Abstract
Mycobacterium avium complex (MAC) infections are increasing annually in various countries, including Japan, but the route of transmission and pathophysiology of the infection remain unclear. Currently, a variable-number tandem-repeat (VNTR) typing method using the Mycobacterium avium tandem repeat (MATR) loci (MATR-VNTR) is employed in Japan for epidemiological studies using clinical isolates of M. avium. In this study, the usefulness of this MATR-VNTR typing method was compared with that of the IS1245-restriction fragment length polymorphism (IS1245-RFLP) typing method and a mycobacterial interspersed repetitive-unit (MIRU)-VNTR typing method reported previously (V. C. Thibault, M. Grayon, M. L. Boschiroli, C. Hubbans, P. Overduin, K. Stevenson, M. C. Gutierrez, P. Supply, and F. Biet, J. Clin. Microbiol. 45:2404-2410, 2007). Seventy clinical isolates identified as M. avium from human immunodeficiency virus-negative patients with MAC infections were used. MATR-VNTR typing using 15 loci distinguished 56 patterns of different allele profiles, yielding a Hunter-Gaston discriminatory index (HGDI) of 0.990. However, IS1245-RFLP and MIRU-VNTR typing yielded HGDIs of 0.960 and 0.949, respectively, indicating that MATR-VNTR has an excellent discriminatory power compared with MIRU-VNTR and IS1245-RFLP typing. Moreover, concomitant use of the MATR-VNTR method and IS1245-RFLP typing increased the HGDI to 0.999. MATR-VNTR typing is inexpensive and easy to perform and could thus be useful in establishing a digital multifacility database that will greatly contribute to the clarification of the transmission route and pathophysiology of M. avium infections.
There are many species of nontuberculous mycobacteria in nature, and they are widely distributed as environmental inhabitants that are found in soil, water, and dust (7). The Mycobacterium avium complex (MAC) is comprised of major pathogenic nontuberculous mycobacteria infecting humans and animals. MAC infection is increasing annually in many countries, including Japan (13, 14). The cause of this increase remains unclear because the route of transmission and the pathophysiology of the disease are not fully understood. The original descriptions of MAC as the Battey bacillus were reported for men with underlying chronic lung disease (18). Thereafter, among species belonging to MAC, M. avium was considered an opportunistic bacterium that causes generalized disseminated diseases in human immunodeficiency virus (HIV)-infected patients and was not considered to cause disease in the absence of a compromised host immune system. However, the emergence of pulmonary disease, especially in women, has been well described for some time (20). Recently, pulmonary MAC infections producing lesions in the lingular segments and middle lobe are increasing in middle-aged and elderly women with no underlying disease, especially in Japan. Also, since the treatment for MAC infections is not as established as that for tuberculosis, patient outcomes are often poor. Patients who exhibit lung lesions characterized by cavities or bronchiectasis on radiography and who show progressive lung destruction may develop fatal dyspnea (4, 13). Since such diseases are also observed in patients showing no immunological abnormalities, we speculate that they may be caused not only by host characteristics but also by the bacteria.
M. avium strains are typed according to their patterns of insertion sequences. IS1245-restriction fragment length polymorphism (RFLP) analysis as reported by Guerrero et al. (5) is regarded as the standard method for the typing of M. avium strains (28). However, the usefulness of this method is limited because of reduced resolution when M. avium strains have few or no IS1245 sequences (1) and because of reduced reproducibility when they have a substantial number of IS1245 sequences, as is the case with the Mycobacterium tuberculosis complex (2). This method is therefore inappropriate for epidemiological analysis performed jointly by multiple facilities. In contrast, variable-number tandem-repeat (VNTR) typing analysis using Mycobacterium avium tandem repeats (MATR-VNTR), which has a high level of reproducibility and allows the digitalization of data for easy comparison among facilities, has begun to be applied not only in the veterinary field but also for the genotyping of human clinical isolates in Japan for further clarification of human M. avium diseases (10, 16). TRs or repetitive units are interspersed throughout chromosomes in which sequences of 10- to 100-bp minisatellites appear repeatedly (24). The number of repetitions in the sequence is known to vary specifically among bacterial strains, allowing for their discrimination. The advantages of this method are as follows. First, the method is based primarily on PCR. Second, it can be performed with minimal training. Third, it can be performed with a small amount of sample. Fourth, a database derived from multiple facilities can easily be created because the results are expressed numerically (23).
In this study, M. avium strains were typed according to IS1245-RFLP patterns, MATR-VNTR patterns, and mycobacterial interspersed repetitive-unit (MIRU)-VNTR patterns, using the MIRUs reported by Thibault et al. in 2007 (25). The usefulness of MATR-VNTR typing analysis employed in Japan was also evaluated.
MATERIALS AND METHODS
Bacterial strains.
Seventy clinical isolates were identified as M. avium by the Cobas Amplicor mycobacterium test (Roche Diagnostic Systems, Inc., Basel, Switzerland) (9) and further identified as M. avium by PCR (17). The standard MAC strains M. avium subsp. avium (GTC00603; derived from M. avium ATCC 25291 and ATCC 35718), M. avium subsp. paratuberculosis (ATCC 19698), M. avium subsp. hominissuis (ATCC 19978), and M. avium subsp. silvaticum (ATCC 49884) were used. Clinical isolates were recovered from the sputa of HIV-negative patients with pulmonary MAC infection who had been examined at a medical organization in Japan between 2004 and 2008. One strain from each patient was analyzed.
