Abstract
Bloodstream infections (BSIs) are a significant cause of morbidity and mortality. Successful patient outcomes are diminished by a failure to rapidly diagnose these infections and initiate appropriate therapy. A rapid and reliable diagnostic platform of high sensitivity is needed for the management of patients with BSIs. The combination of an RNA-dependent nucleic acid sequence-based amplification and molecular beacon (NASBA-MB) detection system in multiplex format was developed to rapidly detect medically important BSI organisms. Probes and primers representing pan-gram-negative, pan-gram-positive, pan-fungal, pan-Candida, and pan-Aspergillus organisms were established utilizing 16S and 28S rRNA targets for bacteria and fungi, respectively. Two multiplex panels were developed to rapidly discriminate bacterial or fungal infections at the subkingdom/genus level with a sensitivity of 1 to 50 genomes. A clinical study was performed to evaluate the accuracy of this platform by evaluating 570 clinical samples from a tertiary-care hospital group using blood bottle samples. The sensitivity, specificity, and Youden's index values for pan-gram-positive detection and pan-gram-negative detection were 99.7%, 100%, 0.997 and 98.6%, 95.9%, 0.945, respectively. The positive predictive values (PPV) and the negative predictive values (NPV) for these two probes were 100, 90.7, and 99.4, 99.4, respectively. Pan-fungal and pan-Candida probes showed 100% sensitivity, specificity, PPV, and NPV, and the pan-Aspergillus probe showed 100% NPV. Robust signals were observed for all probes in the multiplex panels, with signal detection in <15 min. The multiplex real-time NASBA-MB assay provides a valuable platform for the rapid and specific diagnosis of bloodstream pathogens, and reliable pathogen identification and characterization can be obtained in under 3 h.
Bloodstream infections (BSIs) are a significant cause of morbidity and mortality in the United States. Approximately 250,000 nosocomial BSIs occur annually, with a crude mortality rate of 20 to 50% (2, 17, 38). Based on data from death certificates, BSIs are the 10th leading cause of death in the United States (33), and the age-adjusted death rate has risen by 78% during the past two decades (10). The mortality rates of BSI patients are increased with measurable delays in the institution of effective, appropriate antimicrobial therapy (19, 35). Inadequate therapy was an independent determinant of mortality for individual organisms such as Candida species (21). Yu et al., in a U.S. multicenter study, found that failure to start antimicrobial therapy within 24 h of illness onset was strongly correlated with a higher probability of a 28-day mortality (49). A rapid and reliable diagnosis may be the optimal method for avoiding such delays in the treatment of BSIs.
A variety of microorganisms can cause BSIs. A nationwide surveillance study showed that gram-positive bacteria, gram-negative bacteria, and fungi (including yeasts and molds) caused 65, 25, and 9.5% of monomicrobial BSIs, respectively. The most common organisms associated with BSIs are the coagulase-negative staphylococci (CoNS), Staphylococcus aureus, Enterococcus spp., and Candida spp. and the gram-negative bacilli Escherichia coli and Klebsiella spp.(47). The conventional diagnostic test for BSI is blood culture, which usually is specific but can have limited sensitivity and is inherently slow in providing complete diagnostic information (14, 37). Even after the detection of growth in cultured blood (usually not before 6 to 12 h of incubation), conventional blood cultures require at least another 24 to 48 h for the definitive identification of the pathogen and the assessment of resistance to antibiotics (4, 37). In recent years, real-time PCR assays have been developed for bacterial and fungal detection from clinical samples (23, 32, 48). The limit of detection was observed as 40, 50, or 2,000 CFU/ml for E. coli, group B Streptococcus spp., and Listeria monocytogenes, and it was 18 CFU/ml for Candida species. The nucleic acid extraction and the PCR amplification could be done in about 4 h. Compared to current culture-based and biochemical methods, nucleic acid sequence-based amplification (NASBA) assays may be the most promising diagnostic approach, because they offer the ultrasensitive detection of pathogens from blood. Despite this progress, current PCR-based diagnostics have some inherent limitations. A variety of pathogen species-specific PCR-based assays are available; however, the probes developed are species specific, which restricts the range of pathogens that can be detected in a single assay. Therefore, the assays are best applied when a specific infection is suspected (37), which is rarely the case during the early stages of BSIs. In addition, a positive signal does not necessarily reflect viable microorganisms (25), as the amplification of DNA from dead cells can confuse clinical management, especially following therapy. Importantly, some amplified detection methods generally require a growth enrichment step, i.e., inoculation and growth in blood culture bottles, to achieve the sensitivity necessary for pathogen detection and identification.
The universal detection of bacteria or fungi in blood using broad-range PCR assays has been described (1, 16, 22, 42), but interpretations can be difficult, as assay contaminants can cause false-positive results (37). The early detection of BSIs requires reliable sensitivity, around 10 CFU/ml of blood for bacteria and fungi, which is below the level of most existing diagnostic platforms. As an alternative to PCR, NASBA provides the ultrasensitive detection of nucleic acids. It is an isothermal process that selectively amplifies RNA even in the presence of genomic DNA (44). The reaction is highly robust, resulting in 1012 amplified copies of template in a 30-min assay (12), which is comparable to a nested PCR in terms of sensitivity. When used to target rRNA genes, which are present in multiple copies in the genomes and expressed at a high level (thousands of copies per cell) in all known human bacterial and fungal pathogens, a NASBA-based platform is capable of high-level detection. It detects both live and dead cells. The amplified single-stranded RNA can be detected easily in real time by molecular beacon (MB) probes, which are small, self-reporting, single-stranded nucleic acid hairpin probes that brightly fluoresce when bound to their targets (45). MB probes offer a superior detection modality to perform real-time NASBA assays, which are rapid, highly specific, and sensitive.
To address the special considerations of BSIs, we have developed a real-time NASBA-MB platform that rapidly identifies medically important bacterial and fungal pathogens for BSI. A cascade pathogen detection and identification approach was developed that recognizes increasing complexity ranging from kingdom-specific (e.g., pan-fungal) targets to genus-specific (e.g., pan-Candida and pan-Aspergillus) and species-specific targets to drug resistance targets. The bacterial primers and probes target 16S rRNA genes, while the fungal assays target the 28S rRNA genes. This is the first report of a comprehensive NASBA-based platform for the rapid detection at the genus level of the major bacteria and fungi that are responsible for BSIs. This platform is suitable for use in simplex or multiplex assays incorporating a range of probes and primer sets to meet a variety of clinical laboratory detection needs.
This initial proof-of-concept study was designed to assess the technical feasibility of this method to detect a large variety of bloodstream pathogens. Although the final goal of a rapid diagnostic is detection from directly drawn blood samples, these studies are essential to evaluate the potential of the amplification system, primers, and probes. Technical parameters evaluated included assay sensitivity and specificity. To evaluate the overall accuracy of this platform, a clinical study was performed from a tertiary-care hospital group using blood bottle samples.
MATERIALS AND METHODS
Bacterial and fungal strains.
Bacterial strains of Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterobacter cloacae, and Acinetobacter spp. and fungal strains of Candida albicans, Candida glabrata, Candida krusei, Candida tropicalis, Candida parapsilosis, Candida dubliniensis, Candida guilliermondii, Cryptococcus neoformans, Saccharomyces cerevisiae, Aspergillus fumigatus, Aspergillus flavus, Aspergillus terreus, and Aspergillus niger were obtained from the historical stock of the Public Health Research Institute (PHRI), which is collected from many sites across North America and Europe (Table 1).
TABLE 1.
