Abstract
Daptomycin is a novel lipopeptide antibiotic agent approved for the treatment of gram-positive life-threatening infections. Here we report, for the first time, the isolation of a highly daptomycin-resistant strain of Corynebacterium jeikeium causing a life-threatening infection in a neutropenic patient undergoing cord blood transplantation for secondary acute myeloid leukemia.
CASE REPORT
A 46-year-old man with an uneventful past medical history was diagnosed with acute lymphoblastic leukemia in early 2007. He received a first allogeneic stem cell transplant from a matched unrelated donor after successful induction and consolidation therapy. Early after transplantation, he presented with secondary acute myeloid leukemia and was induced with cyclophosphamide and clofarabine, leading to prolonged neutropenia. Without neutrophil recovery, he proceeded to a second allogeneic stem cell transplantation using a double cord blood source. Since vancomycin-resistant Enterococcus faecium (VRE) was detected in earlier stool samples of this patient, two further stool samples were taken after admission to the transplant unit and the presence of VRE was confirmed. Due to this finding, the prior systemic antibiotic chemotherapy and persistent neutropenia, and continuing fever, we treated the patient with a combination of ceftazidime and tigecycline (Fig. 1).
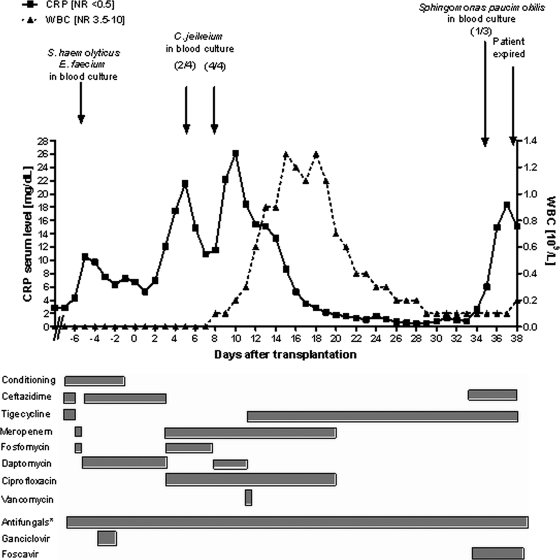
FIG. 1.
Schematic diagram of clinical course at transplant. Ratios in parentheses show the number of cultures positive for the indicated organism versus the number tested. Dosing of antibiotic drugs was as follows: ceftazidime, 2 g intravenously (i.v.) three times daily; tigecycline, 50 mg i.v. twice daily after 100 mg i.v. as a loading dose; meropenem, 1 g i.v. three times daily; fosfomycin, 3 g i.v. three times daily; daptomycin, 500 mg i.v. once daily for three days [7.5 mg/kg of body weight] followed by 350 mg i.v. once daily [5 mg/kg]; ciprofloxacin, 500 mg orally and 400 mg i.v., respectively, twice daily; vancomycin, 1 g i.v. twice daily with pharmacomonitoring. *, Antifungal therapy included one of the following: voriconazole, caspofungin, posaconazole, or liposomal amphotericin B. Abbreviations: WBC, white blood cell count; NR, normal range; L, liter.
After conditioning chemotherapy was initiated, the antibiotic treatment was changed to meropenem and fosfomycin due to recurring febrile temperatures. While fever continued for 2 days, three blood cultures taken on day −6 (before the second transplant) were found to be positive for vancomycin-sensitive E. faecium and Staphylococcus haemolyticus. As both isolates showed susceptibility to daptomycin, the antibiotic regimen was changed to daptomycin and ceftazidime (Fig. 1). The patient recovered from febrile temperatures and received a double cord blood allograft on day 0 (8 January 2008). Blood cultures taken from each of the four central venous catheter lines on days −1 and +1 remained sterile. The clinical course was again complicated on day +3 by increasing C-reactive protein (CRP) levels and fever. The antibiotic regimen was therefore changed to meropenem, fosfomycin, and ciprofloxacin, leading to a short-term decrease of CRP levels and clinical symptoms.