RFLP analysis. (i) DNA extraction.
After the isolated M. avium strains were cultured at 37°C for 1 to 3 weeks in 5 ml of Middlebrook 7H9 liquid medium supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment, 0.5 ml was transferred to Mycobroth liquid medium (Kyokuto, Tokyo, Japan), and enrichment culture was performed until the absorbance (530 nm) reached 0.4. The culture was centrifuged at 2,000 × g for 15 min, the pellets were treated using Isoplant II (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions, and the DNA extract was dissolved in 50 μl of Tris-EDTA buffer (pH 8.0) (Sigma-Aldrich, St. Louis, MO).
(ii) IS1245-RFLP.
IS1245-RFLP typing was performed according to the method proposed by van Soolingen et al. (5, 28). The probe was labeled with digoxigenin, using a PCR DIG probe synthesis kit (Roche Diagnostic Systems), and was confirmed to be IS1245 by sequencing. Two micrograms of DNA extract was digested with the restriction enzyme PvuII, separated by electrophoresis on a 1% agarose gel (Takara Bio, Tokyo, Japan), transferred from the gel by vacuum blotting onto a nylon membrane (Hybond N; GE Healthcare, Buckinghamshire, United Kingdom), and exposed in a UV illuminator. The membrane was hybridized using a DIG luminescent detection kit (Roche Diagnostic Systems) and exposed to X-ray film (GE Healthcare). The gel images were saved in TIFF format and were analyzed with Finger Printing II software (Bio-Rad Laboratories, Hercules, CA). A dendrogram of pattern relatedness among the strains was constructed using the unweighted-pair group method using average linkages and with the different bands of calculation. The calculation was made with a 1.0% tolerance.
For clinical isolates that showed no band on IS1245-RFLP analysis, PCR amplification of IS1245 was performed, using the same primers as those used in probe preparation. Regarding the M. avium subsp. paratuberculosis standard strain of the standard MAC strains, since the strain does not have IS1245 and also since PCR amplification was not observed, it was not used for IS1245-RFLP analysis in this study (8, 26).
VNTR analysis. (i) DNA extraction.
The clinical isolates of M. avium were cultured at 37°C for 1 to 3 weeks in 5 ml of Middlebrook 7H9 liquid medium supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment, and 5 ml was cultured for enrichment in Mycobroth liquid medium (Kyokuto, Tokyo, Japan) to an absorbance (530 nm) of 0.2. DNA was then extracted using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions.
The distinctive features of InstaGene Matrix are its DNA purification reagent for PCR amplification and its extremely simple DNA extraction procedure. Furthermore, DNA extraction is possible within 1 hour (17).
(ii) MATR-VNTR.
In the search for homology with the TRs of M. tuberculosis, MATR-VNTR typing selected a candidate by use of M. avium strain 104, and by comparing this strain with M. avium subsp. paratuberculosis strain K10, 16 VNTR loci were thus identified. Among these, except for MATR-10, 15 loci are presently used for analysis (16). Also, primers were designed so that PCR amplification can be performed under the same conditions for all loci.
The primers used for MATR-VNTR are listed in Table 1. The PCR mixture (50 μl) was composed of 2 μl of DNA solution, 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 5 μl of 2 mM deoxynucleoside triphosphate mix, 5 μl of 10× PCR buffer, and 1 μl of each primer at 25 μM. The reactions were carried out using a GeneAmp 9700 PCR system (Applied Biosystems). The PCR conditions were as follows: 1 cycle of 95°C for 10 min followed by 38 cycles of 98°C for 10 s, 68°C for 30 s, and 72°C for 1 min and then 1 cycle of 72°C for 7 min. The PCR products were electrophoresed with the TrackIt 50-bp DNA ladder (Invitrogen, San Diego, CA) in a 2% agarose gel (E-gel; Invitrogen). The amplification product of M. avium subsp. paratuberculosis (ATCC 19698), for which the number of repetitions of each VNTR locus had been determined by sequence analysis, was used as a positive control. After electrophoresis, the gel was photographed with Gel-Doc (Bio-Rad), and the number of base pairs in the target VNTR loci was estimated using Quantity One (Bio-Rad) analysis software. The numbers of repetitions of various VNTR loci of each strain were determined and regarded as an allele profile. The Manhattan distance was determined on the basis of each obtained allele profile, and a dendrogram was estimated by the Fitch-Margoliash method, which was prepared using Tree View (http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
TABLE 1.
