Abstract
This study reveals diverse-length polymorphisms in long mononucleotide repeats (microsatellites) in several serotypes of epidemic human respiratory adenovirus. The length of one of these microsatellites, a homopolymeric thymidine [poly(T)] repeat, is measured in 68 isolates of adenovirus serotype 14. These isolates were collected during a series of sudden and sometimes fatal outbreaks among both military recruits and civilians as the virus emerged for the first time in the United States in 2006 and 2007. The results demonstrate the usefulness of adenoviral microsatellites as high-resolution molecular strain markers. The described homopolymer is hypervariable in length, varying from 12 to 17 bp in the analyzed sample set. All intermediate lengths were identified in at least one isolate. Furthermore, the specific length of the marker is stable for significant periods of time (up to 7 months) at individual sites where the virus is in consistent circulation. The microsatellite also can maintain specific length identity through site-to-site transmission events, as determined by the analysis of isolates from three advanced training sites that appeared to be subject to pathogen transfer from one of the affected recruit training installations. Public database searches revealed that the polymorphic nature of the microsatellite extends to other species B serotypes, and that other polymorphic microsatellites can be identified readily in a variety of epidemic respiratory adenovirus clades. This study shows that microsatellites are a ubiquitous source of polymorphic markers for human adenoviruses and demonstrates their use through an epidemiological analysis of isolates from a recent North American epidemic.
Adult respiratory adenoviruses cause symptoms ranging from, commonly, febrile respiratory illness (FRI) to, more rarely, pneumonia and death (2, 21, 24). In civilian populations, they occur both sporadically and in local and national epidemics. Epidemics often are associated with the emergence of new variants (genome types) of otherwise common serotypes (8, 19) or the reemergence of recently rare serotypes (4, 11, 16).
Adenoviruses of three species, human adenovirus serotypes B, C, and E (HAdV-B, HAdV-C, and HAdV-E, respectively), are strongly associated with respiratory disease in otherwise healthy people (2, 21, 22, 24). HAdV-C serotypes include HAdV-1, HAdV-2, HAdV-5, and HAdV-6 and are endemic among children and young adults (20). Most adults are immune to these serotypes. The sole HAdV-E serotype, HAdV-4, and several HAdV-B serotypes, including HAdV-3, HAdV-7, and HAdV-21, are commonly associated with epidemic outbreaks of FRI and pneumonia in healthy adults and children throughout the world (16, 17, 24, 21). Two other HAdV-B serotypes, HAdV-11a and HAdV-14, had, until recently, been identified only in association with respiratory disease epidemics in rare (though severe) outbreaks in Eurasia (3, 5, 16). Prior to the turn of the century, HAdV-14 had never been isolated from any case in North or South America (16).
During 2006 and 2007, HAdV-14 was identified in a series of outbreaks across the United States, in association with widespread FRI and at least 10 documented pneumonia fatalities (4, 12, 16). This phenomenon was tracked simultaneously both in civilian populations, by the Centers for Disease Control and Prevention and local public health agencies (4), and in military recruits, by U.S. Department of Defense public health agencies (16). Detailed comparisons of collected strains, including the sequencing of fiber and hexon genes and whole-genome restriction analysis (genome typing), revealed that all of these events were caused by a single, apparently homogeneous, strain (12). The identified strain (genome type HAdV-14a) was significantly diverged from the prototypical Eurasian strain (HAdV-14p) identified in outbreaks in the mid-1900s (12).
The full-genome sequencing of multiple HAdV-14a isolates, including both a fatal pneumonia isolate from a severe outbreak in Texas in 2007 and an isolate from a mild outbreak in California in 2006, which had little effect on local adenoviral illness rates, revealed only two genetic polymorphisms between the two strains. Both polymorphisms were noncoding. One of these was a synonymous base mutation in the fiber gene (H. S. Houng, unpublished data). This high degree of homogeneity (clonality) did not offer a simple way to track different lineages of the emerging virus or suggest a functional genetic etiology of the observed variation in clinical impact at different sites.
The second polymorphism was an insertion/deletion mutation in a mononucleotide microsatellite repeat, consisting of 17 consecutive thymidines [T(17)] in one sequenced isolate and T(13) in the other. This polymorphism was identified in the noncoding region at the 3′ end of the terminal binding protein (TBP) gene. The repeat likely serves as the polyadenylation signal for the TBP mRNA. Microsatellite repeats, also referred to as variable-number tandem repeats, simple sequence repeats, and amplified fragment length polymorphisms, are widely recognized as extremely variable high-resolution strain markers (6, 7, 13, 15, 18, 26). These repetitive sequences experience an extremely high rate of replication slippage, resulting in length changes through the gain or loss of single or, more rarely, multiple repeat units (14, 23, 26). Microsatellites therefore vary by length rather than by sequence content, and resulting changes are readily reversed by further mutation (unlike random base substitutions) (14, 23, 26). For extremely clonal populations such as that of the emerging HAdV-14 strain, microsatellites and other hypervariable repeats can offer the only available source of variable genetic strain markers.