Due to recurrent febrile temperatures, the past infectious history, and positive results for gram-positive rods in two out of four blood cultures taken on day +5, fosfomycin was substituted for daptomycin on day +8. Four further aerobic and anaerobic blood cultures, taken on day +8 and incubated at 37°C in a BacT/Alert 3D instrument (bioMérieux, Marcy-l'Etoile, France), were found to be positive for gram-positive rods after 24 h of cultivation. Subculture testing for another 24 h at 37°C in a 5% CO2-enriched atmosphere revealed small, gray, catalase-positive colonies on chocolate and blood agar. The organism was subsequently identified as Corynebacterium jeikeium (P = 0.963) by using an API Coryne V3.0 kit (bioMérieux, Marcy-l'Etoile, France). This result was confirmed by PCR amplification of a 414-bp segment of the 16S rRNA gene using primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 519r (5′-GWATTACCGCGGCKGCTG-3′) as described in reference 9, followed by standard Sanger sequencing using fluorescently labeled dideoxynucleotides and an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA) for fragment separation and detection, respectively. The isolated strain was designated C. jeikeium strain 1087. Antimicrobial susceptibility testing was performed on Mueller-Hinton agar plates supplemented with 5% sheep blood at 37°C in a 5% CO2-enriched atmosphere using Etest strips (AB Biodisk, Solna, Sweden) as described in reference 6. Testing revealed daptomycin resistance in all strains isolated, as indicated by MIC values of >256 μg/ml (Table 1). Also tested were VRE strain 381, isolated from an earlier stool sample of the same patient; a type strain of C. jeikeium (DSMZ 7171), kindly provided by the German Resource Centre for Biological Material (DSMZ, Braunschweig, Germany); and another C. jeikeium isolate, obtained from a different patient who was suffering from bacterial endocarditis (C. jeikeium strain 957). According to the manufacturer's instructions, strains E. faecalis (ATCC 29212) and Staphylococcus aureus (ATCC 29213) were included as technical controls for the daptomycin Etest (Table 1). As can be further seen in Table 1, in contrast to C. jeikeium strain 1087, all other strains tested were susceptible to daptomycin, with MIC values of <0.25 μg/ml. However, C. jeikeium strain 1087 was clearly susceptible to vancomycin and tigecycline.
TABLE 1.
Results of antimicrobial susceptibility testing using Etests on Mueller-Hinton agar supplemented with 5% sheep blood, done in duplicates
| Drug | MIC (μg/ml)a for:
|
|||||
|---|---|---|---|---|---|---|
| Patient isolate
|
Control strain
|
|||||
| C. jeikeium strain 1087 | VRE strain 381 | C. jeikeium strain 957 | C. jeikeium DSMZ 7171 | E. faecalis ATCC 29212 | S. aureus ATCC 29213 | |
| AMP | >256 | ND | 0.75 | >256 | ND | ND |
| CIP | >32 | ND | 0.125 | 0.125 | ND | ND |
| CLI | >256 | ND | 1.5 | 12 | ND | ND |
| DAP | >256 | 2 | 0.25 | 0.125 | 3 | 0.016 |
| ERY | >256 | ND | 0.094 | 2 | ND | ND |
| GEN | >256 | ND | 0.094 | >256 | ND | ND |
| LVX | >32 | ND | 0.094 | 0.19 | ND | ND |
| MEM | 2 | ND | 0.38 | 0.38 | ND | ND |
| PEN | >32 | ND | 2 | >32 | ND | ND |
| TIG | 0.094 | >32 | 0.094 | 1 | ND | ND |
| VAN | 0.50 | 24 | 0.50 | 0.75 | ND | ND |
AMP, ampicillin; CIP, ciprofloxacin; CLI, clindamycin; DAP, daptomycin; ERY, erythromycin; GEN, gentamicin; LVX, levofloxacin; MEM, meropenem; PEN, penicillin; TIG, tigecycline; VAN, vancomycin; ND, not determined.