Primer sequences, target amplification positions, and sizes of MATR in M. avium strain 104 and M. avium subsp. paratuberculosis strain K10, and position and description of tandem repeats
| VNTR locus | PCR primer sequences | Locus position
|
PCR product size (bp)
|
Tandem repeat position
|
VNTR locus description
|
Reference(s) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| M. avium strain 104a | M. avium subsp. paratuberculosis strain K10b | M. avium strain 104a | M. avium subsp. paratuberculosis strain K10b | M. avium strain 104a | M. avium subsp. paratuberculosis strain K10b | M. avium strain 104a | M. avium subsp. paratuberculosis strain K10b | |||
| MATR-1 | GAACGTTGGGCCGAATGCGA and GTGTCGGACCCCTCCCGTAA | 5145845-5146178 | 4020675-4020956 | 334 | 282 | 5146011-5146136 | 4020841-4020913 | (53 bp × 2) + 20 bp | (53 bp × 1) + 20 bp | |
| MATR-2 | TTGAGCAGCTCGTAAAGCGT and CGCGCTCAAGGAGATGGTTC | 3835243-3835542 | 3253591-3253943 | 300 | 353 | 3835306-3835434 | 3253653-3253834 | (53 bp × 2) + 23 bp | (53 bp × 3) + 23 bp | 3,25 |
| MATR-3 | CCAATCACAACGGCACCATC and TCCTCGACAATCAGCACACT | 4791045-4791504 | 4441728-4442028 | 460 | 301 | 4791067-4791356 | 4441749-4441879 | (53 bp × 5) + 25 bp | (53 bp × 2) + 25 bp | 3, 19, 25 |
| MATR-4 | GACAATGGCATGCCGATCCT and CGCTACGGCCTTCTCCATCT | 3862480-3862753 | 3280891-3281058 | 274 | 168 | 3862530-3862658 | 328940-328962 | (53 bp × 2) + 23 bp | (53 bp × 0) + 23 bp | 3 |
| MATR-5 | CTTGCAGCAGGACGATCAGG and GTGGTCGAAGTCGCTGTTGG | 4296122-4296428 | 3688741-3688989 | 307 | 249 | 4296210-4296370 | 3688828-3688930 | (58 bp × 3) − 13 bp | (58 bp × 2) − 13 bp | 19, 21 |
| MATR-6 | TCGCAGGAAACCAACCTCAA and GCGTGATCGACTCGAAGACC | 3170002-3170385 | 1487154-1487479 | 384 | 326 | 3170123-3170309 | 1487274-1487402 | (57 bp × 3) + 16 bp | (57 bp × 2) + 15 bp | 19, 21 |
| MATR-7 | CCGAGGAAGAGACGAAACCC and TCGTCACCCACAACATGCAG | 736660-737050 | 675974-676311 | 391 | 338 | 736837-737013 | 676154-676273 | (57 bp × 3) + 6 bp | (57 bp × 2) + 6 bp | 19 |
| MATR-8 | CAGGTCCAGGGCATGTTTCC and TCCCGATAATCCGTTGCATGAC | 2423420-2423753 | 2017182-2017401 | 334 | 220 | 2423516-2423722 | 2017277-2017369 | (57 bp × 4) − 21 bp | (57 bp × 2) − 21 bp | 19, 21 |
| MATR-9 | CTGTTGGAGCGCAGCCGTTT and ACCCAGTCGTCGACGGTGTT | 4951928-4952362 | 4279619-4280053 | 435 | 435 | 4952229-4952340 | 4279919-4280030 | (55 bp × 2) + 2 bp | (55 bp × 2) + 2 bp | 25 |
| MATR-11 | TGGCTGCTGTTCAATTGGATG and TCGTCGGTCAATTGCACCTT | 3699292-3699850 | 3124559-3124940 | 559 | 382 | 3699467-3699754 | 3124733-3124843 | (55 bp × 5) + 13 bp | (55 bp × 2) + 1 bp | 21 |
| MATR-12 | TGATGGCGACCACCGACAAGG and TGGATGCGGCCGACCAACA | 4414022-4414563 | 3808216-3808640 | 542 | 425 | 4414147-4414294 | 3808337-3808370 | (57 bp × 3) − 23 bp | (57 bp × 1) − 23 bp | 21 |
| MATR-13 | CCTCGAAGGTGGCGGACTTG and ACCAGGATGGTGCCCAAACC | 4454162-4454508 | 3845472-3845704 | 347 | 233 | 4454300-4454408 | (56 bp × 2) − 4 bp | (56 bp × 0) | ||
| MATR-14 | TGGTCGCCGCACACCTACT and GCCCTTACTGGGCAGGTCCTTC | 525889-526335 | 475536-475867 | 447 | 332 | 526063-526278 | 475710-475809 | (58 bp × 4) − 16 bp | (58 bp × 2) − 16 bp | |
| MATR-15 | GGAAGGCAGCAAGGGTCAAC and TCAGGTCCAGCGACAGCTTC | 372554-372975 | 347148-347398 | 422 | 251 | 372606-372835 | 347199-347257 | (57 bp × 4) + 2 bp | (57 bp × 1) + 2 bp | |
| MATR-16 | GTGGTCAGCACCCGGAGAGT and ACCACCGACTGCTCGACCTT | 383171-383588 | 336466-336883 | 418 | 418 | 383198-383371 | 336492-336665 | (59 bp × 3) − 3 bp | (59 bp × 3) − 3 bp | |
GenBank accession number NC008595.
GenBank accession number NC002944.
(iii) MIRU-VNTR.