The identified microsatellite locus was sequenced from 56 isolates collected during seven recent recruit training center outbreaks, representing 2 years of HAdV-14 surveillance following the initial emergence in the U.S. military. These recruit centers represent the first step in military training and host large numbers of new military recruits from across the United States. Nine duplicate isolates (split aliquots of the same isolate) were sequenced to measure repeatability, because microsatellites can mutate quickly enough to generate mixed populations. When site-specific microsatellite alleles were revealed, the analysis was extended to 11 isolates from three advanced U.S. Air Force training sites. These sites host military personnel for specialized training following initial recruit training. The advanced training sites studied here received large numbers of recruit trainees from Lackland Air Force Base (AFB) in San Antonio, TX, during the time of the most severe HAdV-14 outbreak documented among U.S. recruits, and subsequently suffered from HAdV-14 outbreaks themselves. It was hypothesized that isolates from these advanced Air Force training centers would reflect the microsatellite identity of the Lackland isolates and hence reflect the transmission that was thought to have occurred through trainee movement.
In order to expand the potential set of microsatellite alleles available for use as strain markers in epidemiological studies, public databases were used to identify several other potentially polymorphic microsatellite repeats in respiratory adenoviruses. The lengths of newly identified microsatellites then were compared across all published sequences to identify polymorphisms. Those microsatellites exhibiting a high degree of polymorphism are reported here with the observed distribution of allele lengths.
MATERIALS AND METHODS
Sample collection.
All isolates from recruit training facilities, aside from Lackland AFB, were collected, identified, and analyzed as part of the Naval Health Research Center (NHRC) ongoing population-based FRI surveillance program (16). This surveillance program involves the collection of throat swab samples and symptom data from recruits (new military personnel in initial training programs) who report to sick call with a fever of 38°C or greater and a respiratory symptom such as cough or sore throat. Isolates from this program originated from Fort Benning, GA; Fort Jackson, SC; Fort Leonard Wood, MO (U.S. Army); Naval Recruit Training Command, Great Lakes, IL (U.S. Navy); Marine Corps Recruit Depot, San Diego, CA; and Marine Corps Recruit Depot, Parris Island, SC (U.S. Marine Corps).
Isolates from Lackland AFB, San Antonio, TX, came from both NHRC surveillance samples and samples collected from severely ill patients (mostly recruits, but these also included medical support personnel and other nontrainee employees) at Wilford Hall Medical Center (located at Lackland AFB). Wilford Hall samples and samples from advanced Air Force training centers (including Keesler AFB, Biloxi, MS; Goodfellow AFB, San Angelo, TX; and Sheppard AFB, Wichita Falls, TX) were sent to the Air Force Institute for Operational Health (AFIOH) for diagnostic viral culture and were provided to the NHRC as deidentified viral isolates. All samples collected through the NHRC surveillance activities were provided for research use under written, informed consent as required by institutional review board-approved human use protocols (Department of Defense protocol NHRC.1999.0002). These were cultured at the NHRC and provided to the Walter Reed Army Institute of Research (WRAIR) for microsatellite analysis as deidentified viral isolates.
NHRC samples were collected as oropharyngeal (throat) swabs in VTM (Remel, Lenexa, KS), immediately frozen in either −80°C freezers or on dry ice, and transported on dry ice to the NHRC per collection and transport protocols accredited by the College of American Pathologists (16). AFIOH samples were collected as throat swabs in VTM, cultured in A549 cells, and transported to the NHRC on dry ice as infected tissue culture fluid (isolated virus). All of the described samples and isolates were tested at the NHRC for HAdV-14 (see PCR and sequencing methods below), and positive patient samples from NHRC surveillance collections subsequently were cultured in A549 cells (Diagnostic Hybrids, Athens, OH) and stored frozen as isolated virus. Advanced Air Force training school samples initially were sent as nasal washes to the AFIOH, where they were tested for HAdV-14, cultured in A549 cells, subsequently deidentified, and sent to the NHRC as isolated virus.
Following PCR identification as HAdV-14, a sample set was selected to include two randomly chosen isolates (when possible) from each month at each recruit training site where HAdV-14 was identified. The recruit training sample set came from a set of 502 HAdV-14-positive samples collected from the same sites between March 2006 and October 2007. These in turn were among 3,718 adenovirus-positive samples typed during this period (the rest were mostly HAdV-4, with significant numbers of HAdV-3, HAdV-7, and HAdV-21). While this collection is not inclusive of all FRI cases, it is consistently representative of the temporal and spatial distribution of cases (that is, if cases occur at any surveyed training site, a proportion of them will be collected during any given week, and a representative set of those will be typed). Of the advanced training sites, Keesler and Goodfellow AFB collected five each, of which five and four were HAdV-14 positive, respectively. All of these were studied here. Sheppard AFB collected 40, 20 of which were HAdV-14 positive, and 2 were studied here. The majority of advanced trainees at these sites came directly from recruit training at Lackland AFB. Advanced trainee samples were collected specifically to detect the transmission of HAdV-14 from Lackland AFB.