Due to the unexpected finding of a daptomycin-resistant C. jeikeium strain in the blood samples and the septic presentation, with neutropenia, daptomycin was replaced with tigecycline on day +11. The patient recovered clinically and remained free of fever, and CRP serum levels declined almost to the normal range during the next weeks. Unfortunately, this infectious episode appeared to be associated with a secondary graft failure. The patient never achieved full neutrophil recovery, and from day +34 on, CRP levels reaccelerated despite broad antibacterial, antifungal, and antiviral treatment, probably due to progressive pulmonary infection. Consequently, the patient died from multiorgan failure 38 days after transplant (Fig. 1). Autopsy was refused by his family.
The increasing number of hospital-acquired infections caused by gram-positive bacteria resistant to a large variety of antibiotics, such as methicillin-resistant S. aureus (10, 16) or VRE (1), has been countered by the development of novel antibiotic compounds (2). Among them, the lipopeptide daptomycin has recently been approved by the FDA for the treatment of complicated methicillin-resistant S. aureus skin infections (15) and was successfully used for the treatment of bacteremia (11) and external-ventricular-drain-associated ventriculitis (4) with VRE. Although numerous previous studies showed a broad susceptibility of many different gram-positive pathogens to daptomycin (17), daptomycin-resistant S. aureus (14) and E. faecium (13) have recently been reported. Here, we describe for the first time the isolation of a highly daptomycin-resistant C. jeikeium strain isolated from a neutropenic patient undergoing cord blood transplantation.
Colonization of such indwelling devices as central venous catheters by C. jeikeium is a common problem among immunocompromised patients and may be associated with consecutive bacteremia and septic infections (7). Accordingly, C. jeikeium has already been considered as an opportunistic pathogen which easily acquires resistance to various antibiotic agents (19). In the present case, the changing antibiotic combinations over more than 2 weeks (from days −7 to +8 [Fig. 1]) might have caused a reduction of the commensal microbial flora and a concomitant growth of C. jeikeium, finally resulting in selection of the daptomycin-resistant strain. While high-level resistance to daptomycin has not been described in corynebacteria so far (8, 17), and the genetic basis for daptomycin resistance also remains to be elucidated, daptomycin's activity against C. jeikeium is known to be inferior to its activity against staphylococci, suggesting the presence of intrinsic factors contributing to lower susceptibility (20). Although we cannot exclude the possibility that the genetic determinant conferring resistance to daptomycin in C. jeikeium resides on a low-copy-number plasmid, our attempts to isolate extrachromosomal DNA failed. This suggests that resistance to daptomycin may be encoded on the bacterial chromosome, as is known to be the case for S. aureus and E. faecium. However, the individual mechanism of resistance may differ among gram-positive bacterial species. For example, mutations in the lysylphosphatidylglycerol synthetase gene (mprF) were shown to contribute to daptomycin resistance in S. aureus (5). In contrast, genomic comparisons between C. jeikeium K411 (18) and S. aureus MW 2 (3) showed no mprF homology. Furthermore, we wish to emphasize that in the present case, daptomycin resistance was clearly independent of vancomycin resistance in C. jeikeium (Table 1). This is in line with previous results for the putative mechanisms underlying daptomycin resistance in E. faecium (12). Based on these findings, daptomycin resistance in C. jeikeium may be a consequence of the accumulation of numerous genetic events. It may also involve a set of nonspecific mechanisms, such as changes in the charge, permeability, or composition of the corynebacterial cell wall.
As resistance to daptomycin in gram-positive rods still appears to be a rather rare and therefore unexpected phenomenon, we strongly encourage daptomycin susceptibility testing, particularly for high-risk patients with life-threatening infections.
