty- and 25-bp DNA ladders (Promega) were used as reference standards.
Species identification by DNA sequencing.
At least one randomly selected isolate from each of the 38 hospitals (74 isolates), as well as the isolate recovered from the trocar, the unrelated strain, and isolates that presented distinct PRA-hsp65 patterns were analyzed by the partial sequencing of the hsp65 and/or rpoB genes, as previously described (2, 33).
Partial sequencing of the hsp65 gene.
Primers hsp667FW (5′-GGCCAAGACAATTGCGTACG-3′) and hsp667RV (5′-GGAGCTGACCAGCAGGATG-3′) were used to amplify and sequence a 667-bp region containing Telenti's fragment (positions 145 to 585 of the M. tuberculosis H37Rv genome) of the hsp65 gene. A total of 5 μl of each DNA solution (50 μg/ml) was added to 45 μl of a PCR mixture containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 1% enhancer (Invitrogen), 200 μM each dNTP, 1 μM primers, and 1.25 U of Platinum Taq DNA polymerase (Invitrogen). Amplification conditions were 2 min at 95°C, followed by 30 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 45 s, with a final extension step of 72°C for 5 min. Sequencing was performed as described below.
Partial sequencing of the rpoB gene.
A 764-bp fragment was amplified and sequenced with primers MycoF (5′-GCAAGGTCACCCCGAAGGG-3′) and MycoR (5′-AGCGGCTGCTGGGTGATCATC-3′). A total of 5 μl of each DNA solution (50 μg/ml) was added to 45 μl of a PCR mixture containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 2.5 mM MgCl2, 200 μM each dNTP, 1 μM primers, and 1.0 U of Taq DNA polymerase (Promega). PCR mixtures were heated at 95°C for 1 min and then subjected to 35 cycles of denaturation at 94°C for 30 s, annealing at 64°C for 30 s, and extension at 72°C for 90 s, with a final step of 72°C for 5 min. Amplicons were purified with GFX PCR DNA and a Gel Band purification kit (G&E) and sequenced in an ABI PRISM 3100 sequencer with a BigDye Terminator cycle sequencing kit (Applied Biosystems). The sequences obtained were compared with those deposited in the GenBank database by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST).
Antimicrobial susceptibility testing.
A total of 74 M. massiliense epidemic isolates, including at least one from each hospital, previously identified to the species level by hsp65 or rpoB gene sequencing were evaluated for their susceptibility to amikacin, ciprofloxacin, cefoxitin, clarithromycin, and doxycycline as recommended by the Clinical and Laboratory Standards Institute (10). Staphylococcus aureus ATCC 29213 was used as a quality control strain.
Genomic restriction endonuclease digestion and PFGE.
A total of 65 representative isolates, including at least one isolate from each of the 38 hospitals that had confirmed cases, isolates from previous M. massiliense outbreaks (F1345 and F1832), and epidemiologically unrelated isolates (CRM-0270 and M. massiliense type strain CIP108297) were analyzed by pulsed-field gel electrophoresis (PFGE). Bacterial suspensions were included in agarose plugs, treated as described by Coleman et al. (11) and Sampaio et al. (32), and finally digested with DraI (Promega). Restriction fragments were separated by PFGE in 1% agarose gels in a CHEF-DRIII system (Bio-Rad) with pulse times increasing from 1.6 to 21.3 s for 22 h at 14°C, with a voltage gradient of 6 V/cm. The restriction profiles were analyzed and compared using the Molecular Analyst Fingerprinting Plus software package, version 1.12, of the Image Analysis System (Bio-Rad). The interpretative criteria applied were those proposed by Tenover et al. (37).
Glutaraldehyde tolerance.