Lackland AFB, a recruit training site that experienced a particularly severe outbreak of HAdV-14, provides an example of the public health context in which these collections occurred. During 2007, 1,147 recruits were diagnosed with FRI from a total population of 33,496 recruits who spent 6.5 weeks on site in recruit training, representing an attack rate of 3.4% (25). Of these, 482 were tested for adenovirus by PCR, 267 were positive, and 231 of those tested positive for serotype 14 (25). As noted above, other recruit training sites experienced more mixed outbreaks with greater contributions from other serotypes, including HAdV-4, HAdV-3, HAdV-21, and HAdV-7, but FRI rates and proportional contributions from adenoviruses were similar.
All chosen isolates were extracted and aliquoted at the NHRC and transported frozen on dry ice to the WRAIR for sequence analysis. Following the identification of site-specific microsatellite polymorphisms, all advanced training school isolates were shipped to the WRAIR as extracted viral isolates for analysis.
PCR and sequence analysis.
Before the 2007 Lackland AFB outbreak, the NHRC research protocols were used to identify HAdV-14 isolates in the Clinical Laboratory Improvement Amendment/Clinical Laboratory Improvement Program-accredited NHRC laboratory. The protocol consisted of a series of multiplex and monoplex PCRs designed to inclusively identify all HAdV-B, HAdV-C, and HAdV-E serotypes, as previously described (16). During the outbreak, the NHRC-designed HAdV-14 serotype-specific PCR protocol was formalized and College of American Pathologists accredited for diagnostic use at the AFIOH facility. Sequence analysis was accomplished, using the methods detailed in the next paragraph, at the CLIA/CLIP-accredited virology facility at the WRAIR. The primers used to both amplify and sequence the microsatellite region were 5′ CGGTTCCAAATGTTGCGTAG 3′ (HAdV-14F 10338) and 5′ CGGGCAATCAGGGTTCGCACATGATTA 3′ (HAdV-14R 11166). All reported microsatellite alleles were sequenced in both directions. Two unique clean and verified sequences were deposited in GenBank as EU827616 and EU833993. To reduce redundancy, other alleles were not deposited, because the flanking sequences were invariable and the alleles differed only in the length of the microsatellite (as typified by the deposited representatives).
Isolates (200-μl aliquots) were extracted using the QIAamp 96 DNA blood kit (Qiagen, Valencia, CA) per the manufacturer's instructions and eluted into 200 μl of buffer. PCR amplification reaction mixtures consisted of 2 mM MgCl2, 0.6 mM deoxynucleoside triphosphates (1.5 mM each dATP, dGTP, dCTP, and dTTP), 200 μM each primer (see the previous paragraph), 2.5 U of Platinum Taq Polymerase (Invitrogen, Carlsbad, CA), and 1 μl of extracted sample in 1× ABI Buffer II (Applied Biosystems, Foster City, CA) in 100-μl total reaction volumes. Thermal cycling was carried out on an ABI9700 platform (Applied Biosystems) using the following parameters: initial activation for 2 min at 94°C, then 35 cycles of 20 s at 94°C, 20 s at 53°C, and 2 min at 72°C. The final extension was for 7 min at 72°C. PCR cleanup was performed using the Qiagen PCR cleanup kit (Qiagen) per the manufacturer's instructions. Sequencing reactions were set up per the manufacturer's instructions using the ABI Big Dye Terminator kit (manual version 3.2; Applied Biosystems) and run on an ABI9700 platform. Reaction products were analyzed on an ABI3130XL automated sequencer (Applied Biosystems) per the manufacturer's instructions. Resulting data then were edited and aligned using Mac Sequencer software (Gene Codes Inc., Ann Arbor, MI).
Bioinformatic analysis of secondary microsatellites.
The identified microsatellite and the surrounding 200 bp of genomic nucleotide sequence were used to search GenBank (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) (1). Searches were limited to viral sequences, and the low-complexity filter was turned off to prevent skipping the microsatellite sequences themselves. Unique microsatellite alleles identified for all available adenoviral species and serotypes are reported in Results and Discussion. Several other microsatellites then were identified by a visual search of adenoviral genomic sequences. These then were used to search GenBank. Resulting allele distributions were recorded and are reported in Results and Discussion.
Nucleotide sequence accession numbers.
Two unique clean and verified sequences determined in this study were deposited in GenBank as EU827616 and EU833993.
RESULTS AND DISCUSSION
Epidemiological analysis of a microsatellite marker in clinical HAdV-14 isolates.