Bacterial and fungal strains used in this study
| Species and organism type | No. of strains | Reference strain | Source |
|---|---|---|---|
| Gram-positive bacteria | |||
| Staphylococcus aureus | 8 | MRSA-9897 | Clinical strain |
| Staphylococcus epidermidis | 8 | 10683 | Clinical strain |
| Enterococcus faecium | 8 | 11936 | Clinical strain |
| Enterococcus faecalis | 8 | 9199 | Clinical strain |
| Gram-negative bacteria | |||
| Escherichia coli | 8 | 8781 | Clinical strain |
| Enterobacter cloacae | 8 | 10623 | Clinical strain |
| Pseudomonas aeruginosa | 8 | 10470 | Clinical strain |
| Acinetobacter baumannii | 8 | 10437 | Clinical strain |
| Klebsiella pneumoniae | 8 | 20856 | Clinical strain |
| Candida spp. | |||
| Candida albicans | 8 | 90028 | ATCC strain |
| Candida glabrata | 8 | 90030 | ATCC strain |
| Candida krusei | 8 | 6258 | ATCC strain |
| Candida tropicalis | 8 | 750 | ATCC strain |
| Candida parapsilosis | 8 | 22019 | ATCC strain |
| Candida guilliermondii | 8 | 6260 | ATCC strain |
| Candida lusitaniae | 4 | 200950 | ATCC strain |
| Candida dubliniensis | 8 | 3949 | NCPF strain |
| Aspergillus spp. | |||
| Aspergillus fumigatus | 8 | 293 | ATCC strain |
| Aspergillus flavus | 8 | 204304 | ATCC strain |
| Aspergillus terreus | 8 | 9151 | Merck strain |
| Aspergillus niger | 8 | 9508 | ATCC strain |
| Other fungi | |||
| Cryptococcus neoformans | 1 | 208821 | ATCC strain |
| Saccharomyces cerevisiae | 1 | Y55 | Laboratory strain |
Bacterial and fungal cultures.
All reference bacterial strains were cultured on Luria-Bertani (LB) agar at 37°C, except for S. aureus strains that were grown on mannitol salt agar (MSA) at 37°C. Candida spp. reference strains were grown and maintained on yeast extract-peptone-dextrose (YPD) agar at 30°C, and Aspergillus spp. reference strains were cultured on potato dextrose agar (PDA) at 37°C. Brain heart infusion (BHI) medium, LB broth, or YPD broth was used to prepare overnight cultures for the nucleic acid extraction from bacteria or Candida reference strains, respectively.
Primers and MB design.
16S and 28S rRNA were chosen to be our detection targets for bacteria and fungi, respectively. The 16S rRNA gene sequences from 12 bacterial species were obtained from online databases (Table 2). The sequences were aligned using the online program ClustalW from the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/clustalw/index.html). Similarly, 28S rRNA gene sequences for 15 different fungal species were collected and aligned (Table 2). For bacteria, one region highly conserved for gram-positive bacteria but variable for gram-negative bacteria was selected as the diagnostic target for the pan-gram-positive probe. On the contrary, a region conserved for all of the gram-negative bacteria but variable for other microbes was chosen as the diagnostic target for the pan-gram-negative probe. Based on this principle, pan-fungal, pan-Candida, and pan-Aspergillus targets were selected accordingly. MB probes were designed by adding two artificial arm sequences to both ends of the target sequence. The secondary structure of the MB was evaluated by the software OligoAnalyzer 3.0 (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/). The MBs were labeled with the fluorophores 5-carboxyfluorescein (FAM), 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein (HEX), CAL fluor red (CR610), and Quasar 670 (Q670) at the 5′ end and with benzoic acid succinimidyl ester (dabcyl) or black hole quencher at the 3′ end. The MBs were purchased from Biosearch Technologies, Inc., Novato, CA. The pan-gram-negative MB has a perfect target sequence for most bacterial species tested in this study, except for Acinetobacter spp. and Pseudomonas spp., which both have two mismatches with the pan-gram-negative beacon. Therefore, two individual beacons targeting Acinetobacter spp. and Pseudomonas spp. were designed in the same region covered by the pan-gram-negative primers, and they were mixed with the pan-gram-negative MB for the following experiments. Similarly, three patch probes for pan-gram-positive MB (16S GP v1.21 to 16S GP v1.23) (Table 3) were designed, because three gram-positive species (Corynebacterium sp., Propionibacterium sp., and Micrococcus sp.) from the clinical samples were missed by the pan-gram-positive probe. NASBA primers also were designed based on the sequence alignments. A T7 promoter sequence was added at the 5′ end of each P1 primer. All MBs and NASBA primers used in this study are listed in Table 3.
TABLE 2.
Strains and sequences used for primer and MB design
| Species and organism type | Strain | Accession no. | Region |
|---|---|---|---|
| Gram-positive bacteria | |||
| Staphylococcus aureus | MRSA252 | NC_002952 | 16S rRNA |
| Staphylococcus epidermidis | RP62A | NC_002976 | 16S rRNA |
| Streptococcus pneumoniae | TIGR4 | AAGY02000123 | 16S rRNA |
| Enterococcus faecium | DO ctg565 | AAAK03000115 | 16S rRNA |
| Enterococcus faecalis | V583 | NC_004668 | 16S rRNA |
| Gram-negative bacteria | |||
| Escherichia coli | O157:H7 EDL933 | NC_002655 | 16S rRNA |
| Enterobacter cloacae | CP1 | DQ089673 | 16S rRNA |
| Klebsiella pneumoniae | MGH 78578 | NC_009648 | 16S rRNA |
| Pseudomonas aeruginosa | PAO1 | NC_002516 | 16S rRNA |
| Acinetobacter baumannii | ADP1 | NC_005966 | 16S rRNA |
| Proteus mirabilis | HI4320 | AM942759 | 16S rRNA |
| Haemophilus influenzae | 86-028NP | NC_007146 | 16S rRNA |
| Candida spp. | |||
| Candida albicans | SC5314 | AACQ01000295 | 28S rRNA |
| Candida glabrata | CBS138 | AY198398 | 28S rRNA |
| Candida krusei | NRRL Y-5396 | EF550222 | 28S rRNA |
| Candida tropicalis | MYA-3404 | AAFN01000124 | 28S rRNA |
| Candida parapsilosis | 317 | Sanger Institutea | 28S rRNA |
| Candida guilliermondii | ATCC 6260 | AAFM01000051 | 28S rRNA |
| Candida lusitaniae | ATCC 42720 | AAFT01000066 | 28S rRNA |
| Candida dubliniensis | CD36 | Sanger Instituteb | 28S rRNA |
| Aspergillus spp. | |||
| Aspergillus fumigatus | Af293 | AAHF01000017 | 28S rRNA |
| Aspergillus flavus | NRRL3357 | AAIH01002203 | 28S rRNA |
| Aspergillus terreus | NIH 2624 | Broad Institutec | 28S rRNA |
| Aspergillus niger | CBS 513.88 | NT_166520 | 28S rRNA |
| Other fungal species | |||
| Cryptococcus neoformans | B-3501A | AAEY01000011 | 28S rRNA |
| Saccharomyces cerevisiae | RM11-1a | AAEG01000105 | 28S rRNA |
| Neosartorya fischeri | NRRL 181 | AAKE03000097 | 28S rRNA |
Genome sequence data used in this study were produced by the Pathogen Genomics group at the Wellcome Trust Sanger Institute and are available from http://www.sanger.ac.uk/sequencing/Candida/parapsilosis/.
Genome sequence data used in this study were produced by the Pathogen Genomics group at the Wellcome Trust Sanger Institute and are available from http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/.
Sequence data were produced by the Aspergillus terreus sequencing project at the Broad Institute of Harvard and MIT (http://www.broad.mit.edu).
TABLE 3.