The primers reported by Thibault et al. (25) were used for MIRU-VNTR typing analysis (see Table S1 in the supplemental material). The PCR mixture (25 μl) was composed of 1 μl of DNA solution, 1.25 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 2.5 μl of 2 mM deoxynucleoside triphosphate mix, 2.5 μl of 10× PCR buffer, and 0.5 μl of each primer at 25 μM, with 1 μl of dimethyl sulfoxide (Wako, Osaka, Japan) for TR292, TR25, TR3, TR7, and TR47 and 2 μl of dimethyl sulfoxide for TR32 and TR10. The reactions were carried out using a GeneAmp 9700 PCR system (Applied Biosystems). The PCR conditions were as follows for TR292, TR25, TR3, TR7, and TR47: 1 cycle of 95°C for 10 min followed by 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s and then 1 cycle of 72°C for 7 min. For TR32 and TR10, the PCR conditions were 1 cycle of 95°C for 10 min followed by 40 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min and then 1 cycle of 72°C for 7 min. The PCR products were electrophoresed alongside TrackIt 50-bp DNA ladder size markers (Invitrogen) in a 2% agarose gel (Invitrogen). The amplification product of M. avium subsp. paratuberculosis (ATCC 19698), for which the number of repetitions of each VNTR locus had been determined by Thibault et al. (25), was used as a positive control. Hereafter, the same analytical procedure as that used for MATR-VNTR was performed. For the samples that showed no amplification, PCR amplification was attempted again, using the same method as that reported previously (15, 25).
Sequence and in silico analyses.
The PCR products obtained from various loci of MIRU-VNTR and MATR-VNTR of M. avium subsp. avium (GTC00603; derived from M. avium ATCC 25291 and ATCC 35718) were purified using a GenElute PCR DNA purification kit (Sigma-Aldrich), and direct sequencing analysis was performed using the same primers as those used in the PCRs. The nucleotide sequences were compared with the genomic data for M. avium subsp. paratuberculosis strain K10 (GenBank accession number NC002944) (12) and M. avium strain 104 (GenBank accession number NC008595), using a minisatellite database (http://minisatellites.u-psud.fr/). Alignment of the nucleotide sequences was performed using CLC Sequence Viewer 4.6.2 (CLC Bio, Tokyo, Japan).
VNTR allelic diversity analysis.
The allelic diversity index h was used for the evaluation of allelic diversity of the various VNTR loci. The value of h was calculated using the following formula described by Selander et al. (22): h = 1 − Σxi2[n/(n − 1)], where xi is the frequency of the ith allele at the locus and n is the number of bacterial strains.
Calculation of discriminatory power.
The discriminatory power of each typing method was compared using the Hunter-Gaston discriminatory index (HGDI) (6). The HGDI was calculated using the following formula:
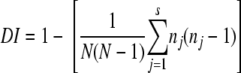 |
where DI is the discriminatory index, N is the total number of strains in the typing scheme, s is the number of clusters obtained, and nj is the number of strains belonging to the jth pattern cluster.
RESULTS
IS1245-RFLP typing.
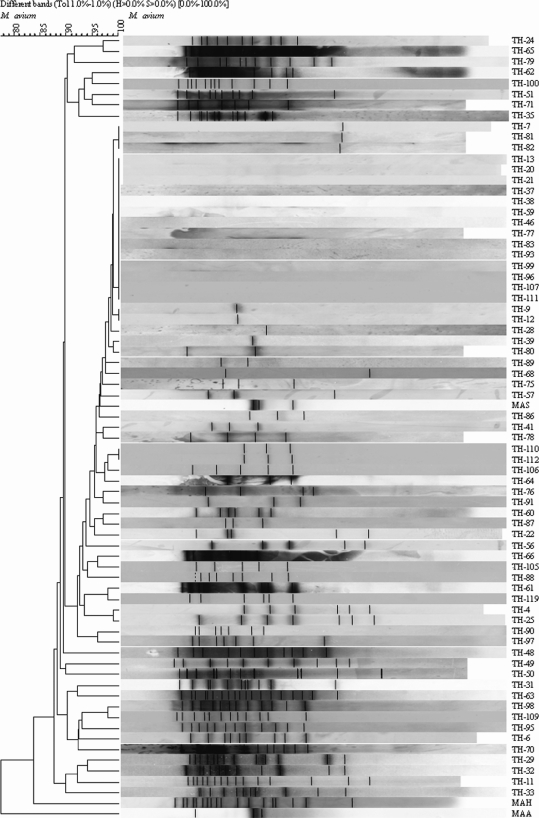
Of the 70 clinical isolates analyzed, 14 strains (20.0%) showed no band corresponding to IS1245 (Fig. 1). The failure of amplification of IS1245 by PCR suggested that the strains do not have IS1245. The number of bands was one to six for 28 strains (40.0%) and seven or more for 28 strains (40.0%). RFLP profiles were similar for strains TH-7, TH-81, and TH-82, for strains TH-9 and TH-12, and for strains TH-110 and TH-112, all of which had one to six bands, indicating that no differentiation could be made using IS1245-RFLP typing.
FIG. 1.
Dendrogram of IS1245-RFLP patterns of M. avium isolates, including 70 clinical strains and reference strains, as determined by the unweighted-pair group method using average linkages, using the different band calculation and a 1.0% position tolerance. The scale at the top shows the genetic similarity among the profiles, expressed as percentages. Faint bands were included in the comparison. MAA (M. avium subsp. avium), strain GTC00603; MAH (M. avium subsp. hominissuis), strain ATCC 19978; MAS (M. avium subsp. silvaticum), strain ATCC49884.
MATR-VNTR typing.