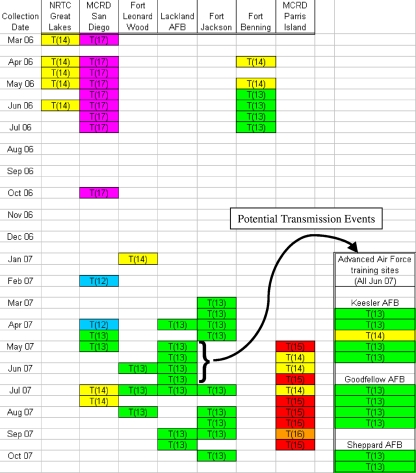
Results for all isolates are shown in the Fig. 1. The sequenced microsatellite (HAdV MicrosatI) varied from T(12) to T(17) among the tested isolates. All intermediate lengths were seen at least once. Eight of the nine tested duplicate pairs gave matched results, while one yielded a variable result of T(13) and T(14) (replicates are not shown in the figure). The results suggest that the microsatellite varies at an appropriate rate for use as a short-term strain-tracking marker, insofar as the identified alleles are diverse between outbreaks, much less diverse within outbreaks, and repeatable for specific isolates.
FIG. 1.
Geographic and temporal variation in MicrosatI reveals the stable maintenance of site-specific strains in U.S. military recruit training centers. This figure shows the allele identity distribution of MicrosatI among 68 febrile respiratory illness patients at seven Army, Navy, Air Force, and Marine Corps recruit training centers and three Air Force advanced training centers, collected during the outbreaks of 2006 and 2007, by site and date (the full names of these sites appear in Materials and Methods). The black arrow represents the flow of trainees from recruit training to advanced training centers, the most likely route of adenoviral transmission to those sites. NRTC, Naval Recruit Training Command; MCRD, Marine Corps Recruit Depot.
Several sites’ sample sets were completely homogeneous; for example, of the five isolates collected during 4 months of HAdV-14 outbreaks at the Naval Recruit Training Command in Great Lakes, IL, all yielded identical T(14) alleles, while nine isolates collected during 8 months at Lackland AFB in Texas all yielded identical T(13) alleles. Some alleles were seen only at one site. For example, the T(17) allele was seen only at the Marine Corps Recruit Depot in San Diego, CA, and was maintained there exclusively throughout 2006.
At sites with variable allele distributions across contiguous months, different alleles isolated in succession always differed in length by a single base pair, suggesting that they were the evolved descendants of the previous resident strain. If they had been imported randomly from other sites, they would be expected to represent a random distribution of the existing allele pool rather than tending so strongly toward similarity to the previous strain.
At sites where HAdV-14 disappeared for several months and then reappeared, there did not appear to be an immediate relationship between the strains isolated in the different outbreaks. The Marine Corps Recruit Depot, San Diego, CA, site offers an excellent example: the T(17) allele was dominant for several months but was replaced by the T(12) allele when HAdV-14 reemerged after a 4-month absence. This pattern is suggestive of a secondary importation event.
Microsatellite variation can play a functional role if the variability affects fitness or the phenotype. This sort of functional variation has been observed in a wide variety of organisms but most commonly is seen in pathogenic prokaryotes in which the variability can play a key role in virulence by generating diversity in traits like antigen presentation (14). Given the high degree of variation in the clinical impact of HAdV-14 emergence at different sites and the apparent clonality throughout the majority of the genome, the possibility that this microsatellite was responsible for variation in pathogenicity or virulence was considered.
The results suggested that the length of the studied microsatellite was not correlated with outbreak or disease severity. One recruit training site affected by HAdV-14 carrying the T(13) allele (Lackland AFB, TX) experienced severe outbreaks that required active intervention and investigation (4) and attracted national media attention and a civilian public health investigation. At other sites, such as Fort Jackson, SC, and Fort Leonard Wood, MO (both of which had preexisting adenovirus epidemics of other serotypes), HAdV-14 carrying the T(13) allele had little effect on disease rates and did not cause excess cases of pneumonia or any fatalities (16). From the data collected here, there appears to be no correlation between microsatellite allele identity [the length of the poly(T) tract] and the clinical character of the HAdV-14 outbreaks; that is, there is no evidence that the microsatellite variability observed here contributes to the pathogenic nature of the adenovirus.
The observed distribution shows that strains are maintained at individual sites, at least across periods of continuous infection, rather than being constantly imported by incoming recruits. This was seen previously for much more divergent strains of another serotype, HAdV-4 (9). The hypothesis that the three advanced Air Force training sites (Sheppard AFB, Goodfellow AFB, and Keesler AFB) were infected through transmission by trainees transferring from Lackland AFB is consistent with the fact that the most common allele at all three advanced sites (10/11 isolates) was identical to the sole allele observed at Lackland [T(13)]. However, this allele was both the most common and the most widely distributed of all observed alleles; hence, it is possible that it came from other sites or from repetitive reintroduction from nonmilitary sources.
The limited number of alleles found for this microsatellite, combined with its high rate of mutation, suggests that the sequence will be highly homoplastic; that is, independent evolutionary lineages often will arrive at the same allelic identity by convergent mutation. Therefore, it cannot be assumed that two strains are related simply because they share the same allele (13, 15). Despite this limitation, consistent sampling can reveal site-specific identities (as it does here) and thereby allow strain tracking. If all sites yielded mixed populations of apparently unrelated alleles, it would be impossible to tell if that situation resulted from a rapidly mutating local strain retained for long periods and derived from a single source or from an influx of diverse strains from multiple sources. In such situations, or in situations where irregular or sporadic collection does not allow a clear demonstration of site specificity and allele stability, multiple microsatellites or more complex polymorphic repeats (like compound microsatellites) may be used to more effectively track strains (15).