Primers and MBs for each assay
| Oligonucleotide or primer and organism type | Type | Sequence (5′-3′)h | Position |
|---|---|---|---|
| Gram-positive bacteria | |||
| PLK1 | Forward primer | TACGGGAGGCAGCAGT | 351-366a |
| PLK2N v1.4 | Reverse primer | AATTCTAATACGACTCACTATAGGGGCTGCTGGCACGTAGTTAGCCGTGGCTTTC | 533-504a |
| 16S GrP MB | MB | CGAGCTAGCAACGCCGCGTGAGTGAAGCTCG | 401-419a |
| 16S GP v1.21 | MB | CGAGCTAGCAACGCCGCGTGGGTGAAGCTCG | 401-419a |
| 16S GP v1.22 | MB | CGAGCTAGCAACGCCGCGTGAGGGAAGCTCG | 401-419a |
| 16S GP v1.23 | MB | CGAGCTAGCGACGCCGCGTGGGGGAAGCTCG | 401-419a |
| Gram-negative bacteria | |||
| Grm neg 16S P2 | Forward primer | CCTGATGCAGCCATGCCGCGTG | 385-406b |
| Grm neg 16S P1 | Reverse primer | AATTCTAATACGACTCACTATAGGGCACGGAGTTAGCCGGTGCTT | 517-498b |
| Grm neg 16S MB | MB | CGAGCTTGAAGAAGGCCTTCGGGTTGTAAAGAGCTCG | 409-433b |
| Grm neg16S-Acin | MB | CGAGCTTGAAGAAGGCCTTATGGTTGTAAAGAGCTCG | 397-421c |
| Grm neg16S-Pseu | MB | CGAGCTAGCACTTTAAGTTGGGAGGAAGCTCG | 427-446d |
| Bacteria | |||
| GP/GN P1g | Reverse primer | AATTCTAATACGACTCACTATAGGGGTATTACCGCGGCTGCTGGCAC | 544-523a |
| Fungi | |||
| 28SPF P2 | Forward primer | CGGCTCTTCCTATCATACCG | 2850-2869e |
| 28SPF P1 | Reverse primer | AATTCTAATACGACTCACTATAGGGCTAAACCCAGCTCACGTTCC | 2931-2912e |
| 28SPF MB | MB | CGCGATATTCGGTAAGCGTTGGATTGATCGCG | 2877-2896e |
| Candida species | |||
| 28SPC P2 | Forward primer | GGAATCCGCTAAGGAGTGTG | 1246-1265e |
| 28SPC P1 | Reverse primer | AATTCTAATACGACTCACTATAGGGCCATCCATTTTCAGGGCTAGT | 1309-1289e |
| 28SPC MB | MB | CGCGATTAACAACTCACCGGCCGAATATCGCG | 1266-1285e |
| Aspergillus species | |||
| 28SPAS | Forward primer | CAGCAGTTGGACATGGGTTA | 1503-1522f |
| 28SPAASN | Reverse primer | AATTCTAATACGACTCACTATAGGGGAGAATCCACATCCAGGTGC | 1625-1606f |
| 28SPAMB | MB | CGACCGGCATAGGGAAGTTCCGTTTGGTCG | 1534-1553f |
The position is based on S. aureus MRSA252 NC_002952.
The position is based on E. coli O157:H7 EDL933 NC_002655.
The position is based on A. baumannii ADP1 NC_005966.
The position is based on P. aeruginosa PAO1 NC_002516.
The position is based on C. albicans SC5314 AACQ01000295.
The position is based on A. fumigatus Af293 AAHF01000017.
The primer was used for bacterial multiplex detection.
T7 promoter sequences are labeled in italics, and beacon arm sequences are underlined.
Extraction of TNA and RNA.
A 200-μl aliquot of test culture was mixed with 800 μl NucliSENS lysis buffer (bioMérieux, Durham, NC) in lysing matrix tubes (MP, Biomedicals, Inc., Solon, OH) and vigorously vortexed on a FastPrep instrument (MP, Biomedicals, Inc., Solon, OH) for 45 s, followed by a 30-min incubation at 37°C. Cell lysates were centrifuged at 16,000 × g for 10 min, and then 800 μl of supernatant was transferred and mixed with 2 ml NucliSENS lysis buffer in the NucliSENS easyMAG (bioMérieux) sample cartridge for the total nucleic acid (TNA) extraction. Magnetic silica solution was added, and the TNA extraction was performed according to the easyMAG user manual. The final elution volume of the TNA for each dilution was 110 μl. For Aspergillus spp., conidia were collected from an Aspergillus fumigatus culture grown on a PDA plate at 37°C for 3 days, using sterile saline with 0.01% Tween 20. The conidial suspension was counted under a microscope using a disposable cell counting chamber, and 200 μl of 107 CFU/ml conidial suspension was extracted for total nucleic acids. Tenfold serial dilutions were made from the extracts. In some experiments, pure RNA was isolated using the RNeasy kit according to the procedure of the manufacturer (Qiagen, Valencia, CA). The RNA concentration was determined by spectrophotometry using a Nanodrop DN1000 device (Nanodrop Techonologies, DE), and the 260-nm wavelength/280-nm wavelength and 260-nm wavelength/230-nm wavelength ratios were calculated.
Real-time NASBA.
The NASBA reactions were performed with the NucliSENS basic kit version 2 (bioMérieux). Reactions were carried out in a total volume of 20 μl containing reagent mix, 0.2 μM each primer, 0.1 μM MB, 5 μl of RNA or TNA template, and 5 μl of NASBA enzyme mix (nucleic acid-free enzyme; provided by bioMérieux, France). The enzyme mixture contained T7 RNA polymerase, avian myeloblastosis virus reverse transcriptase, RNase H, and bovine serum albumin, and the mixture was added to the reaction mixture after a two-step incubation of 2 min at 65°C and 2 min at 41°C. For simplex NASBA assays, amplification and real-time detection were performed on the NucliSENS EasyQ analyzer (bioMérieux) at 41°C for 90 min. The fluorescence signal was measured with an interval time of 30 s for each independent reaction at two wavelengths using the accompanying NucliSENS EasyQ Director software (version 2). For multiplex NASBA assays, reactions were carried out on a Stratagene Mx3005P real-time PCR system (Stratagene, La Jolla, CA) with the setting of 180 cycles at 41°C with 30 s for each cycle. The fluorescence signal was measured once for each cycle. The cutoff threshold for the positive signal was set as 10% higher than the end point signal from the no-template control, which used 5 μl of nuclease-free water instead of nucleic acids in the reaction mixture. The time to positive was defined as the time point when the positive signal crossed the threshold. Two multiplex real-time NASBA assays (pan-gram positive/pan-gram negative and pan-fungal/pan-Candida/pan-Aspergillus) were developed for bacterial and fungal detection, respectively.
Preliminary in vitro specificity and sensitivity. (i) Quantifying target template.
An artificial rRNA fragment for each pathogen was synthesized by in vitro transcription from a PCR product containing the corresponding genomic DNA using a specific primer set (Table 4), of which the forward primer was linked with a T7 promoter sequence. The specific PCR products either were purified by paramagnetic beads (Agencourt AMPure, Beverly, MA) or extracted from agarose gels (Montáge; Millipore, Billerica, MA). After purification, the PCR product was transcribed in vitro for 1 h at 37°C using an RNAMaxx High Yield Transcription kit (Stratagene) and then treated by RNase-free DNase I (Ambion, Austin, TX) for 30 min at 37°C. RNA products were purified using the RNeasy kit according to the RNA cleanup protocol (Qiagen). Purified RNA targets were quantified by a Quant-iT RiboGreen RNA quantification kit (Molecular Probes, Invitrogen, Carlsbad, CA). For TNA quantification, overnight cultures were subcultured in the same broth and grown for 3 to 4 h until log phase and then were harvested by centrifugation at 4,000 rpm for 5 min at 4°C. The cells were washed twice with sterile phosphate-buffered saline (PBS). The numbers of structurally intact cells were determined microscopically using a disposable cell counting chamber. Cultures were serially diluted from 107 to 100 CFU/ml accordingly. TNA was extracted from each dilution as described above. The accurate CFU was enumerated by plating 100 μl of each dilution onto LB agar for bacteria (MSA for S. aureus) or YPD agar for Candida and incubating it at 37°C for 24 h.
TABLE 4.