Five randomly selected isolates obtained from patients from hospitals H28 (CRM 0018, CRM 0019, and CRM 0020) and H34 (CRM 0034 and CRM 0035) belonging to the clonal group BRA100, as well as two unrelated isolates cultivated from sputum samples (CRM 0270 and CRM 0273), were evaluated concerning their ability to survive after 30 min or 10 h of exposure to 2% commercial glutaraldehyde solutions using the suspension method, as recommended by the manufacturers for high-level disinfection and sterilization (6, 12). Each assay was repeated three times and also included Mycobacterium smegmatis PRD no.1 00061, Mycobacterium bovis INCQS 00062, M. abscessus ATCC 19977, and M. chelonae ATCC 35752 strains as quality control strains. Mycobacterial suspensions with a turbidity equivalent to a McFarland 1 standard were prepared from cultures that had been grown on LJ medium for no more than 7 days. Five hundred microliters was added to 4.5 ml of each activated glutaraldehyde solution and incubated at 25°C. After 30 min and 10 h, an aliquot of 0.5 ml of each mixture containing bacteria and glutaraldehyde was transferred to a new vial containing the same volume of 1% sodium sulfite (1:1, vol/vol). They were mixed, seeded on LJ slants, and incubated at 35°C in ambient air for 60 days.
Nucleotide sequence accession numbers.
The GenBank database accession numbers generated in this study are EU009467.1, EU009468.1 EU009469.1, FJ515915.1, FJ515916.1, FJ516384.1, FJ516386.1, FJ531482.1, FJ531483.1, and FJ531484.1.
RESULTS
Outbreak description and clinical findings.
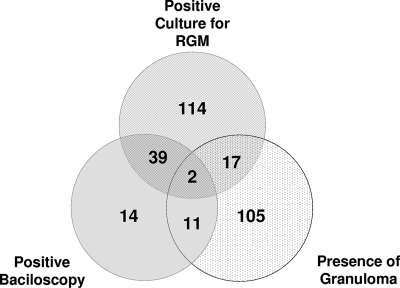
Among the 1,051 possible cases of postsurgery infections reported to the Secretary of Health of Rio de Janeiro state between August 2006 and July 2007, 302 presented positive reported laboratory tests. One hundred thirty were classified as probable cases, and 172 were considered confirmed cases (Fig. 1). Among the confirmed cases, 148 isolates were available for further characterization. Information regarding the gender and age of the patient, type of surgery, surgeon name, and more were obtained for 126 (86%) patients who had a positive culture for mycobacteria. The most frequent type of surgery was cholecystectomy (56.0%), followed by diagnostic laparoscopy (7.8%), appendicectomy (7.1%), arthroscopy (5.5%), oophorectomy/oophoroplasty (3.1%), hernioplasty (2.3%), tubal ligation (2.3%), myomectomy (2.3%), gastroplasty (1.5%), rectosigmoidectomy (1.5%), and other video-assisted procedures (10.6%). The median age was 44 years old (range, 14 to 89), and most patients (73.0%) were female. All patients had visible cutaneous lesions (single or multiple), and most of them (61.1%) presented serous, bloody, or purulent drainage from infected wounds. Other clinical signs observed included erythema (25.3%), subcutaneous nodules (23.0%), local pain (21.4%), subcutaneous abscesses (19.8%), fever (11.1%), edema (11.1%), fistulas (6.3%), and ulcers (3.9%). The median time from surgery to the manifestation of clinical signs was 31 days (range, 2 to 187). A significant portion of the infections (58.7%) occurred in patients that submitted to surgical procedures at private institutions. A total of 38 hospitals, most of them located around the urban area of the city of Rio de Janeiro, had 1 to 15 confirmed cases of surgical site infection caused by RGM.
FIG. 1.
Distribution of positive laboratory tests among 302 probable cases with notified laboratory diagnosis during the epidemic of RGM postsurgical infections in Rio de Janeiro, Brazil (2006 to 2007).
The initial empirical treatment proposed by the federal health authority included clarithromycin plus ethambutol and terizidon or clarithromycin plus ethambutol and amikacin. As soon as antimicrobial susceptibility tests and species identification were available, ethambutol and terizidon were removed from treatment regimens, and most of the patients were treated with clarithromycin plus amikacin or clarithromycin plus amikacin and imipenem or cefoxitin with good clinical outcomes. Besides antimicrobial therapy, most of the patients underwent surgical debridement and abscess drainage.