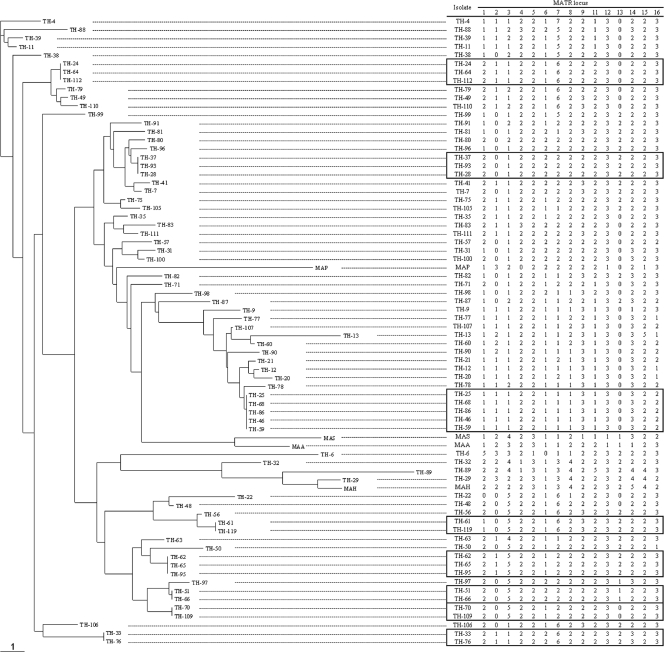
As shown in Fig. 2, the 70 clinical isolates showed allele profiles representing 56 different patterns. Of these isolates, 48 showed distinct patterns and the remaining 22 formed eight clusters. Each of the standard MAC strains showed a unique pattern.
FIG. 2.
Dendrogram and allele profiles constructed from MATR-VNTR typing results for M. avium isolates, including 70 clinical strains and reference strains. The dendrogram was created from distance matrix files by Fitch-Margoliash analysis according to MATR-VNTR markers. Allele profiles analyzed using MATR-VNTR loci from isolates showing 100% similarity are boxed. The Manhattan distance is indicated at the bottom. MAA (M. avium subsp. avium), strain GTC00603; MAH (M. avium subsp. hominissuis), strain ATCC 19978; MAS, (M. avium subsp. silvaticum) strain ATCC 49884; MAP (M. avium subsp. paratuberculosis), strain ATCC 19698.
Table 2 shows the allelic diversity in various MATR-VNTR loci. Among the 15 loci, the highest allelic diversity index was 0.718, for MATR-7, while the allelic diversity index was ≥0.50 for MATR-1, MATR-2, MATR-3, and MATR-13. The allelic diversity index was low (≤0.1) for 4 of the 15 loci, and no allelic diversity was observed in MATR-12 in this analysis.
TABLE 2.
VNTR typing of MATR allelic distribution in M. avium clinical isolates
| Locus | No. of isolates with specified MATR alleleb
|
Allelic diversity (h)a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| MATR-1 | 1 | 30 | 38 | 1 | 0.514 | ||||
| MATR-2 | 29 | 34 | 5 | 2 | 0.581 | ||||
| MATR-3 | 42 | 10 | 1 | 3 | 14 | 0.571 | |||
| MATR-4 | 2 | 66 | 2 | 0.096 | |||||
| MATR-5 | 1 | 66 | 3 | 0.096 | |||||
| MATR-6 | 1 | 49 | 20 | 0.420 | |||||
| MATR-7 | 20 | 27 | 3 | 5 | 14 | 1 | 0.718 | ||
| MATR-8 | 14 | 53 | 3 | 0.376 | |||||
| MATR-9 | 44 | 26 | 0.459 | ||||||
| MATR-11 | 21 | 48 | 1 | 0.431 | |||||
| MATR-12 | 71 | 0.000 | |||||||
| MATR-13 | 38 | 3 | 29 | 0.525 | |||||
| MATR-14 | 1 | 45 | 22 | 2 | 0.480 | ||||
| MATR-15 | 67 | 2 | 1 | 0.070 | |||||
| MATR-16 | 5 | 13 | 52 | 0.400 | |||||
Calculated as described by Selander et al. (22).
There was a total of 70 clinical isolates tested.
MIRU-VNTR typing.
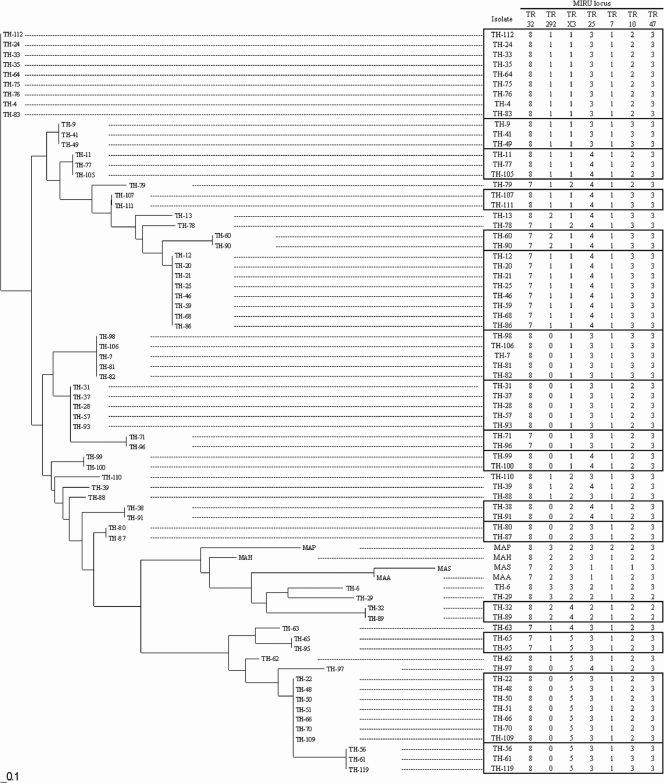
As shown in Fig. 3, the 70 clinical isolates showed allelic profiles representing 27 patterns. Eleven formed unique patterns, and the remaining 59 formed 16 clusters. Each of the standard MAC strains formed an individual pattern.