Bioinformatic analyses of microsatellites from diverse HAdVs.
Public database searches demonstrated that the observed variability of the identified microsatellite (HAdV MicrosatI) extends to other microsatellite loci, to other species B serotypes, and to other adenoviral species (Table 1). HAdV MicrosatI becomes compound in many species B1 serotypes (that is, it is interrupted by another nucleotide base, and both ends begin to act as independent length-variable microsatellites). This sort of microsatellite can offer a greater range of alleles than a simple microsatellite, as long as sequence analysis is used to define alleles and not length alone (15).
TABLE 1.
Alleles of HAdV MicrosatI identified in GenBanka
| Accession no. | Serotype (species) | Microsatellite sequence | Allele (including extensions) |
|---|---|---|---|
| AY599834 | 3 (B1) | CCCttTTTTTTTTTTTTTTgTTTTTGTCGC | T(16)GT(5) |
| AY599836 | 3 (B1) | CCCttTTTTTTTTTTgTTTTTTGTTGA | T(12)GT(6) |
| AY594255 | 7 (B1) | GCCCtTTTTTTTTTTTgTTTTTGTCGC | T(12)GT(5) |
| AY601634 | 7 (B1) | CCCTCTTTTTTTTTTTgTTTTTTGTTGC | T(11)GT(7) |
| AY495969 | 7 (B1) | CCCTCTTTTTTTTTTTTgTTTTTTGTTGC | T(12)GT(6) |
| AY601636 | 16 (B1) | CCCTCTTTTTTTTgTTTTTTgTTGCCCA | T(8)GT(6)GT(2) |
| AY601633 | 21 (B1) | CCCTCTTTTTTTgTTTTTTTgTTGCCCA | T(7)GT(7)GT(2) |
| AF532578 | 11 (B2) | TCCTATTTTTTTTTTTTTGCCGC | T(13) |
| AY598970 | 11 (B2) | TCCTATTTTTTTTTTTTGCCGC | T(12) |
| AY803294 | 14b (B2) | TCCTATTTTTTTTTTTGCCGC | T(11) |
| AY737797 | 34 (B2) | TCCTATTTTTTTTTTTTGCCGC | T(12) |
| AY271307 | 35 (B2) | TCCTATTTTTTTTTTTTTTTTGCCGC | T(16) |
| AY128640 | 35 (B2) | TCCTATTTTTTTTTTTTTTTGCCGC | T(15) |
| AY737798 | 50 (B) | CCCTCTTTTTTTgTTTTTTTgTTGCCCA | T(7)GT(7)GT(2) |
| AY601635 | 5c (C) | GCCCCTTTTTTgcTTTTCCCAG | T(6)GCT(4) |
| DQ393829 | 49c (D) | CCCCCTTTTTTcTTTTTGCCAG | T(6)CT(5) |
| AY487947 | 4 (E) | CCCTCTTTTgTTTTgTTTTTTGCCAG | T(4)GT(4)GT(6) |
| AY599837 | 4 (E) | CCCTCTTTTgTTTTgTTTTTGCCAG | T(4)GT(4)GT(5) |
| L19443 | 40 (F) | GAGTCTTTTTgTTgTTTTTTGTAGA | T(5)GT(2)T(6) |
This table shows unique microsatellite alleles found in GenBank (BLASTN version 2.2.18; accessed 3 February 2008) for various strains of HAdV from BLASTN alignments with 5 bp of flanking sequence on each side identified using a 200-bp fragment surrounding an HAdV-14 T(17) allele as the query. Apparent base mutations that either shorten the microsatellite by interruption or elongate it by extension are shown in lowercase type. The 5′ flanking sequence of MicrosatI differs between genomic DNA and RNA transcript sequences, but the microsatellite itself does not. This suggests that the end of the microsatellite is a splice point. The expressed RNA, in which the microsatellite appears at the 3′ end, codes for the terminal binding protein. The position analyzed is bp 10780 in AY601634, from the complete genome of HAdV-7 (10).
From the prototype deWit strain of HAdV-14, isolated from The Netherlands recruits in the 1950s. Note that it is different from all alleles seen in the current outbreak of HAdV-14.
Many other species D serotypes share the same allele as serotype 49, and all four species C serotypes (HAdV-1, HAdV-2, HAdV-5, and HAdV-6) share the same allele. The microsatellite generally appears to be invariable in these two species.
A visual search for other potential mononucleotide microsatellites revealed several candidates, and these also exhibited widespread length diversity when alleles were compared across multiple isolates of the same serotype, multiple serotypes, and multiple species. Unique alleles for these microsatellites are reported in Tables 2 through 5. This toolbox of highly variable markers should provide a rich source of strain-specific sequence polymorphisms for tracking and identifying strains of adenovirus during future outbreaks and investigations.
TABLE 2.