Primers used for in vitro transcription
| Primer | Sequence (5′-3′) | Position |
|---|---|---|
| SA16S F | AATTCTAATACGACTCACTATAGGGTAACGGCTTACCAAGGCAAC | 269-288 |
| SA/Enterc16S R | ACCTTCCGATACGGCTACCT | 1533-1514a |
| Enterc16S F | AATTCTAATACGACTCACTATAGGGCACATTGGGACTGAGACACG | 303-322 |
| SE16S F | AATTCTAATACGACTCACTATAGGGCAAGTCGAGCGAACAGACGA | 59-78 |
| SE16S R | TGTCACCGGCAGTCAACTTA | 1181-1162 |
| Ecoli/Kleb16S F | AATTCTAATACGACTCACTATAGGGGATCATGGCTCAGATTGAACG | 15-35b |
| Ecoli/Kleb16S R | GCTGGCAACAAAGGATAAGG | 1132-1113b |
| 28SPF F | AATTCTAATACGACTCACTATAGGGGAGGCTAGAGGTGCCAGAAA | 2756-2775c |
| 28SPF R | TCTGACTTAGAGGCGTTCAGC | 3101-3081c |
| Calb28S F | AATTCTAATACGACTCACTATAGGGCGATACTGCCAGCCTAGACC | 566-585 |
| Calb28S R | CGCTCTTCCAGCCATAAGAC | 1751-1732 |
| Afum28S-F | AATTCTAATACGACTCACTATAGGGGTGAAGCTCCTTCGACGAGT | 250-269 |
| Afum28S-R | TCTCCGCGTTCAGTTACGTT | 1651-1632 |
The position is based on the S. aureus 16S sequence from NC_002952.
The position is based on the E. coli 16S sequence from NC_002655.
The position is based on the C. albicans 28S sequence from AACQ01000295.
(ii) Sensitivity and specificity testing.
The sensitivities of the real-time NASBA assays were assessed by amplifying both in vitro RNA transcripts and TNAs isolated from serial dilutions of the target organism cultures. As described above, the in vitro RNA transcripts were synthesized, purified, and quantified, and the number of RNA copies was calculated according to the concentration and the size of the RNA transcript. Tenfold serial dilutions were prepared covering a range of 8-log (107 to 100) copies of RNA molecules per NASBA reaction. These dilutions of RNA were added directly into the NASBA mix. For the TNA isolated from the bacterial or fungal culture serial dilutions, the NASBA assay was performed on each of these extracts. To mimic the clinical environment, blood culture bottles containing 8 ml of whole blood were spiked with 10-fold serially diluted bacterial cultures of Enterobacter cloacae and yeast cultures of Candida glabrata and were subjected to TNA extraction. A 200-μl aliquot of the spiked blood bottle culture was used for TNA extraction at a concentration ranging from 4 × 106 to 4 × 100 CFU/ml. Blood culture bottles lacking the spiking pathogen were subjected to extraction in parallel as the negative control. All of the real-time NASBA assays described above were repeated at least three times. Preliminary probe specificity was evaluated by testing the nucleic acids from the target organisms, as well as the nucleic acids from other related microbes. For instance, the pan-gram-positive probe was tested against nucleic acids from four gram-positive bacterial species as well as a panel of five gram-negative species, eight Candida spp., C. neoformans, S. cerevisiae, and four Aspergillus spp., as listed in Table 1. The no-template control also was tested in parallel, and at least eight TNA samples from different clinical isolates of the same species were tested for each probe.
Clinical evaluation: sample collection, preparation, and NASBA detection.
To assess the clinical performance of the NASBA assays, aerobic and anaerobic blood culture bottles submitted for routine clinical testing were processed according to the manufacturer's instructions in either the BacT/Alert 3D (bioMérieux) or the BacTec 9240 (Becton Dickinson, Sparks, MD) automated blood culture system. Five-milliliter aliquots from 520 randomly selected positive blood culture bottles (BacTec Aerobic Plus, n = 250; BacTec Anaerobic Plus, n = 232; BacTec Anaerobic Lytic, n = 6; BacTec Peds Plus, n = 12; and BacT/Alert Aerobic, n = 20) were collected at the time blood bottles were flagged as positive by the automated blood culture systems and processed for routine microbiologic testing. The identification of the organism(s) present in the positive blood culture bottles was not known at the time of selection. They were selected in this manner so as not to bias results and to mimic the pattern of clinical isolates routinely encountered in our laboratory. The blood culture aliquots were deidentified by the North Shore-Long Island Jewish Health System Laboratories and stored at 4°C until shipment to PHRI on a weekly basis. Gram stain and culture on solid media were performed according to standard laboratory procedures for microbe identification and the determination of sensitivity to antibiotics. Microbe identification and the methicillin susceptibility profiles for S. aureus isolates were not provided to PHRI until the completion of the study. In addition, 5-ml aliquots from 50 negative blood culture bottles, collected on day 5 at the completion of routine culture, also were deidentified. The blood aliquots were stored at 4°C and transported within 7 days to PHRI for real-time NASBA identification. Briefly, 200 μl of the blood culture was mixed with 800 μl of a mixture of lysis buffer and proteinase K (25 μg/ml). FastPrep for 45 s, as described above, was followed by a 1-h incubation at 55°C and then a hard spin of the lysates at 16,000 × g. The supernatants were processed as described for TNA extraction from laboratory cultures. The final volume of each TNA sample eluate was 110 μl, 5 μl of which was used for each real-time NASBA assay. Blood bottle samples were tested by multiplex real-time NASBA assays. Studies were performed under an Institutional Review Board-approved protocol.
Statistical evaluation.
After breaking the code of the blinded blood bottle samples, the accuracy of the real-time multiplex NASBA assay was assessed by calculating the sensitivity, specificity, Youden's index (6), and predictive values for each probe.
RESULTS
Specificity of the broad-range real-time NASBA platform.
The specificity of the assay was defined as the ability to exclusively detect the target of interest. Like PCR-based methods, broad-range NASBA assays also are more vulnerable to trace amounts of nucleic acid contamination than the species-specific assays, which can lead to false-positive results. To overcome this potential problem, a nucleic acid-free enzyme mix (R&D Group, bioMérieux, France) was used for amplification in the broad-range bacterial and fungal pathogen detection system. All probes showed 100% specificity in initial tests using reference strains. Specific signal was observed for all target species but not for any other nontarget organism. Pan-gram-positive and pan-gram-negative probes were able to detect four (S. aureus, S. epidermidis, E. faecium, and E. faecalis) and five (E. coli, P. aeruginosa, A. baumannii, E. cloacae, and K. pneumoniae) medically important species, respectively, and no cross-reactivity was observed between each group or with fungi. Pan-fungal, pan-Candida, and pan-Aspergillus probes also demonstrated 100% specificity. Eight Candida species (C. albicans, C. glabrata, C. krusei, C. tropicalis, C. parapsilosis, C. guilliermondii, C. lusitaniae, and C. dubliniensis), four Aspergillus species (A. fumigatus, A. flavus, A. niger, and A. terreus), Cryptococcus neoformans, and Saccharomyces cerevisiae (Table 1) were detected easily by the pan-fungal probe without any cross-reactivity with the bacterial probes. Pan-Candida and pan-Aspergillus probes selectively recognized all of the Candida or Aspergillus species among all of the fungal species listed in Table 1. All probes were validated with at least eight clinical isolates.
Sensitivity of real-time NASBA assays: simplex assays.
C. albicans and A. fumigatus were selected as representative species for the pan-fungal, pan-Candida, and pan-Aspergillus sensitivity evaluations. As shown in Table 5, the analytical sensitivity of the assay was 104 copies of target RNA transcripts for pan-fungal and pan-Candida assay and 103 copies of target RNA for pan-Aspergillus detection. To assess the effect and the efficiency of the extraction process, the TNAs isolated from serially diluted fungal cultures were used to evaluate the limit of detection of the assay. All fungal probes were able to detect fewer than 2 CFU of target organisms. Similar sensitivities were observed for both pan-gram-positive and pan-gram-negative probes, with a detection limit ranging from 103 to 104 RNA copies and around 1 to 5 CFU of target organisms (Table 5). To investigate the applicability of our pan-NASBA assay, specificity and sensitivity tests were performed using TNAs extracted from blood culture bottles containing whole blood that had been spiked with serially diluted C. glabrata or E. cloacae cultures. Robust signals were detected at as low as 0.28 CFU of C. glabrata by the pan-fungal and the pan-Candida probes, and the pan-gram-negative probe could detect 3.1 CFU of E. cloacae in a single NASBA reaction.