FIG. 3.
Dendrogram and allele profiles constructed from MIRU-VNTR typing results for M. avium isolates, including 70 clinical strains and reference strains. The dendrogram was created from distance matrix files by Fitch-Margoliash analysis according to MIRU-VNTR markers. Allele profiles analyzed using MIRU-VNTR loci from isolates showing 100% similarity are boxed. The Manhattan distance is indicated at the bottom. MAA (M. avium subsp. avium), strain GTC00603; MAH (M. avium subsp. hominissuis), strain ATCC 19978; MAS (M. avium subsp. silvaticum), strain ATCC 49884; MAP (M. avium subsp. paratuberculosis), strain ATCC 19698.
No amplification of the TR3 locus was observed by MIRU-VNTR typing for 49 of the 70 isolates. Recent reports using the same methods also showed no amplification of this locus (15, 25). In the 21 strains that showed amplification, the number of repetitions was 1, similar to the report of Thibault et al. (25). Therefore, even when amplification was observed, this locus lacked allelic diversity and was excluded from subsequent analyses.
The same 191-bp products were amplified from the TR7 loci of GTC00603 and all clinical isolates. For the TRX3 locus, multiple bands were confirmed for all strains, with two or more repetitions.
Table 3 shows the allelic diversity in various loci by MIRU-VNTR typing. The allelic diversity index was >0.50 for three of the seven loci other than TR3, was <0.10 for two loci, and was 0 for TR7.
TABLE 3.
VNTR typing of MIRU allelic distribution in M. avium clinical isolates
| Locus | No. of isolates with specified MIRU allelec
|
Allelic diversity (h)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 7 | 8 | NPa | ||
| TR32 | 17 | 53 | 0.359 | |||||||
| TR292 | 29 | 34 | 5 | 2 | 0.581 | |||||
| TRX3 | 42 | 10 | 1 | 3 | 14 | 0.571 | ||||
| TR25 | 4 | 42 | 24 | 0.512 | ||||||
| TR3 | 21 | 49 | ||||||||
| TR7 | 70 | 0.000 | ||||||||
| TR10 | 44 | 26 | 0.459 | |||||||
| TR47 | 3 | 67 | 0.069 | |||||||
NP, no PCR product.
Calculated as described by Selander et al. (22).
There was a total of 70 clinical isolates tested.
Sequence and in silico analyses.
When theoretically calculated with only one repetition, the size of TR7 in M. avium subsp. paratuberculosis strain K10 was 181 bp, while that in M. avium strain 104 was shown to be 191 bp by in silico analysis. For TRX3, the primer we used is highly homologous to the sequence in the TR. TRX3 and MATR-3, TR292 and MATR-2, and TR10 and MATR-9 were found to be located at the same loci by VNTR typing. These loci were also in agreement upon sequence analysis using GTC00603. Table 1 shows the MATR-VNTR loci that were in agreement with those reported in references 3, 19, and 21.
Comparison of IS1245-RFLP, MIRU-VNTR, and MATR-VNTR typing.
All strains that formed clusters using IS1245-RFLP typing could be discriminated by MATR-VNTR analysis. The 14 strains that showed no band on IS1245-RFLP typing could still be discriminated using MATR-VNTR analysis, except for strains TH-37 and TH-93 and strains TH-46 and TH-59, which could not be discriminated using this method. Also, all strains that formed the same clusters using MATR-VNTR typing could be discriminated by IS1245-RFLP analysis, except for strains TH-37, TH-46, TH59, and TH-93, which did not have IS1245.
Comparing MATR-VNTR with MIRU-VNTR typing, all of the strains that formed clusters by MIRU-VNTR typing could be discriminated by MATR-VNTR analysis. However, strains TH-62, TH-65, and TH-95 formed the same cluster on MATR-VNTR typing but could be divided into two clusters by MIRU-VNTR typing. The number of repetitions of TR32 was eight for strain TH-62 and seven for strains TH-65 and TH-95.
HGDI.
Table 4 shows the HGDIs of the different typing methods. The discriminatory power of MATR-VNTR typing yielded an HGDI of 0.990, that of IS1245-RFLP typing yielded an HGDI of 0.960, and that of MIRU-VNTR typing yielded an HGDI of 0.949, indicating that MATR-VNTR typing has an excellent discriminatory power. Moreover, a combination of the MATR-VNTR method with IS1245-RFLP typing increased the HGDI to 0.999.
TABLE 4.
Discriminatory index of VNTR and IS1245-RFLP typing of M. avium clinical isolatesb
| Typing method | No. of different patterns | No. of clusters | No. of clustered isolates | No. of unique isolates | HGDIa |
|---|---|---|---|---|---|
| IS1245-RFLP | 53 | 4 | 21 | 49 | 0.960 |
| MATR-VNTR | 56 | 8 | 22 | 48 | 0.990 |
| MIRU-VNTR | 27 | 16 | 59 | 11 | 0.949 |
| IS1245-RFLP plus MATR-VNTR | 68 | 2 | 4 | 66 | 0.999 |
Calculated as described by Hunter and Gaston (6).
