Abstract
CHROMagar Acinetobacter was used to screen stool and perineal swabs for enteric carriage of multidrug-resistant Acinetobacter baumannii in samples from critically ill patients. Results were compared with a molecular assay resulting in sensitivity and specificity of culture compared to PCR of 91.7% and 89.6%, respectively.
Acinetobacter baumannii has emerged as an important nosocomial pathogen, impacting considerably on patient care (6, 9). In many parts of the world, strains resistant to almost all available antimicrobial classes (multidrug-resistant Acinetobacter baumannii [MDRAB]) have been reported and implicated in numerous hospital outbreaks, usually in intensive care units (2). Despite the implementation of infection control measures involving isolation of patients, rigorous hand hygiene, and aggressive environmental decontamination, many centers have continued to have ongoing problems, with rapid reemergence of the organism unless high standards of cleaning are maintained (4, 10).
Although environmental cleaning is of paramount importance, it is likely that patients themselves also represent a significant reservoir of the organism. For many gram-negative bacteria, colonization of the digestive tract is an important prerequisite for the development of nosocomial infections such as ventilator-associated pneumonia (3, 7). Rapid identification of individuals with enteric carriage of MDRAB could therefore be an important component in strategies aimed at limiting the transmission and dissemination of the organism.
CHROMagar Acinetobacter (CHROMagar, Paris, France) is a recently developed selective agar for the rapid identification of MDRAB. It contains agents which inhibit the growth of most gram-positive organisms as well as carbapenem-susceptible gram-negative bacilli. It incorporates substrates enabling color-based preliminary identification of colonies recovered within 18 to 24 h of inoculation. In this study, we evaluated the ability of CHROMagar Acinetobacter to detect the carriage of MDRAB in enteric samples from critically ill patients in a unit experiencing problems with MDRAB, compared with a molecular method.
(This study was presented in part at the 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, May 2009.)
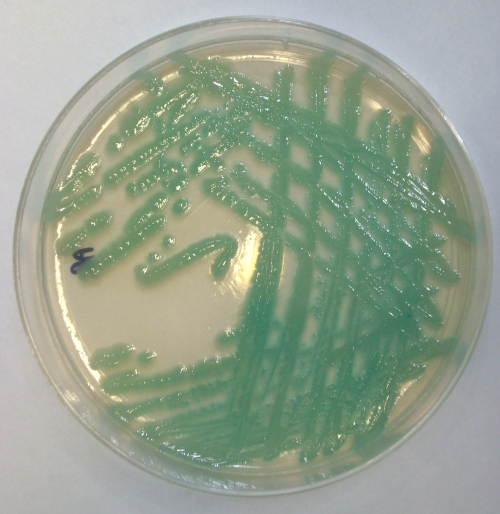
CHROMagar Acinetobacter was prepared from dehydrated powder according to the manufacturer's instructions. Each batch of media was quality controlled using a representative isolate of the MDRAB OXA-23 clone 1, obtained from the Health Protection Agency, Colindale, United Kingdom, previously characterized as resistant to carbapenems (2). Stool samples and/or perineal swabs collected from intensive care unit patients as part of routine screening for Clostridium difficile-associated diarrhea or methicillin-resistant Staphylococcus aureus carriage were used to inoculate CHROMagar Acinetobacter plates and then 3-ml peptone water broths. Plates were incubated in air at 37°C and examined after 24 h for aqua blue colonies indicative of MDRAB (Fig. 1). The peptone broths were also incubated for 24 h and used to generate templates for an MDRAB-specific molecular assay. One hundred microliters of turbid broth was heated at 96°C for 5 min and then centrifuged at 13,000 rpm to pellet lysed bacterial cells. Two microliters of supernatant was used in a multiplex PCR for the A. baumannii species-specific OXA-51-like gene and the OXA-23, -24, and -58-like genes, which mediate carbapenem resistance, using the primers and conditions described by Woodford et al. (11). Isolates presumed to be A. baumannii based on their growth and appearance on CHROMagar Acinetobacter were confirmed as such by Gram staining, oxidase testing, and multiplex PCR assay. The clonal lineage of each isolate was also determined using another multiplex PCR assay, based on sequence variations in the csuE, ompA, and blaOXA-51 alleles. This enables strains to be assigned to clonal complexes with a reliability equivalent to that obtained by pulsed-field gel electrophoresis (8).
FIG. 1.
Appearance of MDRAB on a CHROMagar Acinetobacter plate.
Sixty-six stool samples and 37 perineal swabs obtained from a total of 70 patients were ultimately included in the analysis. Thirty-three of these were positive by both culture and PCR on peptone broths (Table 1). Seven were culture positive but PCR negative, while three were found to be culture negative yet positive by PCR. The gene profiles detected in the PCR assay were identical in every case: OXA-51+, OXA-23+, OXA-24−, and OXA-58−.
TABLE 1.
Comparison of culture and molecular tests for the identification of MDRAB
| Culture result | PCR result | No. of specimens examined | No. of specimens known to be colonized |
|---|---|---|---|
| Negative | Negative | 60 | 20 |
| Positive | Positive | 33 | 28 |
| Positive | Negative | 7 | 6 |
| Negative | Positive | 3 | 1 |
| Total | 103 | 55 |
Biochemical and molecular testing of colonies identified as MDRAB by CHROMagar Acinetobacter confirmed that all of the organisms were A. baumannii strains resistant to β-lactams (including carbapenems), quinolones, and aminoglycosides. Multiplex PCR typing placed them as members of European clone II. The only other organisms recovered from samples inoculated onto CHROMagar Acinetobacter were Enterococcus spp., which were easily distinguishable by their dark blue color and small colonial morphology.