TABLE 5.
Sensitivity of real-time NASBA
| Probe and species | Sensitivity in simplex assay
|
Sensitivity in multiplex assay
|
||
|---|---|---|---|---|
| Copy no. | CFU/reaction | Copy no. | CFU/reaction | |
| GrP16S | ||||
| E. faecium | 103∼104 | 0.2 | 105∼106 | 2 |
| E. faecalis | 103∼104 | 0.56 | 105 | 5.6 |
| S. epidermidis | 104 | 4.5 | 106 | 45 |
| S. aureus | 104 | 1.3 | 105∼106 | 13 |
| GrN16S | ||||
| E. coli | 104 | 5.5 | 106 | 55 |
| A. baumannii | 104 | 1.1 | 105∼106 | 11 |
| K. pneumoniae | 104 | 1.1 | 105∼106 | 11 |
| E. cloacae | 104 | 1.7 | 106 | 17∼170 |
| P. aeruginosa | 104 | 1.1 | 105∼106 | 11 |
| 28SPF | ||||
| C. albicans | 103∼104 | 1.8 | 104 | 1.8 |
| A. fumigatus | 103∼104 | 0.14 | 104 | 1.4 |
| 28SPC | ||||
| C. albicans | 104 | 1.8 | 105 | 1.8 |
| 28SPA | ||||
| A. fumigatus | 103 | 0.14 | 104 | 1.4 |
Multiplex assays.
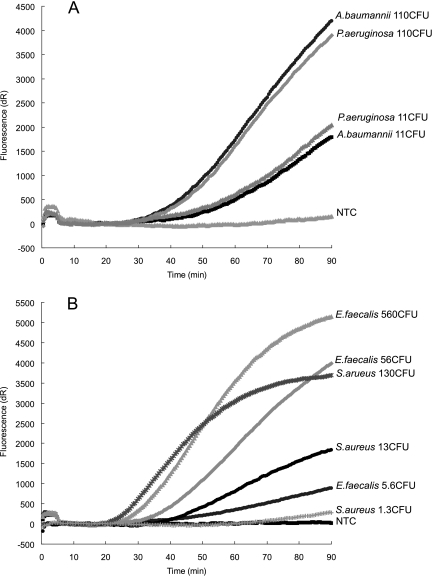
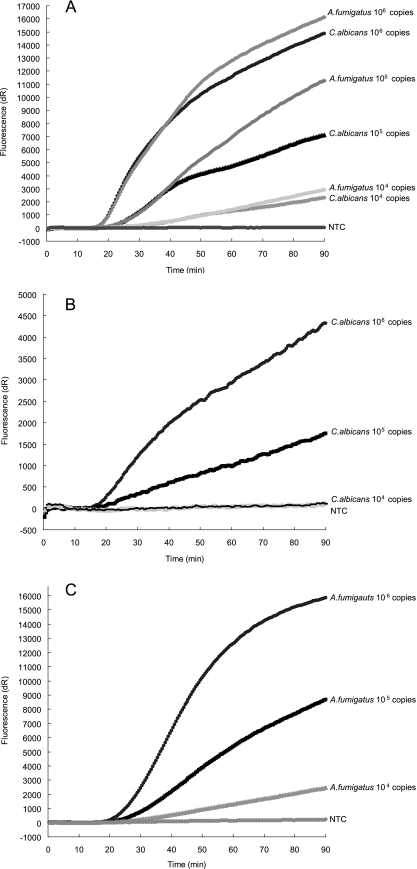
Compared to the ultrasensitive simplex NASBA reactions, multiplex NASBA assays were less sensitive (10- to 100-fold) for most of the probes. Interactions among different MBs and primer sets were expected to occur in these multiplex detections, which accounts for the diminished sensitivity. To improve the sensitivity, one common P1 (GP/GN P1) was designed and used for the bacterial panel. The common P1 demonstrated that approximately 105 to 106 copies of target RNA molecules or 5 to 50 CFU of organisms (Table 5, Fig. 1) could be detected by the pan-gram-positive/pan-gram-negative assays. Additional patch probes for gram-positive bacteria did not change the sensitivity of the multiplex assay. In contrast, less competition was seen in the fungal panel, of which the detection limit ranged from 104 to 105 copies of C. albicans or A. fumigatus RNA or around 1 CFU of target organism (Table 5, Fig. 2).
FIG. 1.
Sensitivity of the multiplex real-time NASBA assay for bacteria. Sensitivity of pan-gram negative (FAM) (A) and pan-gram positive (HEX) (B) probes in multiplex format using a 10-fold dilution series of TNA isolated from cells of A. baumannii and P. aeruginosa (A) and S. aureus and E. faecalis (B). Oligonucleotides 16S GrP MB and Grm neg 16S MB and the primer set of PLK1, primer Grm neg 16S P2, and primer GP/GN P1 were used in this multiplex assay.
FIG. 2.
Sensitivity of the multiplex real-time NASBA assay for fungi. A 10-fold dilution series of in vitro 28S rRNA transcripts for C. albicans and A. fumigatus were used for the evaluation. Shown is the sensitivity, in multiplex format, of pan-fungal probe (CR610) (A), pan-Candida probe (FAM) (B), and pan-Aspergillus probe (Q670) (C). The same primer-MB sets for pan-fungal, pan-Candida, and pan-Aspergillus simplex detection (Table 3) were used in combination for this multiplex assay.
Clinical evaluation.
A total of 520 randomly selected samples from positive blood bottles were identified to the subkingdom or genus level by multiplex NASBA, and the results were compared to those obtained by conventional culture systems. Based on the culture identification, 362 samples contained gram-positive bacteria, 149 samples contained gram-negative bacteria, and 10 samples contained Candida spp. A wide BSI pathogen spectrum was seen in these samples, consisting of eight gram-positive bacterial genera and 21 species (Corynebacterium spp. [including C. jeikeium], Enterococcus spp. [E. casseliflavus, E. faecalis, E. faecium, and E. gallinarum], Lactobacillus casei, Micrococcus sp., Propionibacterium acnes, Staphylococcus aureus [methicillin sensitive and resistant], CoNS [including S. lugdunensis], and Streptococcus spp. [S. agalactiae, S. bovis, S. milleri group, S. mitis group, S. pneumoniae, S. pyogenes, S. sanguis, and S. salivarius]), 17 gram-negative bacterial genera and 20 species (Acinetobacter baumannii, Alcaligenes sp., Bacteroides sp., Burkholderia cepacia, Citrobacter spp. [C. freundii and C. koseri], Enterobacter spp. [E. aerogenes and E. cloacae], Escherichia coli, Fusobacterium nucleatum, Haemophilus influenzae, Klebsiella spp. [K. oxytoca and K. pneumoniae], Morganella morganii, Prevotella loescheii, Proteus mirablis, Providencia stuartii, Pseudomonas aeruginosa, Salmonella sp. group B, and Serratia marcescens), and 3 Candida species (C. albicans, C. parapsilosis, and C. tropicalis). High-level specificity was observed for all probes in the multiplex NASBA detections (Table 6). In a set of controlled experiments, 50 culture-negative blood bottle samples were evaluated and failed to elicit any detectable fluorescent signals. In addition, full sensitivity as described above was unaltered with blood bottle media used as the background matrix.
TABLE 6.