Commercial 2% glutaraldehyde solution was used for the disinfection of the surgical instruments (15 to 30 min of exposure) in all institutions that had confirmed cases. At least 58 different surgeons reported suspected cases during the epidemic period. From April 2008, no more cases have been reported in the state of Rio de Janeiro. The decrease in the number of cases was observed either after the discontinuation of the use of 2% glutaraldehyde solution in high-level disinfection or during temporary interruptions of surgeries until the resolution of epidemics at some care centers. No surveillance studies were performed for mycobacterial isolation from medical devices. No public health measures were suggested or implemented by local authorities before September 2008.
Species identification.
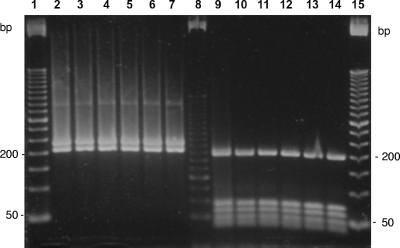
A total of 148 acid-fast-positive isolates were recovered from clinical samples and available for further characterization. Most of them (n = 144; 97.2%) were nonpigmented rapid growers, with a PRA-hsp65 pattern compatible with that of M. massiliense or M. bolletii (PRA-MmMb), with BstEII restriction fragments of 235 and 210 bp and HaeIII restriction fragments of 200, 70, 60, 50, and 40 bp (Fig. 2). All isolates with the PRA-hsp65 pattern PRA-MmMb submitted to sequencing (n = 74) had identical rpoB sequences. Such sequences (GenBank accession numbers EU009467.1, EU009468.1, and EU009469.1) are 99.71% (686/688) similar to those of the M. massiliense type strain CIP 108297 (EU254721.1 and AY593981.2). These two substitutions, located at positions 2563 and 2754 of rpoB from M. tuberculosis H37Rv (GenBank accession BX842574.1), are silent, since the amino acid sequences are identical. Two (1.3%) nonpigmented, rapidly growing isolates presented identical PRA-hsp65 patterns, with BstEII fragments of 230, 125, and 80 bp and HaeIII fragments of 145, 120, 60, and 40 bp, which match the patterns of the Mycobacterium fortuitum group. One of these isolates (CRM-0264) had hsp65 and rpoB partial sequences that were identical to those of accession numbers AY458072.1 and AY147165.1, respectively, which belong to the type strain of M. fortuitum CIP104534. The second isolate (CRM-0267) had hsp65 (GenBank accession no. FJ531484.1) and rpoB (GenBank accession no. FJ531483.1) partial sequences that were 100% (424/424) and 99.56% (694/697) similar to GenBank accession numbers AY458064.1 and AY262743.2, respectively, which correspond to the Mycobacterium wolinskyi type strain ATCC700010. The two yellow-pigmented scotochromogenic isolates F1672 and F1694 had hsp65 partial sequences (GenBank accession numbers FJ515915.1 and FJ515916.1) that had the highest similarity indexes (99.8% [423/424] and 99.5% [422/424], respectively) compared to the hsp65 sequence of the Mycobacterium neoaurum type strain CIP 105387 (GenBank accession no. AF547860.1). Each of these M. neoaurum isolates were recovered together with M. massiliense colonies from two patients with mixed mycobacterial infection.
FIG. 2.
PRA-hsp65 profiles of six representative M. massiliense epidemic isolates. Lanes 1 and 15, 50-bp DNA ladder; lane 8, 25-bp DNA ladder; lanes 2 to 7, amplicons digested with BstEII; lanes 9 to 14, amplicons digested with HaeIII. Each lane represents a different strain: lane 2, CRM-0006; 3, CRM-0018; 4, CRM-0029; 5, CRM-0181; 6, CRM-0189; 7, CRM-0195; 9, CRM-0006; 10, CRM-0018; 11, CRM-0029; 12, CRM-0181; 13, CRM-0189; and 14, CRM-0195.
The unrelated isolate (CRM-0270) obtained from sputum and the isolate cultivated from a trocar during the outbreak period (CRM-0029) had rpoB sequences with the same similarity indexes as those described above for the M. massiliense epidemic isolates.