Alleles of HAdV MicrosatII identified in GenBanka
| Accession no. | Serotype (species) | Microsatellite sequence | Allele (including extensions) |
|---|---|---|---|
| AY599834 | 3 (B1) | AGTCGTTTTTgctgcTTTTTCCTGG | T(5)GCTGCT(5) |
| AY599836 | 3 (B1) | AGTCGTTTTTgctgctttgttgTTTTTCCTGG | T(5)GCTGCTTTGTTGT(5) |
| AD7001 | 7 (B1) | AGTCGTTTTTgctgcTTTTTTCCTGG | T(5)GCTGCT(6) |
| AY601634 | 7 (B1) | GGTCGTTTTTTTgcTTTTTCCTGG | T(7)GCT(5) |
| AY601636 | 16 (B1) | GGTCGTTTTTTTTTTgcTTTTTCCTGG | T(10)GCT(5) |
| HAU52565 | 21 (B1) | GGTCGTTTTTTTTTTgcTTTTTCCTGG | T(10)GCT(5) |
| AY601633 | 21 (B1) | GGTCGTTTTTTTTTTTTTTTgcTTTTTCCTGG | T(15)GCT(5) |
| AY737798 | 50 (B1) | GGTCGTTTTTTTTTTTTTggcTTTTTCCTGG | T(13)GGCT(5) |
This microsatellite does not appear to exist as a variable repeat in species other than B1. Apparent base mutations that either shorten the microsatellite by interruption or elongate it by extension are shown in lowercase type. The position analyzed is bp 10582 in AY601634, from the complete genome of HAdV-7 (10).
TABLE 5.
Alleles of HAdV MicrosatV identified in GenBanka
| Accession no. | Serotype (species) | Microsatellite sequence | Allele (including extensions) |
|---|---|---|---|
| AY599837 | 4 (E) | GGTCCAAAAAAAAAAAAAGCTAG | A(13) |
| AY599835 | 4 (E) | GGTCCAAAAAAAAAAGCTAG | A(10) |
| HAU10681 | 4 (E) | GGTCCAAAAAAAAGCTAG | A(8) |
| AY594253 | 4 (E) | GGTCGAAAAAAGCTAG | A(6) |
This microsatellite locus position is very close to the locus position of MicrosatI in species B adenoviruses. MicrosatV does not exist in species B, much as MicrosatI does not appear to exist in species E (serotype 4), at least not in a polymorphic state. This suggests that either there is an adaptive reason to have microsatellites in this genomic region (but little selection for what kind) or that this entire region is under very little constraint in terms of variability. The position analyzed is bp 10538 in AY599837, from the complete genome of HAdV-4 (10).
TABLE 3.
Alleles of HAdV MicrosatIII identified in GenBanka
| Accession no. | Serotype (species) | Microsatellite sequence | Allele (including extensions) |
|---|---|---|---|
| AY599836 | 3 (B1) | TTTCTAAAAAAAAAAAAAAAATGTCC | A(16) |
| DQ086466 | 3 (B1) | TTTCTAAAAAAAAAAAAAATGTCC | A(14) |
| AY601634 | 7 (B1) | TTTCTAAAAAAAAAAAAAATGTCC | A(14) |
| AY594255 | 7 (B1) | TTTTTAAAAAAcAAAAAATGTCC | A(6)CA(6) |
| AY495969 | 7 (B1) | TTTCTAAAAAAAAAAAATGTCC | A(12) |
| AY601636 | 16 (B1) | TTTCTAAAAAAAAAAAAAAATGTCC | A(15) |
| AY601633 | 21 (B1) | TTTCTAAAAAAAAAAATGTCC | A(11) |
| AY598970 | 11 (B2) | TTTCTAAAAAAAAAAAAAATGTCC | A(14) |
| AY803294 | 14 (B2) | TTTCTAAAAAAATGTCC | A(7) |
| AY737797 | 34 (B2) | TTTCTAAAAAAAAAAAAAAAATGTCC | A(16) |
| AY271307 | 35 (B2) | TTTCTAAAAAAAAAAAATGTCC | A(12) |
| AY737798 | 50 (B1) | TTTCTAAAAAAAAATGTCC | A(9) |
| AJ854486 | 9 (D) | CTTCTAAAAAAATGTCT | A(7) |
| EF153474 | 26 (D) | CTTCTAAAAAAATGTCT | A(7) |
| AY875648 | 46 (D) | CTTCTAAAAAATGTCT | A(6) |
| EF153473 | 48 (D) | CTTCTAAAAAATGTCT | A(6) |
| DQ393829 | 49 (D) | CTTCTAAAAAATGTCT | A(6) |
| AY599837 | 4 (E) | CTTCTAAAAAATGTCC | A(6) |
This microsatellite does not exist in species C adenoviruses [the allele is A(0)]. The position analyzed is bp 15488 in AY601634, from the complete genome of HAdV-7 (10).
TABLE 4.