Comparison of results from real-time multiplex NASBA detection and blood culture system
| Probe type and NASBA IDc | No. with culture ID:
|
Sensitivity (%) | Specificity (%) | Youden's indexa | PPV (%) | NPV (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram+ | Non-gram+ | Gram− | Non-gram− | Fungal/Candida spp. | Non-fungi/non-Candida spp. | Aspergillus spp. | Non-Aspergillus spp. | Total | ||||||
| Pan-gram positive (with patch pan-gram-positive MBs)d | ||||||||||||||
| Gram+ | 348 (361) | 0 | 348 (361) | 100.00 (100.00) | ||||||||||
| Non-gram+ | 14 (1) | 156 | 170 (157) | 91.76 (99.36) | ||||||||||
| Total | 362 | 156 | 518 | 96.13 (99.72) | 100.00 (100.00) | 0.961 (0.997) | ||||||||
| Pan-gram negative | ||||||||||||||
| Gram− | 147 | 15 | 162 | 90.74 | ||||||||||
| Non-gram− | 2 | 354 | 356 | 99.44 | ||||||||||
| Total | 149 | 369 | 518 | 98.66 | 95.93 | 0.946 | ||||||||
| Pan-fungal/pan-Candida spp. | ||||||||||||||
| Fungal/Candida spp. | 10 | 0 | 10 | 100.00 | ||||||||||
| Non-fungal/non-Candida spp. | 0 | 260 | 260 | 100.00 | ||||||||||
| Total | 10 | 260 | 270 | 100.00 | 100.00 | 1 | ||||||||
| Pan-Aspergillus | ||||||||||||||
| Aspergillus spp. | 0 | 0 | 0 | NAb | ||||||||||
| Non-Aspergillus spp. | 0 | 270 | 270 | 100.00 | ||||||||||
| Total | 0 | 270 | 270 | NAb | 100.00 | NAb | ||||||||
Youden's index is calculated as sensitivity plus specificity minus one.
NA indicates that sensitivity, Youden's index, and PPV for the pan-Aspergillus spp. probe were not applicable because of no positive blood culture.
ID, identification.
Values obtained by using the mixture of the original pan-gram-positive probe and the patch probes are shown in parentheses when those values are different from those obtained by using the original pan-gram-positive probe only.
One blood culture, flagged as positive by the automated blood culture instrument, was culture negative, and the NASBA assays identified a gram-negative organism. A second blood culture collected from the same patient several hours later grew Pseudomonas aeruginosa. A blood culture from another patient had very poor growth that did not initially allow for the identification of the two pathogens present in the sample. The NASBA assays were positive for both a gram-positive and a gram-negative organism. Conventional culture took more than 1 week to finally confirm the presence of a gram-positive rod in the same bottle. A gram-negative rod was not recovered from this bottle, but another blood culture drawn on the same day contained E. coli.
It was observed that the uncommon organisms Corynebacterium spp., Propionibacterium sp., and Micrococcus sp. were falsely identified as gram-negative bacteria. To address this issue, three patch probes were designed for pan-gram-positive detection (Table 3). The use of a mixture of four pan-gram-positive MBs increased the specificity of pan-gram-positive detection to >99%. A total of 13 out of 14 missed gram-positive samples subsequently were picked up by the new pan-gram-positive detection, and no cross-reaction was seen from gram-negative bacteria and fungi. In addition, the pan-gram-negative probe failed to recognize the anaerobe Prevotella loescheii in the clinical blood culture, and no detectable signal was seen from other probes as well. In control experiments, no organisms were detected in the 50 negative blood cultures submitted for testing.
The overall performance of multiplex NASBA assays was measured by sensitivity, specificity, Youden's index, and predictive values for each primer/probe set (Table 6). High sensitivity (96.1 and 98.6%), specificity (100 and 95.9%), and Youden's indices for pan-gram-positive and pan-gram-negative detection demonstrate the great accuracy of this multiplex assay. The positive predictive values (PPV) for pan-gram-positive and pan-gram-negative probes were 100 and 90.7%, respectively, and the corresponding negative predictive values (NPV) for these two probes were 91.8 and 99.4% (Table 6). The relatively low NPV for pan-gram-positive probe and low PPV for pan-gram-negative probe were due largely to the false identifications for three gram-positive organisms, Corynebacterium spp., Propionibacterium acnes, and Micrococcus sp., which counted for 13/14 samples falsely identified as gram-negative pathogens. However, with the addition of the patch probes, the NPV for the pan-gram positive assay was increased to 99.4%. Meanwhile, the fungal panel was observed to be highly reliable. Both pan-fungal and pan-Candida probes demonstrated 100% sensitivity, specificity, Youden's index, PPV, and NPV, and the pan-Aspergillus probe also showed a 100% negative predictability (Table 6). It should be noted that the high PPV needs to be viewed in context with the limited number of positive fungal cultures obtained in this study. Robust signals were observed for all probes in the multiplex panels, with signal detection in less than 15 min.
DISCUSSION
Conventional diagnostic methods for BSIs require several days to complete, and both false-positive (15, 26, 34) and false-negative results (24, 40, 41) have been reported. Delayed diagnosis correlates strongly with the high mortality of BSIs (19, 35, 49). Thus, there is an urgent need for a rapid and reliable diagnostic method that will allow clinicians to make faster and better-informed treatment choices for BSI patients. In this study, we describe a rapid, highly specific, and sensitive diagnostic platform for BSI, using NASBA and MB technology to perform real-time assays. Five broad-range MBs and the corresponding primer sets were developed and evaluated in this study. The primer/probe sets were able to rapidly and accurately identify most medically important BSI pathogens to the genus level. Furthermore, the simultaneous detection and characterization of the BSI pathogen at the subkingdom or genus level could be done by the two multiplex real-time NASBA detection panels presented in this study. Reliable pathogen identification and characterization can be obtained in under 3 h. This approach holds promise for the rapid diagnosis of BSIs.
As a powerful diagnostic tool, real-time NASBA has been used widely for detecting RNA viruses such as enterovirus and human immunodeficiency virus (27, 31, 39), certain microbial pathogens including Legionella species, Vibrio cholerae, etc. (18, 30), and pathogens from food and environmental samples (11, 36). However, only a few studies have reported using conventional NASBA to detect Candida spp. and Aspergillus spp. (7, 29, 46), and no real-time NASBA methods had been reported for BSIs. Conversely, during the past decade, many PCR-based assays were developed and dominated the methods used for the identification of microorganisms and the diagnosis of infections. Compared to PCR, a major advantage of NASBA is the fast amplification kinetics. It is an isothermal process, and the single-stranded RNA amplicons produced can be used directly for the subsequent rounds of amplification or probe detection without the need for denaturation or strand separation. When using the multicopy rRNA as the target, NASBA results in high sensitivity and allows for a faster identification of the pathogens.
The universal PCR-based detection of bacteria or fungi has been attempted because of its powerful potential to rapidly detect and identify a broad range of pathogens, but the clinical value of this testing still is debatable (8). The most significant limitation to date is the potential for DNA or RNA contamination from a variety of uncertain sources, including enzymes, reagents, nucleic acid extraction kits, columns, water, the environment, and so on. Attempts to decontaminate PCR materials (9, 13) are not entirely effective or reproducible. Some real-time PCR assays were not able to detect pathogens presenting at low levels without reporting false positives caused by low-level endogenous contamination. In our preliminary experiments, the availability of nucleic acid-free enzyme preparations and high-quality nucleic acid-free reagents has helped to eliminate the contamination problem. In this context, five broad-range real-time NASBA assays were developed to detect a wide variety of BSI pathogens. Several medically important pathogens, including 9 bacterial species and 12 fungal species, were used to evaluate these real-time NASBA detections. All of these assays have great selectivity for their target organisms, and no cross-reaction was observed in tested species. The great diversity of bacterial species represented in the clinical study was extremely helpful in highlighting false-negative results for three gram-positive bacterial organisms, Corynebacterium sp., Propionibacterium sp., and Micrococcus sp., when tested using the original pan-gram-positive probe. With the exception of immune-compromised persons or persons with indwelling devices, these bacterial species generally do not cause invasive disease, can be found on human skin, and usually are present in blood cultures as a result of contamination during sample collection. However, since there are circumstances in which these organisms are clinically relevant, several new pan-gram-positive probes were designed and tested. The mixture of these new probes and the old pan-gram-positive probe then could easily detect all species. Similarly, the pan-gram-negative probe failed to identify Prevotella spp. from the blood cultures, although a patch probe was not designed against this genus.