M. massiliense isolates from the outbreaks that occurred in the cities of Goiânia (F1345 and F1334; GenBank accession nos. EU031904.1 and EU031905.1) and Belém (F1832; GenBank accession no. FJ531482.1) had the same rpoB sequence as those described for the isolates from Rio de Janeiro.
Antimicrobial susceptibility.
All 74 isolates tested were susceptible to amikacin (MIC at which 90% of the tested isolates are inhibited [MIC90], 8 μg/ml) and clarithromycin (MIC90, 0.25 μg/ml) but resistant to ciprofloxacin and doxycycline. Cefoxitin MICs were high for most isolates (Table 1).
TABLE 1.
Antimicrobial susceptibility of 74 RGM M. massiliense epidemic isolates recovered from wound infections after video laparoscopic surgery during an outbreak in Rio de Janeiro, Brazil (2006 to 2007)
| Antimicrobial agent | MIC (μg/ml)
|
||
|---|---|---|---|
| Range | MIC50 | MIC90 | |
| Amikacin | 4-16 | 8 | 8 |
| Cefoxitin | 16-128 | 64 | 128 |
| Ciprofloxacin | 8->16 | >16 | >16 |
| Clarithromycin | 0.06-0.5 | 0.12 | 0.25 |
| Doxycycline | 16->32 | >32 | >32 |
Clonal diversity.
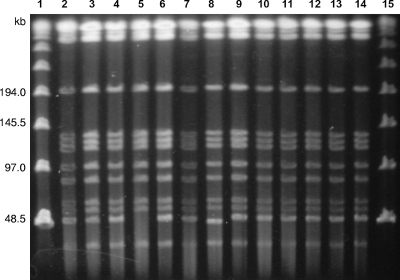
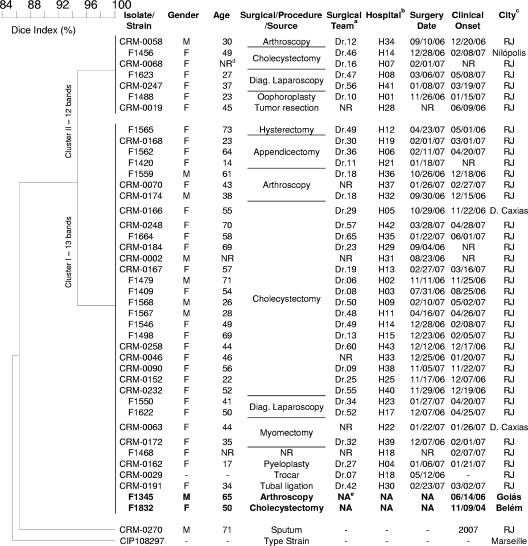
PFGE fingerprinting using DraI as the restriction enzyme generated profiles with 12 or 13 fragments without the use of thiourea in the running buffer. Clinical isolates from the Rio de Janeiro epidemic and the isolate cultivated from the trocar had identical PFGE patterns or patterns that differed by a single fragment (Fig. 3 and 4). Their PFGE patterns also were identical to those obtained for the isolates from the previously reported outbreaks but differed from PFGE patterns of the unrelated isolates CRM-0270 and CIP 18297 (Fig. 4). The predominant clonal group was named BRA100 to express its interhospital and interstate dissemination.
FIG. 3.
PFGE profiles of genomic DNA from M. massiliense isolates after digestion with DraI. Lanes 1 and 15, molecular size markers (Lambda DNA concatemers ranging from 48.5 to 1,018.5 kb); 2, CRM-0006; 3, CRM-0013; 4, CRM-0018; 5, CRM-0019; 6, CRM-0020; 7, CRM-0029; 8, CRM-0169, 9, CRM-0172; 10, CRM-0181; 11, CRM-0185; 12, CRM-0189; 13, CRM-0191; and 14, CRM-0195.
FIG. 4.
Dendrogram resulting from computer-assisted analysis of the PFGE profiles of 43 representative M. massiliense isolates. The Dice coefficient was used for calculating the percentages of similarity among the profiles. M. massiliense isolates recovered during outbreaks in other Brazilian cities are marked in bold. Diag., diagnosis. Footnotes: a, the mycobacterial isolate was recovered from a patient who submitted to video laparoscopic surgery headed by the Dr.n team; b, the mycobacterial isolate was obtained from a patient who submitted to surgery at hospital (H) n; c, RJ, Rio de Janeiro city; d, NR, not reported; e, NA, not applicable.