Alleles of HAdV MicrosatIV identified in GenBanka
| Accession no. | Serotype (species) | Microsatellite sequence | Allele (including extensions) |
|---|---|---|---|
| AY599834 | 3 (B1) | AGATCTTTTTTTTTTTCTCTT | T(11) |
| AY599836 | 3 (B1) | AGATCTTTTTTTTTTCTCTT | T(10) |
| ADRE3AA | 3 (B1) | AGATCTTTTTTTTTCTCTT | T(9) |
| AY748817 | 7 (B1) | AGATCTTTTTTTTTTTTCTCTT | T(12) |
| AY601634 | 7 (B1) | AGATCTTTTTTTTTTTCTCTT | T(11) |
| AY594255 | 7 (B1) | AGATCTTTTTTTTTTCTCTT | T(10) |
| MAV7HE3 | 7 (B1) | AGATCTTTTTTTTTCTCTT | T(9) |
| HAD315931 | 16 (B1) | AGATCTTTTTTTTTTTTCTCTT | T(12) |
| AY601636 | 16 (B1) | AGATCTTTTTTTTTCTCTT | T(9) |
| AY601633 | 21 (B1) | AGATCTTTTTTTTTCTTTT | T(9) |
| AY598970 | 11 (B2) | TGTTCTTTTTTTTTTTATTTA | T(11) |
| AY803294 | 14 (B2) | TGTTCTTTTTTTTTTTATTTA | T(11) |
| AB070505 | 14 (B2) | TGTTCTTTTTTTTTTAATTT | T(10) |
| AB079724 | 34 (B2) | TGTTCTTTTTTTTTTTATTTA | T(11) |
| AY271307 | 35 (B2) | ATTTGTTcTTTTTTTTTATTTA | T(2)CT(9) |
| AY737798 | 50 (B1) | AGATCTTTTTTTTTCTTTT | T(9) |
This microsatellite is broken up into small fragments in species D adenoviruses and does not offer length variability. It does not appear to exist at all in other species. It is worth noting that these adenoviral microsatellites are unstable at the interspecies level. Hence, efforts to identify new microsatellites should begin with the identification of promising repetitive regions in the targeted species rather than attempting to rely on identified microsatellites from other species. Apparent base mutations that either shorten the microsatellite by interruption or elongate it by extension are shown in lowercase type. The position analyzed is bp 29895 in AY601634, from the complete genome of HAdV-7 (10).
Acknowledgments
For the permissions, access, and assistance necessary to conduct these studies, we acknowledge the clinic commanders and medical staff at Fort Benning, GA; Fort Jackson, SC; Fort Leonard Wood, MO (U.S. Army); Lackland AFB, TX; Keesler AFB, MS; Goodfellow AFB, TX; Sheppard AFB, TX; AFIOH; Wilford Hall Medical Center (U.S. Air Force); Naval Recruit Training Command, Great Lakes, IL (U.S. Navy); Marine Corps Recruit Depot, San Diego, CA; and Marine Corps Recruit Depot, Parris Island, SC (U.S. Marine Corps).
This work represents NHRC report number 08-31 and was supported by the Department of Defense Global Emerging Infections Surveillance and Response System under research work unit no. 6609, done in collaboration with the WRAIR and the Air Force Surgeon General's Directorate of Modernization.
We acknowledge the administrative support of the Henry M. Jackson Foundation for Military Medicine, SpecPro, Inc., and Eagle Applied Sciences, L.L.C.
The views expressed in this work are those of the authors and do not reflect the official policy or position of the Department of the Navy, Department of the Army, Department of the Air Force, Department of Defense (DoD), or U.S. government. This research has been conducted in compliance with all applicable federal and international regulations governing the protection of human subjects in research (DoD protocol NHRC.1999.0002). We declare that no conflict of interest exists.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baum, S. 2005. Adenovirus, p. 1835-1840. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 6th ed., vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 3.Bruj, J., J. Farnik, and V. Sedmidubsky. 1966. Epidemic of acute respiratory disease due to adenovirus type 14. Cesk. Epidemiol. Mikrobiol. Imunol. (The Czech Republic) 15165-171. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2007. Acute respiratory disease associated with adenovirus serotype 14-four states, 2006-2007. MMWR 561181-1184. [PubMed] [Google Scholar]
- 5.Chmielewicz, B., J. Benzler, G. Pauli, G. Krause, F. Bergmann, and B. Schweiger. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, C. L., D. Field, D. Metzgar, R. Saiz, P. A. Morin, I. L. Smith, S. A. Spector, and C. Wills. 1999. Numerous length polymorphisms at short tandem repeats in human cytomegalovirus. J. Virol. 736265-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field, D., and C. Wills. 1996. Long, polymorphic microsatellites in simple organisms. Proc. R. Soc. Lond. 263209-215. [DOI] [PubMed] [Google Scholar]