The analytical sensitivity tests of the real-time NASBA assays were performed. All five probes were able to detect approximately 103 to 104 copies of artificial RNA targets in one NASBA reaction, as well as genomic RNA extracted from around 1 to 5 CFU of bacterial or fungal cultures, assuming 100% efficiency of the extraction. In comparison, the limit of broad-range bacterial detection was 10 to 100 CFU per reaction for E. coli and 100 to 1,000 CFU per reaction for S. aureus (20, 50) in regular PCR, and 1 to 10 CFU per reaction was observed in real-time PCR (20, 50). NASBA fungal assays showed limits of detection comparable to those of pan-fungal PCR assays (3, 43). The detection limits of pan-Candida and pan-Aspergillus assays are in agreement with the results reported by other studies that used traditional NASBA detection (electrochemiluminescence) for Candida spp. (28, 46) or Aspergillus spp. (29).
The clinical applicability of our real-time multiplex NASBA assays was evaluated by testing blinded blood cultures. To our knowledge, this is the first study to compare real-time NASBA assays with the capability to detect a wide range of potential pathogens to the classical cultural approach for the diagnosis of BSIs. Since this pilot study was conducted using samples derived from positive blood culture bottles, one might argue that there is little benefit to be obtained by using multiple molecular tests instead of a simple and inexpensive gram stain as the first diagnostic step. This is true for the majority of the samples tested in this pilot study. However, an example of a notable exception with a clear diagnostic benefit would be the identification of Candida krusei, since this yeast is intrinsically resistant to a first-line treatment drug, fluconazole. The ultimate goal in the development of such assays is to significantly reduce the time to initial pathogen detection, not to completely replace blood cultures. In the majority of cases, susceptibility studies still must be performed, and the detection of all pathogens must be guaranteed. With improved nucleic acid isolation and concentration methods, detection from directly drawn specimens may be possible. However, if this is shown not to be sufficiently sensitive, reducing the time to blood culture testing, i.e., sampling the bottle before it is flagged as positive, may be a viable alternative approach.
Although our testing was performed on positive blood cultures, in fact, our real-time NASBA assays demonstrated several advantages that could make it a valuable complement to traditional blood culture for the diagnosis of BSIs. First, the NASBA assays demonstrated a high predictability to rule out a BSI. The data also suggest that it can be used for accurate diagnosis when a pathogen is present. For example, one blood culture, reported as positive but from which no organism was grown, was positive by NASBA for a gram-negative organism and was confirmed by the isolation of P. aeruginosa from another blood culture bottle that became positive hours after the first blood bottle. The earlier detection of the gram-negative organism in the first bottle suggests that the increased sensitivity of the NASBA assay accurately identified a BSI in this patient much sooner than traditional culture methods. Second, the assays allow for a faster identification of the pathogen(s). This was demonstrated in particular by the blood culture that grew too poorly for timely identification using conventional test methods, yet the NASBA assays identified both a gram-positive and a gram-negative organism in the blood culture at the time the sample was flagged as positive. Third, in consideration of the high sensitivity of the assay, provided that pathogen nucleic acid extraction from the whole blood is sufficiently efficient, the assays could be used for detecting BSI pathogens from a direct blood draw without the need for a primary culture step. Studies to evaluate the potential of testing direct blood draws and blood culture bottles at various time points after inoculation but prior to instrument detection currently are in progress. Additionally, studies to include other blood culture bottle types and a more extensive testing of yeast and fungal isolates are indicated to comprehensively assess the performance of the assays. Finally, there is the possibility that amplified methods lead to an increase in false-positive results due to the detection of dead organisms or low-level contaminants. In such cases, as we currently must do with routine blood cultures, the information must be evaluated in the clinical context of the patient's symptoms, disease, and underlying comorbidities.
Wisplinghoff and colleagues reported that polymicrobial infection constituted 13% of all episodes of nosocomial BSIs in the United States (47), which is associated with higher mortality than monomicrobial BSI and tends to be more resistant to empirical therapy. The early diagnosis of polymicrobial infection is valuable to the patient's management, because the adjustment of empirical treatments often were required due to polymicrobial episodes (5). With the capability to simultaneously detect multiple targets, multiplex real-time NASBA could be not only more suitable to meet the need of rapidly recognizing polymicrobial infections but also more efficient and cost-effective than a simplex assay. In fact, the NASBA assays detected several gram-positive/gram-negative mixed infections, including CoNS with Corynebacterium spp., CoNS with E. coli and P. stuartii, E. faecalis with P. mirabilis, CoNS with E. cloacae and A. baumannii, and CoNS with Alcaligenes sp. These data suggest that the multiplex assays are highly suitable for the primary screening of BSI pathogens.
In conclusion, this study demonstrates the feasibility of using an NASBA-MB approach as a broad-range rapid diagnostic method suitable for detecting BSIs.
Acknowledgments
This work was supported by NIH grant AI066561 to D.S.P. and by in-kind support from bioMèrieux.
We sincerely thank the technical staff of the North Shore-LIJ Health System Microbiology and Molecular Diagnostics Laboratories.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Ammann, R. A., F. Zucol, C. Aebi, F. K. Niggli, T. Kuhne, and D. Nadal. 2007. Real-time broad-range PCR versus blood culture. A prospective pilot study in pediatric cancer patients with fever and neutropenia. Support. Care Cancer 15637-641. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, S. N., T. G. Emori, D. H. Culver, R. P. Gaynes, W. R. Jarvis, T. Horan, J. R. Edwards, J. Tolson, T. Henderson, and W. J. Martone. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. Am. J. Med. 9186S-89S. [DOI] [PubMed] [Google Scholar]
- 3.Basková, L., C. Landlinger, S. Preuner, and T. Lion. 2007. The Pan-AC assay: a single-reaction real-time PCR test for quantitative detection of a broad range of Aspergillus and Candida species. J. Med. Microbiol. 561167-1173. [DOI] [PubMed] [Google Scholar]
- 4.Beekmann, S. E., D. J. Diekema, K. C. Chapin, and G. V. Doern. 2003. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J. Clin. Microbiol. 413119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berild, D., A. Mohseni, L. M. Diep, M. Jensenius, and S. H. Ringertz. 2006. Adjustment of antibiotic treatment according to the results of blood cultures leads to decreased antibiotic use and costs. J. Antimicrob. Chemother. 57326-330. [DOI] [PubMed] [Google Scholar]
- 6.Biggerstaff, B. J. 2000. Comparing diagnostic tests: a simple graphic using likelihood ratios. Stat. Med. 19649-663. [DOI] [PubMed] [Google Scholar]
- 7.Borst, A., M. A. Leverstein-Van Hall, J. Verhoef, and A. C. Fluit. 2001. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39155-160. [DOI] [PubMed] [Google Scholar]
- 8.Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Bottger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37167-172. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, N. M., P. Adamson, and N. Okhravi. 1999. Elimination of bacterial DNA from Taq DNA polymerases by restriction endonuclease digestion. J. Clin. Microbiol. 373402-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2000. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992-April 2000. Am. J. Infect. Control 28429-448. [DOI] [PubMed] [Google Scholar]