Glutaraldehyde tolerance.
All five M. massiliense isolates tested belonging to the clonal group BRA100 survived after 30 min and 10 h of exposure to 2% glutaraldehyde solution. Colonies were visible in LJ medium after 7 days. They were confirmed to be acid-fast bacilli and had the same PRA-hsp65 pattern as isolates exposed to 2% glutaraldehyde. M. smegmatis PRD no.1, M. bovis BCG INCQ 00062, M. abscessus ATCC 19977, M. chelonae ATCC 35752, and two unrelated M. massiliense isolates (CRM-0270 and CRM-0273) did not survive after 30 min of exposure.
DISCUSSION
This work reports the high frequency of a single clone of M. massiliense associated with an epidemic of postsurgical infections caused by RGM. Although only five randomly chosen isolates from this clonal group (BRA100) were tested, they were reproducibly tolerant to 2% glutaraldehyde solution, while controls and isolates not belonging to the epidemic clone were not. This evidence suggests that glutaraldehyde tolerance is a hallmark of the clonal group BRA100 and contributed to the occurrence of the epidemic. Outbreaks due to RGM have been detected in Brazil since 1998, but until 2004 none of them was associated with a member of the M. abscessus group presenting a PRA-hsp65 pattern that could correspond to M. massiliense or M. bolletii (5, 16, 30-32). The first two reports of outbreaks associated with M. massiliense in Brazil recently were published and describe surgical-site infections following video-assisted surgeries that occurred in states other than Rio de Janeiro (9, 40). The exact number of video-assisted procedures performed monthly in Brazil is unknown, but this technology has been available in Brazil since 1990, and it is estimated that 70% of abdominal surgeries currently are performed using video equipment. No reports of infections following video-assisted medical procedures had emerged before 2004 in Brazil. Postlaparoscopy infections caused by mycobacteria also have been described worldwide as isolated cases or limited outbreaks (26, 27, 29, 41) and linked to possible sterilization pitfalls, as is probably the case in the present epidemic. In this study, all hospitals that had cases of infections due to M. massiliense used 2% alkaline glutaraldehyde solution to disinfect surgical instruments that cannot be sterilized by autoclaving, and this routine has long been employed. On the other hand, some hospitals located in the state of Rio de Janeiro that had used the same commercial glutaraldehyde solutions have not had cases of surgical infections caused by RGM to date. This may be explained by the well-known concept that adequate mechanical cleaning may prevent biofilm formation and bacterial adhesion (17). Our findings of a single clone (BRA100) of glutaraldehyde-tolerant M. massiliense may partially explain the occurrence of temporally clustered outbreaks in three different regions in Brazil. Although biofilm formation has been demonstrated to contribute to tolerance to biocidal solutions (35), biofilm formation alone could not explain the occurrence of a single clone in three distant cities. Two hypotheses arise from our observations: (i) this clone has been spread in aquatic environments in Brazil for a long time, and both the selective pressure of 2% glutaraldehyde use and the inadequate mechanical cleaning of surgical instruments have facilitated the occurrence of outbreaks, or (ii) this clone was disseminated inside commercially available nonactivated glutaraldehyde solutions, and the inadequate mechanical cleaning of surgical instruments has facilitated the occurrence of outbreaks. Some surgical teams carried their own instruments and performed surgeries at different hospitals and even different cities and states. The reuse of disposable or improperly sterilized trocars by some of these teams also may have contributed to the spread of a single clone and the large number of infected patients.