There was a total of 70 clinical isolates tested.
DISCUSSION
In this study, three different typing methods, IS1245-RFLP typing, MATR-VNTR typing, used in Japan, and MIRU-VNTR typing, were compared using 70 M. avium clinical isolates derived from HIV-negative patients with pulmonary MAC infection.
One problem with IS1245-RFLP typing was that only 80.0% of the M. avium isolates had IS1245 and the copy numbers were six or less for one-half of the M. avium isolates used in this study. Upon IS6110-RFLP typing of the Mycobacterium tuberculosis complex (27), the reproducibility among laboratories has been reported to decline with an increase in the number of bands, and the discriminatory power decreases when there are fewer bands (1, 2). To overcome these problems, the addition of another typing method, such as spoligotyping or VNTR typing, is recommended (1). This may also be true for typing of M. avium strains with small numbers of bands that cannot be discriminated using RFLP typing. For 14 (20.0%) strains that did not have IS1245, IS1245-RFLP typing had no discriminatory power at all, whereas 10 of these 14 M. avium isolates could be distinguished using MATR-VNTR typing. Since all M. avium subsp. paratuberculosis strains do not have IS1245, IS1245-RFLP analysis cannot be used for these strains. Furthermore, cross-hybridization of IS1311 has been reported with the use of IS1245 as a probe (8, 26). However, it is possible that VNTR analysis is applicable to these strains. To evaluate the in vivo stability of the IS1245 and VNTR loci, Kazumi et al. reported that whereas strains collected on 26 October 1998 and on 19 February 1999 had different RFLP patterns and four strains collected between 20 February 1999 and 17 March 1999 also had different RFLP patterns, all six strains showed the same VNTR pattern. Including these cases, changes in IS1245-RFLP patterns were observed in a short period for 2 of 16 strains, while no changes in the number of repetitions were reported using VNTR (10). Therefore, VNTR analysis has a higher level of reproducibility than IS1245-RFLP analysis. For RFLP analysis, highly purified DNA is required. On the other hand, for VNTR analysis, since PCR is the base for the method, detection is possible as long as DNA is extracted, even though its purity is not high. Therefore, for DNA used for VNTR analysis in this study, a DNA purification reagent for PCR amplification was used. By use of this reagent, the DNA extraction procedure is very simple and easy, and DNA extraction is possible within 1 hour. This procedure is considered a DNA extraction method useful for performing VNTR analysis.
Several shortcomings were noted in the use of MIRU-VNTR typing. First, discrimination by electrophoresis of MIRU-VNTR TR32, whose TR length is 18 bp long, was difficult. Le Fleche et al. showed that TRs larger than 50 bp were easier to analyze by electrophoresis (11). In MATR-VNTR typing, since all VNTR loci were selected to be 53 to 58 bp in TR length, size discrimination was much easier. Second, PCR of the TRX3 locus produced multiple bands by MIRU-VNTR analysis because of the presence of sequences inside the TR locus that are highly homologous to the primer. However, the MATR3 primers used in MATR-VNTR analysis, which are targeted to the same locus as the TRX3 primers, were designed to be nonhomologous to the sequences of the TR locus, and a single band was always obtained. Third, for all clinical isolates studied, the size of the PCR products from the TR7 locus was 191 bp, 10 bp longer than that shown originally for M. avium subsp. paratuberculosis by Thibault et al. (25). In silico analysis of the TR7 locus in GTC00603 and M. avium strain 104 confirmed this difference. Similarly, it is presumed that this result could also be obtained even by MATR-VNTR analysis of M. avium subsp. paratuberculosis. Since in silico analysis of M. avium subsp. paratuberculosis strains K10 and ATCC 19698 showed that MATR-11 was 10 bp shorter than the theoretically calculated value, it was suggested that MATR-11 could not be used to discriminate M. avium subsp. paratuberculosis strains. Finally, no amplification of the TR3 locus was noted for 49 (70.0%) of the 70 strains, indicating that this locus may not be present in these isolates. A similar phenomenon also occurred using VNTR typing of the Mycobacterium tuberculosis complex (23), probably due to the difference in endemic strains in various geographic areas.
As for features that MATR-VNTR and MIRU-VNTR analyses have in common, the standard strains were all shown to form a single pattern by either analysis. Since MATR-VNTR typing was superior to MIRU-VNTR typing in classification, it was presumed that the standard strains were spotted, not concentrated. MATR-VNTR loci and VNTR loci reported by Thibault et al. (25) and other groups (3, 19, 21), which turned out to be the same loci as MATR-VNTR loci by in silico analysis, were found to show diversity. Therefore, since MATR-VNTR and MIRU-VNTR typing methods are based on the same principle, MATR-VNTR typing could also be a useful tool in molecular epidemiology in countries other than Japan.
VNTR typing is a very simple procedure which can be performed with a very small amount of DNA, regardless of whether the bacteria are alive or dead. PCR analysis and electrophoresis can be completed in just 1 day. VNTR typing is also a promising analytical method in molecular epidemiology that can be performed with strains that show a small number of bands on RFLP typing. This study showed that the discriminatory power of MATR-VNTR analysis was higher than that of IS1245-RFLP analysis. However, since there were strains that could be discriminated using IS1245-RFLP but not by MATR-VNTR typing, the concomitant use of RFLP typing, when necessary, is recommended.