- 8.Kajon, A. E., and G. Waddell. 1996. Sequence analysis of the E3 region and fiber gene of human adenovirus genome type 7h. Virology 215190-196. [DOI] [PubMed] [Google Scholar]
- 9.Kajon, A. E., J. M. Moseley, D. Metzgar, H. S. Houng, A. Wadleigh, M. A. Ryan, and K. L. Russell. 2007. Molecular epidemiology of adenovirus type 4 infections in US military recruits in the postvaccination era (1997-2003). J. Infect. Dis. 19667-75. [DOI] [PubMed] [Google Scholar]
- 10.Lin, B., Z. Wang, G. J. Vora, J. A. Thornton, J. M. Schnur, D. C. Thach, K. M. Blaney, A. G. Ligler, A. P. Malanoski, J. Santiago, E. A. Walter, B. K. Agan, D. Metzgar, D. Seto, L. T. Daum, R. Kruzelock, R. K. Rowley, E. H. Hanson, C. Tibbetts, and D. A. Stenger. 2006. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 16527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, K. H., Y. C. Lin, H. L. Lin, G. M. Ke, C. J. Chiang, K. P. Hwang, P. Y. Chu, J. H. Lin, D. P. Lin, and H. Y. Chen. 2004. A two decade survey of respiratory adenovirus in Taiwan: the reemergence of adenovirus types 7 and 14. J. Med. Virol. 73274-279. [DOI] [PubMed] [Google Scholar]
- 12.Louie, J. K., A. E. Kajon, M. Holodniy, L. Guardia-LaBar, B. Lee, A. M. Petru, J. K. Hacker, and D. P. Schnurr. 2008. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin. Infect. Dis. 46421-425. [DOI] [PubMed] [Google Scholar]
- 13.Metzgar, D. 2003. The use of noncoding microsatellite length analysis for bacterial strain typing, p. 34-40. In M. Blot, (ed.), Methods and tools in biosciences and medicine: prokaryotic genomics. Berkhäuser Verlag, Basel, Switzerland.
- 14.Metzgar, D., and C. Wills. 2000. Evidence for the adaptive evolution of mutation rates. Cell 101581-584. [DOI] [PubMed] [Google Scholar]
- 15.Metzgar, D., D. Field, R. Haubrich, and C. Wills. 1998. Sequence analysis of a compound coding-region microsatellite in Candida albicans resolves homoplasies and provides a high-resolution tool for genotyping. FEMS Immunol. Med. Microbiol. 20103-109. [DOI] [PubMed] [Google Scholar]
- 16.Metzgar, D., M. Osuna, A. E. Kajon, A. W. Hawksworth, M. Irvine, and K. L. Russell. 2007. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 1961465-1473. [DOI] [PubMed] [Google Scholar]
- 17.Metzgar, D., M. Osuna, S. Yingst, M. Rakha, K. Earhart, D. Elyan, H. Esmat, M. D. Saad, A. Kajon, J. Wu, G. C. Gray, M. A. K. Ryan, and K. L. Russell. 2005. PCR analysis of Egyptian respiratory adenovirus isolates, including identification of species, serotype, and coinfections. J. Clin. Microbiol. 435743-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura, Y., M. Leppert, P. O'Connell, R. Wolff, T. Holm, M. Culver, C. Martin, E. Fujimoto, M. Hoff, E. Kumlin, and R. White. 1987. Variable number tandem repeat (VNTR) markers for human gene mapping. Science 2351616-1622. [DOI] [PubMed] [Google Scholar]
- 19.Noda, M., T. Yoshida, T. Sakeguchi, Y. Ikeda, K. Yamaoka, and T. Ogino. 2002. Molecular and epidemiological analysis of human adenovirus type 7 strains isolated from the 1995 nationwide outbreak in Japan. J. Clin. Microbiol. 40140-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter, C. W., and W. I. H. Shedden. 1963. The distribution of adenovirus antibodies in normal children. J. Hyg. (London) 61155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin, B. A. 1993. Clinical picture and epidemiology of adenovirus infections. Acta Microbiol. Hung. 40303-323. [PubMed] [Google Scholar]
- 22.Sambrook, J., M. Sleigh, J. A. Engler, and T. R. Broker. 1980. The evolution of the adenoviral genome. Ann. N. Y. Acad. Sci. 354426-452. [DOI] [PubMed] [Google Scholar]
- 23.Schlötterer, C., and D. Tautz. 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz, H., R. Wigand, and W. Heinrich. 1983. Worldwide epidemiology of adenoviruses. Am. J. Epidemiol. 117455-466. [DOI] [PubMed] [Google Scholar]
- 25.Tate, J. E., M. L. Bunning, L. Lott, X. Lu, J. Su, D. Metzgar, L. Brosch, C. A. Panozzo, V. C. Marconi, D. J. Faix, M. Prill, B. Johnson, D. D. Erdman, V. Fonseca, L. J. Anderson, and M.-A. Widdowson. 2009. Outbreak of severe respiratory disease associated with emergent adenovirus type 14 at a US Air Force training facility. J. Infect. Dis. 1991419-1426. [DOI] [PubMed] [Google Scholar]
- 26.Tautz, D., and C. Schlotterer. 1994. Simple sequences. Curr. Opin. Genet. Dev. 4832-837. [DOI] [PubMed] [Google Scholar]