Our work, as with the two previous reports of M. massiliense outbreaks, has a limitation; namely, the small number of unrelated strains and the unknown discriminatory power of PFGE for this bacterial species. M. massiliense has phenotypic and genotypic characteristics that are very similar to those of M. abscessus. Although Adékambi et al. described some phenotypic tests (nitrate reductase, β-glucuronidase, β-galactosidase, N-acetyl-β-glucosaminidase, and indole production) that could differentiate these species, their conclusions were based on two isolates and should not be applied to clinical laboratories before validation with a large collection of unrelated isolates (4). Although there is not a consensus on the best target for mycobacterial species discrimination, the partial sequencing of hsp65 and/or rpoB genes has been used to differentiate RGM species (2). Currently, species that have a similarity index equal to or less than 98.1% for the rpoB fragment, as proposed by Adékambi et al. (3), should be considered different. In this work, we obtained the highest similarity index (99.7%) for the comparison of our sequences to those of the M. massiliense type strain. The hsp65 partial sequence of the epidemic isolates had 100% similarity to that of the M. massiliense type strain, while the minimal similarity index for the same species has been proposed as ≥97% (21). These data indicate that under the current classification of mycobacteria, the epidemic was caused by M. massiliense.
In this report, all of the M. massiliense isolates tested were susceptible to amikacin and clarithromycin but resistant to cefoxitin, ciprofloxacin, and doxycycline. Our results are in accordance with those reported by Adékambi et al. (4), except that the doxycycline MICs for our isolates was higher, which is in agreement with results reported by other authors (34, 40). This evidence reinforces that susceptibility to doxycycline cannot be used as a marker for differentiation between M. abscessus and M. massiliense.
In summary, to our knowledge, this report describes the largest surgical-site infection epidemic caused by a unique clone of M. massiliense reported to date and reinforces the risk of using 2% glutaraldehyde solution for disinfecting surgical devices.
Acknowledgments
The DNA Sequencing Platform (PDTIS/FIOCRUZ), Rio de Janeiro, Brazil, is acknowledged for performing the rpoB sequencing procedures.
The Instituto Nacional de Controle de Qualidade em Saúde (INCQS/FIOCRUZ) is acknowledged for providing Mycobacterium smegmatis PRD no.1 00061 and Mycobacterium bovis INCQS 00062 strains.
This study was supported in part by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ; processes no. 110.347/2007 and 18123/2008), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (MCT/CNPq; Edital Universal, process no. MCT/CNPq-no. 15/2007), and PDTIS-FIOCRUZ.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Adekambi, T., P. Berger, D. Raoult, and M. Drancourt. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56133-143. [DOI] [PubMed] [Google Scholar]
- 2.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 415699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adékambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 542095-2105. [DOI] [PubMed] [Google Scholar]
- 4.Adékambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 425493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarenga, L., D. Freitas, A. L. Hofling-Lima, R. Belfort, Jr., J. Sampaio, L. Sousa, M. Yu, and M. Mannis. 2002. Infectious post-LASIK crystalline keratopathy caused by nontuberculous mycobacteria. Cornea 21426-429. [DOI] [PubMed] [Google Scholar]
- 6.Best, M., S. A. Sattar, V. S. Springthorpe, and M. E. Kennedy. 1990. Efficacies of selected disinfectants against Mycobacterium tuberculosis. J. Clin. Microbiol. 282234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickman, M., A. A. Parsa, and F. D. Parsa. 2005. Mycobacterium chelonae infection after breast augmentation. Aesthetic Plast. Surg. 29116-118. [DOI] [PubMed] [Google Scholar]