Elucidation of the transmission routes and pathophysiology of M. avium infection will likely be considerably advanced by construction of a molecular epidemiological database by use of VNTR typing, using clinical data from multiple facilities. This technique is inexpensive and easy to perform and has a high HGDI.
Supplementary Material
Footnotes
Published ahead of print on 29 April 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bauer, J., A. B. Andersen, K. Kremer, and H. Miörner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 372602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden, C. R., J. T. Crawford, and B. A. Schable. 2002. Quality assessment of Mycobacterium tuberculosis genotyping in a large laboratory network. Emerg. Infect. Dis. 81210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, T. J., K. Sidi-Boumedine, E. J. McMinn, K. Stevenson, R. Pickup, and J. Hermon-Taylor. 2003. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17157-164. [DOI] [PubMed] [Google Scholar]
- 4.Griffith, D. E., T. Aksamit, B. A. Brown-Elliott, A. Catanzaro, C. Daley, F. Gordin, S. M. Holland, R. Horsburgh, G. Huitt, M. F. Iademarco, M. Iseman, K. Olivier, S. Ruoss, C. F. von Reyn, R. J. Wallace, Jr., and K. Winthrop. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175367-416. [DOI] [PubMed] [Google Scholar]
- 5.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansen, T. B., B. Djønne, M. R. Jensen, and I. Olsen. 2005. Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J. Clin. Microbiol. 432500-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katila, M. L., P. Katila, and R. Erkinjuntti-Pekkanen. 2000. Accelerated detection and identification of mycobacteria with MGIT 960 and COBAS AMPLICOR systems. J. Clin. Microbiol. 38960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazumi, Y., T. Udagawa, S. Maeda, Y. Murase, I. Sugawara, M. Okumura, Y. Azuma, M. Goto, and N. Tsunematsu. 2007. Comparison of usefulness between variable numbers of tandem repeats (VNTR) analysis and restriction fragment length polymorphism (RFLP) in the genotyping of Mycobacterium avium. Kekkaku 82741-748. [PubMed] [Google Scholar]
- 11.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 10212344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maekura, R., Y. Okuda, A. Hirotani, S. Kitada, T. Hiraga, K. Yoshimura, I. Yano, K. Kobayashi, and M. Ito. 2005. Clinical and prognostic importance of serotyping Mycobacterium avium-Mycobacterium intracellulare complex isolates in human immunodeficiency virus-negative patients. J. Clin. Microbiol. 433150-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maras, T. K., and C. L. Daley. 2002. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 23553-567. [DOI] [PubMed] [Google Scholar]
- 15.Möbius, P., G. Luyven, H. Hotzel, and H. Köhler. 2008. High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J. Clin. Microbiol. 46972-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriyama, M., K. Ogawa, K. Nishimori, K. Uchiya, T. Ito, T. Yagi, I. Nakashima, T. Nakagawa, O. Tarumi, and T. Nikai. 2006. Usefulness of variable numbers of tandem repeats typing in clinical strains of Mycobacterium avium. Kekkaku 81559-566. [PubMed] [Google Scholar]
- 17.Nishimori, K., M. Eguchi, Y. Nakaoka, Y. Onodera, T. Ito, and K. Tanaka. 1995. Distribution of IS901 in strains of Mycobacterium avium complex from swine by using IS901-detecting primers that discriminate between M. avium and Mycobacterium intracellulare. J. Clin. Microbiol. 332102-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Brien, R. J. 1989. The epidemiology of nontuberculous mycobacterial disease. Clin. Chest Med. 10407-418. [PubMed] [Google Scholar]
- 19.Overduin, P., L. Schouls, P. Roholl, A. van der Zanden, N. Mahmmod, A. Herrewegh, and D. van Soolingen. 2004. Use of multilocus variable-number tandem-repeat analysis for typing Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 425022-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reich, J. M., and R. E. Johnson. 1992. Mycobacterium avium complex pulmonary disease presenting as an isolated lingular or middle lobe pattern. The Lady Windermere syndrome. Chest 1011605-1609. [DOI] [PubMed] [Google Scholar]
- 21.Romano, M. I., A. Amadio, F. Bigi, L. Klepp, I. Etchechoury, M. N. Llana, C. Morsella, F. Paolicchi, I. Pavlik, M. Bartos, S. C. Leão, and A. Cataldi. 2005. Further analysis of VNTR and MIRU in the genome of Mycobacterium avium complex, and application to molecular epidemiology of isolates from South America. Vet. Microbiol. 110221-237. [DOI] [PubMed] [Google Scholar]
- 22.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rüsch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36762-771. [DOI] [PubMed] [Google Scholar]
- 25.Thibault, V. C., M. Grayon, M. L. Boschiroli, C. Hubbans, P. Overduin, K. Stevenson, M. C. Gutierrez, P. Supply, and F. Biet. 2007. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J. Clin. Microbiol. 452404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turenne, C. Y., R. Wallace, Jr., and M. A. Behr. 2007. Mycobacterium avium in the postgenomic era. Clin. Microbiol. Rev. 20205-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leão, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 363051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.