Footnotes
Published ahead of print on 6 May 2009.
REFERENCES
- 1.Abele-Horn, M., U. Vogel, I. Klare, C. Konstabel, R. Trabold, R. Kurihara, W. Witte, W. Kreth, P. G. Schlegel, and H. Claus. 2006. Molecular epidemiology of hospital-acquired vancomycin-resistant enterococci. J. Clin. Microbiol. 444009-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelbaum, P. C., and M. R. Jacobs. 2005. Recently approved and investigational antibiotics for treatment of severe infections caused by Gram-positive bacteria. Curr. Opin. Microbiol. 8510-517. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Elvy, J., D. Porter, and E. Brown. 2008. Treatment of external ventricular drain-associated ventriculitis caused by Enterococcus faecalis with intraventricular daptomycin. J. Antimicrob. Chemother. 61461-462. [DOI] [PubMed] [Google Scholar]
- 5.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 502137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funke, G., and K. A. Bernhard. 2007. Coryneform gram-positive rods, p. 485-514. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 7.Funke, G., A. von Graevenitz, J. E. Clarridge III, and K. A. Bernard. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein, E. J., D. M. Citron, C. V. Merriam, Y. A. Warren, K. L. Tyrrell, and H. T. Fernandez. 2004. In vitro activities of the new semisynthetic glycopeptide telavancin (TD-6424), vancomycin, daptomycin, linezolid, and four comparator agents against anaerobic gram-positive species and Corynebacterium spp. Antimicrob. Agents Chemother. 482149-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 14967-75. [DOI] [PubMed] [Google Scholar]
- 10.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 11.Kvirikadze, N., M. Suseno, T. Vescio, L. Kaminer, and K. Singh. 2006. Daptomycin for the treatment of vancomycin resistant Enterococcus faecium bacteremia. Scand. J. Infect. Dis. 38290-292. [DOI] [PubMed] [Google Scholar]
- 12.Montero, C. I., F. Stock, and P. R. Murray. 2008. Mechanisms of resistance to daptomycin in Enterococcus faecium. Antimicrob. Agents Chemother. 521167-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz-Price, L. S., K. Lolans, and J. P. Quinn. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41565-566. [DOI] [PubMed] [Google Scholar]
- 14.Skiest, D. J. 2006. Treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J. Clin. Microbiol. 44655-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenbergen, J. N., J. Alder, G. M. Thorne, and F. P. Tally. 2005. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 55283-288. [DOI] [PubMed] [Google Scholar]
- 16.Stefani, S., and P. E. Varaldo. 2003. Epidemiology of methicillin-resistant staphylococci in Europe. Clin. Microbiol. Infect. 91179-1186. [DOI] [PubMed] [Google Scholar]
- 17.Streit, J. M., R. N. Jones, and H. S. Sader. 2004. Daptomycin activity and spectrum: a worldwide sample of 6737 clinical Gram-positive organisms. J. Antimicrob. Chemother. 53669-674. [DOI] [PubMed] [Google Scholar]
- 18.Tauch, A., O. Kaiser, T. Hain, A. Goesmann, B. Weisshaar, A. Albersmeier, T. Bekel, N. Bischoff, I. Brune, T. Chakraborty, J. Kalinowski, F. Meyer, O. Rupp, S. Schneiker, P. Viehoever, and A. Puhler. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 1874671-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traub, W. H., U. Geipel, B. Leonhard, and D. Bauer. 1998. Antibiotic susceptibility testing (agar disk diffusion and agar dilution) of clinical isolates of Corynebacterium jeikeium. Chemotherapy 44230-237. [DOI] [PubMed] [Google Scholar]
- 20.Van der Auwera, P. 1989. Ex vivo study of serum bactericidal titers and killing rates of daptomycin (LY146032) combined or not combined with amikacin compared with those of vancomycin. Antimicrob. Agents Chemother. 331783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]