- 8.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso, A. M., E. Martins de Sousa, C. Viana-Niero, F. Bonfim de Bortoli, Z. C. Pereira das Neves, S. C. Leao, A. P. Junqueira-Kipnis, and A. Kipnis. 2008. Emergence of nosocomial Mycobacterium massiliense infection in Goias, Brazil. Microbes Infect. 101552-1557. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard. Document M24-A. CLSI, Wayne, PA. [PubMed]
- 11.Coleman, N. V., and J. C. Spain. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 696041-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, F. M., and V. Montalbine. 1976. Mycobactericidal activity of glutaraldehyde solutions. J. Clin. Microbiol. 4408-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Costa Cruz, J. C. 1938. Mycobacterium fortuitum: um novo bacilo ácido-resistente patogênico para o homem (new acid fast bacillus pathogenic for man). Acta Med. (Rio de Janeiro) 1298-301. [Google Scholar]
- 14.De Groote, M. A., and G. Huitt. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 421756-1763. [DOI] [PubMed] [Google Scholar]
- 15.Dytoc, M. T., L. Honish, C. Shandro, P. T. Ting, L. Chui, L. Fiorillo, J. Robinson, A. Fanning, G. Predy, and R. P. Rennie. 2005. Clinical, microbiological, and epidemiological findings of an outbreak of Mycobacterium abscessus hand-and-foot disease. Diagn. Microbiol. Infect. Dis. 5339-45. [DOI] [PubMed] [Google Scholar]
- 16.Freitas, D., L. Alvarenga, J. Sampaio, M. Mannis, E. Sato, L. Sousa, L. Vieira, M. C. Yu, M. C. Martins, A. Hoffling-Lima, and R. Belfort, Jr. 2003. An outbreak of Mycobacterium chelonae infection after LASIK. Ophthalmology 110276-285. [DOI] [PubMed] [Google Scholar]
- 17.Goller, C. C., and T. Romeo. 2008. Environmental influences on biofilm development. Curr. Top. Microbiol. Immunol. 32237-66. [DOI] [PubMed] [Google Scholar]
- 18.Kim, H. Y., Y. Kook, Y. J. Yun, C. G. Park, N. Y. Lee, T. S. Shim, B. J. Kim, and Y. H. Kook. 2008. Proportions of Mycobacterium massiliense and Mycobacterium bolletii strains among Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J. Clin. Microbiol. 463384-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macadam, S. A., B. M. Mehling, A. Fanning, J. A. Dufton, K. T. Kowalewska-Grochowska, P. Lennox, A. Anzarut, and M. Rodrigues. 2007. Nontuberculous mycobacterial breast implant infections. Plast. Reconstr. Surg. 119337-344. [DOI] [PubMed] [Google Scholar]
- 20.Manzoor, S. E., P. A. Lambert, P. A. Griffiths, M. J. Gill, and A. P. Fraise. 1999. Reduced glutaraldehyde susceptibility in Mycobacterium chelonae associated with altered cell wall polysaccharides. J. Antimicrob. Chemother. 43759-765. [DOI] [PubMed] [Google Scholar]
- 21.McNabb, A., D. Eisler, K. Adie, M. Amos, M. Rodrigues, G. Stephens, W. A. Black, and J. Isaac-Renton. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 423000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nomura, K., M. Ogawa, H. Miyamoto, T. Muratani, and H. Taniguchi. 2004. Antibiotic susceptibility of glutaraldehyde-tolerant Mycobacterium chelonae from bronchoscope washing machines. Am. J. Infect. Control 32185-188. [DOI] [PubMed] [Google Scholar]
- 23.Padoveze, M. C., C. M. Fortaleza, M. P. Freire, D. Brandao de Assis, G. Madalosso, A. C. Pellini, M. L. Cesar, V. Pisani Neto, M. M. Beltramelli, E. Chimara, L. Ferrazoli, M. A. da Silva Telles, J. L. Sampaio, and S. C. Leao. 2007. Outbreak of surgical infection caused by non-tuberculous mycobacteria in breast implants in Brazil. J. Hosp. Infect. 67161-167. [DOI] [PubMed] [Google Scholar]
- 24.Pfyffer, G. E. 2007. Mycobacterium: general characteristics, laboratory detection and staining procedures. In P. R. Murray (ed.), Manual of clinical microbiology, 9th ed., vol. 1. American Society for Microbiology, Washington DC.