Of the 40 patients with positive MDRAB cultures, 33 were known to be colonized at other sites. In seven patients, MDRAB was isolated from stool alone and not from any other clinical specimens. The sensitivity and specificity of culture compared with the PCR assay were calculated as 91.7% and 89.6%, respectively.
For the seven samples found to be culture positive by CHROMagar Acinetobacter, but negative in PCRs performed on peptone broths, follow-up testing of the colonies confirmed that these were all MDRAB. In six of these, the patients had already had MDRAB isolated from other clinical specimens. As the broths were inoculated after the CHROMagar plates, it is possible that either insufficient material remained on the swabs, or in the case of the stool samples, there may have been significant inhibition of the PCR.
Our findings support the use of CHROMagar Acinetobacter as a means of identifying enteric carriage of MDRAB, but are limited by the relatively small numbers and the involvement of a single MDRAB clone. Also, as the stool samples were those sent for C. difficile testing, only semiformed or liquid stools were obtained. The use of the media with formed stools would therefore require further evaluation. A comparison between stool and perineal swab cultures would also help to determine whether perineal swabs can be used as a proxy marker of gut carriage. These are easier to process and are already routinely obtained to screen for other nosocomial pathogens in many clinical settings.
The study also highlights a role for the gut as a significant reservoir for MRAB in addition to environmental contamination. It raises the question of whether selective decontamination of the digestive tract using nonabsorbable antibiotics such as polymyxin B, to which MDRAB strains remain susceptible, could be a suitable method of control. The approach involving selective decontamination of the digestive tract has been shown to be effective in reducing the carriage and incidence of nosocomial infections caused by gram-negative bacteria but has not been widely adopted (1, 5). The availability of a selective culture media such as CHROMagar Acinetobacter, enabling those who may benefit from such a regimen to be rapidly identified, is clearly an important step toward addressing this issue.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Agustí, C., M. Pujol, M. J. Argerich, J. Ayats, M. Badía, M. A. Domínguez, X. Corbella, and J. Ariza. 2002. Short-term effect of the application of selective decontamination of the digestive tract on different body site reservoir ICU patients colonized by multi-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 49205-208. [DOI] [PubMed] [Google Scholar]
- 2.Coelho, J. M., J. F. Turton, M. E. Kaufmann, J. Glover, N. Woodford, M. Warner, M.-F. Palepou, R. Pike, T. L. Pitt, B. C. Patel, and D. M. Livermore. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 443623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbella, X., M. Pujol, J. Ayats, M. Sendra, C. Ardanuy, M. A. Domínguez, J. Liñares, J. Ariza, and F. Gudiol. 1996. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin. Infect. Dis. 23329-334. [DOI] [PubMed] [Google Scholar]
- 4.Denton, M., M. H. Wilcox, P. Parnell, D. Green, V. Keer, P. M. Hawkey, I. Evans, and P. Murphy. 2004. Role of environmental cleaning in controlling an outbreak of Acinetobacter baumannii on a neurosurgical intensive care unit. J. Hosp. Infect. 56106-110. [DOI] [PubMed] [Google Scholar]
- 5.De Smet, A. M., J. A. Kluytmans, B. S. Cooper, E. M. Mascini, R. F. Benus, T. S. van der Werf, J. G. van der Hoeven, P. Pickkers, D. Bogaers-Hofman, N. J. van der Meer, A. T. Bernards, E. J. Kuijper, J. C. Joore, M. A. Leverstein-van Hall, A. J. Bindels, A. R. Jansz, R. M. Wesselink, B. M. de Jongh, P. J. Dennesen, G. J. van Asselt, L. F. te Velde, I. H. Frenay, K. Kaasjager, F. H. Bosch, M. van Iterson, S. F. Thijsen, G. H. Kluge, W. Pauw, J. W. de Vries, J. A. Kaan, J. P. Arends, L. P. Aarts, P. D. Sturm, H. I. Harinck, A. Voss, E. V. Uijtendaal, H. E. Blok, E. S. Thieme Groen, M. E. Pouw, C. J. Kalkman, and M. J. Bonten. 2009. Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 36020-31. [DOI] [PubMed] [Google Scholar]
- 6.Maragakis, L. L., and T. M. Perl. 2008. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 151254-1263. [DOI] [PubMed] [Google Scholar]
- 7.Silvestri, L., H. K. van Saene, A. Casarin, G. Berlot, and A. Gullo. 2008. Impact of selective decontamination of the digestive tract on carriage and infection due to Gram-negative and Gram-positive bacteria: a systematic review of randomised controlled trials. Anaesth. Intensive Care 36324-338. [DOI] [PubMed] [Google Scholar]
- 8.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kaufmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13807-815. [DOI] [PubMed] [Google Scholar]
- 9.Wareham, D. W., D. C. Bean, P. Khanna, E. M. Hennessey, D. Krahe, E. Ely, and M. Millar. 2008. Bloodstream infection due to Acinetobacter spp: epidemiology, risk factors and impact of multi-drug resistance. Eur. J. Clin. Microbiol. Infect. Dis. 27607-612. [DOI] [PubMed] [Google Scholar]
- 10.Wilks, M., A. Wilson, S. Warwick, E. Price, D. Kennedy, A. Ely, and M. R. Millar. 2006. Control of an outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus colonization and infection in an intensive care unit (ICU) without closing the ICU or placing patients in isolation. Infect. Control Hosp. Epidemiol. 27654-658. [DOI] [PubMed] [Google Scholar]
- 11.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27351-353. [DOI] [PubMed] [Google Scholar]