- 11.Churruca, E., C. Girbau, I. Martinez, E. Mateo, R. Alonso, and A. Fernandez-Astorga. 2007. Detection of Campylobacter jejuni and Campylobacter coli in chicken meat samples by real-time nucleic acid sequence-based amplification with molecular beacons. Int. J. Food Microbiol. 11785-90. [DOI] [PubMed] [Google Scholar]
- 12.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 35091-92. [DOI] [PubMed] [Google Scholar]
- 13.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 381747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26781-803. [DOI] [PubMed] [Google Scholar]
- 15.Doern, G. V., A. Barton, and S. Rao. 1998. Controlled comparative evaluation of BacT/Alert FAN and ESP 80A aerobic media as means for detecting bacteremia and fungemia. J. Clin. Microbiol. 362686-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Mahallawy, H. A., H. H. Shaker, H. Ali Helmy, T. Mostafa, and A. Razak Abo-Sedah. 2006. Evaluation of pan-fungal PCR assay and Aspergillus antigen detection in the diagnosis of invasive fungal infections in high risk paediatric cancer patients. Med. Mycol. 44733-739. [DOI] [PubMed] [Google Scholar]
- 17.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fykse, E. M., G. Skogan, W. Davies, J. S. Olsen, and J. M. Blatny. 2007. Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 731457-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbarth, S., J. Garbino, J. Pugin, J. A. Romand, D. Lew, and D. Pittet. 2003. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am. J. Med. 115529-535. [DOI] [PubMed] [Google Scholar]
- 20.Harris, K. A., and J. C. Hartley. 2003. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J. Med. Microbiol. 52685-691. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim, E. H., G. Sherman, S. Ward, V. J. Fraser, and M. H. Kollef. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118146-155. [DOI] [PubMed] [Google Scholar]
- 22.Iwen, P. C., A. G. Freifeld, T. A. Bruening, and S. H. Hinrichs. 2004. Use of a panfungal PCR assay for detection of fungal pathogens in a commercial blood culture system. J. Clin. Microbiol. 422292-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan, J. A., and M. B. Durso. 2005. Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. J. Mol. Diagn. 7575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kami, M., N. Murashige, T. Fujihara, N. Sakagami, and Y. Tanaka. 2005. The mechanism for low yield of blood culture in invasive aspergillosis; the clinical importance of antigen detection tests revisited. Bone Marrow Transplant. 3685-86. [DOI] [PubMed] [Google Scholar]
- 25.Keer, J. T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53175-183. [DOI] [PubMed] [Google Scholar]
- 26.Kirkley, B. A., K. A. Easley, and J. A. Washington. 1994. Controlled clinical evaluation of Isolator and ESP aerobic blood culture systems for detection of bloodstream infections. J. Clin. Microbiol. 321547-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landry, M. L., R. Garner, and D. Ferguson. 2005. Real-time nucleic acid sequence-based amplification using molecular beacons for detection of enterovirus RNA in clinical specimens. J. Clin. Microbiol. 433136-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeffler, J., C. Dorn, H. Hebart, P. Cox, S. Magga, and H. Einsele. 2003. Development and evaluation of the nuclisens basic kit NASBA for the detection of RNA from Candida species frequently resistant to antifungal drugs. Diagn. Microbiol. Infect. Dis. 45217-220. [DOI] [PubMed] [Google Scholar]
- 29.Loeffler, J., H. Hebart, P. Cox, N. Flues, U. Schumacher, and H. Einsele. 2001. Nucleic acid sequence-based amplification of Aspergillus RNA in blood samples. J. Clin. Microbiol. 391626-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loens, K., T. Beck, H. Goossens, D. Ursi, M. Overdijk, P. Sillekens, and M. Ieven. 2006. Development of conventional and real-time NASBA for the detection of Legionella species in respiratory specimens. J. Microbiol. Methods 67408-415. [DOI] [PubMed] [Google Scholar]
- 31.McClernon, D. R., C. Vavro, and M. St Clair. 2006. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. J. Clin. Microbiol. 442280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metwally, L., D. J. Fairley, P. V. Coyle, R. J. Hay, S. Hedderwick, B. McCloskey, H. J. O'Neill, C. H. Webb, W. Elbaz, and R. McMullan. 2008. Improving molecular detection of Candida DNA in whole blood: comparison of seven fungal DNA extraction protocols using real-time PCR. J. Med. Microbiol. 57296-303. [DOI] [PubMed] [Google Scholar]
- 33.Minino, A. M., M. P. Heron, S. L. Murphy, and K. D. Kochanek. 2007. Deaths: final data for 2004. Natl. Vital Stat. Rep. 551-119. [PubMed] [Google Scholar]
- 34.Mirrett, S., K. E. Hanson, and L. B. Reller. 2007. Controlled clinical comparison of VersaTREK and BacT/ALERT blood culture systems. J. Clin. Microbiol. 45299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrell, M., V. J. Fraser, and M. H. Kollef. 2005. Delaying the empiric treatment of candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 493640-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadal, A., A. Coll, N. Cook, and M. Pla. 2007. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: role of target mRNA secondary structure on NASBA design. J. Microbiol. Methods 68623-632. [DOI] [PubMed] [Google Scholar]
- 37.Peters, R. P., M. A. van Agtmael, S. A. Danner, P. H. Savelkoul, and C. M. Vandenbroucke-Grauls. 2004. New developments in the diagnosis of bloodstream infections. Lancet Infect. Dis. 4751-760. [DOI] [PubMed] [Google Scholar]
- 38.Pittet, D., N. Li, R. F. Woolson, and R. P. Wenzel. 1997. Microbiological factors influencing the outcome of nosocomial bloodstream infections: a 6-year validated, population-based model. Clin. Infect. Dis. 241068-1078. [DOI] [PubMed] [Google Scholar]
- 39.Rutjes, S. A., R. Italiaander, H. H. van den Berg, W. J. Lodder, and A. M. de Roda Husman. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 713734-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar, S., I. Bhagat, T. E. Wiswell, and A. R. Spitzer. 2007. Role of multiple site blood cultures to document the clearance of bacteremia in neonates. J. Perinatol. 27101-102. [DOI] [PubMed] [Google Scholar]
- 41.Sautter, R. L., A. R. Bills, D. L. Lang, G. Ruschell, B. J. Heiter, and P. P. Bourbeau. 2006. Effects of delayed-entry conditions on the recovery and detection of microorganisms from BacT/ALERT and BACTEC blood culture bottles. J. Clin. Microbiol. 441245-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schabereiter-Gurtner, C., M. Nehr, P. Apfalter, A. Makristathis, M. L. Rotter, and A. M. Hirschl. 2008. Evaluation of a protocol for molecular broad-range diagnosis of culture-negative bacterial infections in clinical routine diagnosis. J. Appl. Microbiol. 1041228-1237. [DOI] [PubMed] [Google Scholar]
- 43.Schabereiter-Gurtner, C., B. Selitsch, M. L. Rotter, A. M. Hirschl, and B. Willinger. 2007. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 45906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpkins, S. A., A. B. Chan, J. Hays, B. Popping, and N. Cook. 2000. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett. Appl. Microbiol. 3075-79. [DOI] [PubMed] [Google Scholar]
- 45.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14303-308. [DOI] [PubMed] [Google Scholar]
- 46.Widjojoatmodjo, M. N., A. Borst, R. A. Schukkink, A. T. Box, N. M. Tacken, B. Van Gemen, J. Verhoef, B. Top, and A. C. Fluit. 1999. Nucleic acid sequence-based amplification (NASBA) detection of medically important Candida species. J. Microbiol. Methods 3881-90. [DOI] [PubMed] [Google Scholar]
- 47.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39309-317. [DOI] [PubMed] [Google Scholar]
- 48.Yang, S., S. Lin, G. D. Kelen, T. C. Quinn, J. D. Dick, C. A. Gaydos, and R. E. Rothman. 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J. Clin. Microbiol. 403449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, D. T., E. Black, K. E. Sands, J. S. Schwartz, P. L. Hibberd, P. S. Graman, P. N. Lanken, K. L. Kahn, D. R. Snydman, J. Parsonnet, R. Moore, R. Platt, and D. W. Bates. 2003. Severe sepsis: variation in resource and therapeutic modality use among academic centers. Crit. Care 7R24-R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zucol, F., R. A. Ammann, C. Berger, C. Aebi, M. Altwegg, F. K. Niggli, and D. Nadal. 2006. Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J. Clin. Microbiol. 442750-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]