- 25.Rahav, G., S. Pitlik, Z. Amitai, A. Lavy, M. Blech, N. Keller, G. Smollan, M. Lewis, and A. Zlotkin. 2006. An outbreak of “Mycobacterium jacuzzii” infection following insertion of breast implants. Clin. Infect. Dis. 43823-830. [DOI] [PubMed] [Google Scholar]
- 26.Rajini, M., S. R. Prasad, R. R. Reddy, R. V. Bhat, and K. R. Vimala. 2007. Postoperative infection of laparoscopic surgery wound due to Mycobacterium chelonae. Indian J. Med. Microbiol. 25163-165. [DOI] [PubMed] [Google Scholar]
- 27.Ramesh, H., K. Prakash, V. Lekha, G. Jacob, A. Venugopal, and B. Venugopal. 2003. Port-site tuberculosis after laparoscopy: report of eight cases. Surg. Endosc. 17930-932. [DOI] [PubMed] [Google Scholar]
- 28.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues, C., A. Mehta, U. Jha, M. Bharucha, F. D. Dastur, and T. E. Udwadia. 2001. Nosocomial Mycobacterium chelonae infection in laparoscopic surgery. Infect. Control Hosp. Epidemiol. 22474-475. [DOI] [PubMed] [Google Scholar]
- 30.Sampaio, J. L., E. Chimara, L. Ferrazoli, M. A. da Silva Telles, V. M. Del Guercio, Z. V. Jerico, K. Miyashiro, C. M. Fortaleza, M. C. Padoveze, and S. C. Leao. 2006. Application of four molecular typing methods for analysis of Mycobacterium fortuitum group strains causing post-mammaplasty infections. Clin. Microbiol. Infect. 12142-149. [DOI] [PubMed] [Google Scholar]
- 31.Sampaio, J. L., D. N. Junior, D. de Freitas, A. L. Hofling-Lima, K. Miyashiro, F. L. Alberto, and S. C. Leao. 2006. An outbreak of keratitis caused by Mycobacterium immunogenum. J. Clin. Microbiol. 443201-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampaio, J. L., C. Viana-Niero, D. de Freitas, A. L. Hofling-Lima, and S. C. Leao. 2006. Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates. Diagn. Microbiol. Infect. Dis. 55107-118. [DOI] [PubMed] [Google Scholar]
- 33.Selvaraju, S. B., I. U. Khan, and J. S. Yadav. 2005. A new method for species identification and differentiation of Mycobacterium chelonae complex based on amplified hsp65 restriction analysis (AHSPRA). Mol. Cell Probes. 1993-99. [DOI] [PubMed] [Google Scholar]
- 34.Simmon, K. E., J. I. Pounder, J. N. Greene, F. Walsh, C. M. Anderson, S. Cohen, and C. A. Petti. 2007. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J. Clin. Microbiol. 451978-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simões, M., M. O. Pereira, and M. J. Vieira. 2005. Effect of mechanical stress on biofilms challenged by different chemicals. Water Res. 395142-5152. [DOI] [PubMed] [Google Scholar]
- 36.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tortoli, E., R. Gabini, I. Galanti, and A. Mariottini. 2008. Lethal Mycobacterium massiliense sepsis, Italy. Emerg. Infect. Dis. 14984-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Klingeren, B., and W. Pullen. 1993. Glutaraldehyde resistant mycobacteria from endoscope washers. J. Hosp. Infect. 25147-149. [DOI] [PubMed] [Google Scholar]
- 40.Viana-Niero, C., K. V. Lima, M. L. Lopes, M. C. Rabello, L. R. Marsola, V. C. Brilhante, A. M. Durham, and S. C. Leao. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J. Clin. Microbiol. 46850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijayaraghavan, R., R. Chandrashekhar, Y. Sujatha, and C. S. Belagavi. 2006. Hospital outbreak of atypical mycobacterial infection of port sites after laparoscopic surgery. J. Hosp. Infect. 64344-347. [DOI] [PubMed] [Google Scholar]
- 42.Vinh, D. C., and J. M. Embil. 2005. Infection in breast implants. Lancet Infect. Dis. 5462-463. [DOI] [PubMed] [Google Scholar]
- 43.Vinh, D. C., A. Rendina, R. Turner, and J. M. Embil. 2006. Breast implant infection with Mycobacterium fortuitum group: report of case and review. J. Infect. 52e63-e67. [DOI] [PubMed] [Google Scholar]
- 44.Waddington, E. 1967. An outbreak of swimming pool granuloma. Trans. St. Johns Hosp. Dermatol. Soc. 53122-124. [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52453-490. [DOI] [PubMed] [Google Scholar]
- 46.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. A. Tschen, and M. S. Stone. 1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5657-679. [DOI] [PubMed] [Google Scholar]