Abstract
Variations in proteins related to bacterial diversity may affect species identification performed using matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry. Using this method, we identified 110 Streptococcus agalactiae isolates characterized by serotyping and multilocus sequence typing. Serotype III and sequence type 23 strains expressed the widest variation in molecular weight of putative “species-identifying” biomarker ions. Recognition of the diversity of MALDI patterns observed in strains that represent all major intraspecies lineages assists in the constitution of an optimal reference database.
Streptococcus agalactiae is the main cause of neonatal infections and has emerged as an increasingly frequent pathogen in nonpregnant humans (15). Several studies have identified diversity among S. agalactiae isolates, first by serotyping and then extensively by multilocus enzyme electrophoresis (12, 13). A large number of different sequence types (STs) distributed over several major phylogenetic lineages or clonal complexes (CCs) have recently been identified by multilocus sequence typing (MLST) (http://pubmlst.org/sagalactiae/). ST17 and ST19 account for the majority of cases of S. agalactiae meningitis in infants (7). CC1, CC12, CC17, CC19, and CC23 are mostly associated with infections in adults (7).
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has emerged as a new technique for species identification. By measuring the exact molecular masses of many peptides and small proteins, it is possible to determine the species within a few minutes, whether the analysis is started with whole cells, cell lysates, or crude bacterial extracts (4, 6, 9, 11). Nevertheless, wide variations in protein expression have been reported, specifically according to the distribution of strains in various phylogenetic lineages that make up many species. For example, we have shown such variations in the expression of metabolic enzymes (12, 13), in catabolic functions (3), and in the expression of surface-exposed bacterial proteins involved in adhesion and/or invasion of host cells (14). In addition, the diversity of the rRNA gene regions found in S. agalactiae isolates (2) may induce variations in the expression of the ribosomal proteins detected by MALDI-TOF MS, a method based mainly on detection of ribosomal protein fractions of bacteria (1, 4, 11).
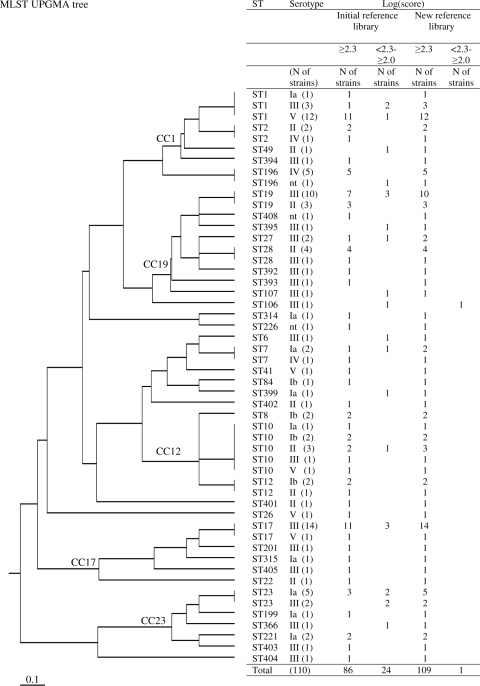
The aim of this study was therefore to determine whether variations in protein expression related to the phylogenetic positions of strains affect the results obtained by MALDI-TOF MS when used to identify S. agalactiae isolates. One hundred ten strains were selected from an epidemiologically unrelated national collection (10, 13) isolated from the vagina, the anatomical site at which the genetic diversity of strains is the highest. As usually performed in medical laboratories, strains were plated on 5% sheep blood agar (Laboratoire bioMérieux, Marcy l'Etoile, France). Serotyping was performed with a Pastorex rapid latex agglutination test (Bio-Rad, Hercules, CA) and by a previously reported PCR serotype identification method (8). Forty-five isolates were from serotype III (40.9% of isolates), 17 from serotype II (15.5%), 16 from serotype V (14.5%), 15 from serotype Ia (13.6%), 7 from serotype Ib (6.4%), and 7 from serotype IV (6.4%). Three isolates were not typeable (2.7%). MLST analysis, carried out with the standard MLST scheme (7), identified 38 STs for the 110 strains (Fig. 1). The relationships between STs were defined by the unweighted-pair group method using average linkages and represented as a tree generated from allelic profile data by using Phylodendron (http://pubmlst.org/sagalactiae/) (Fig. 1). When data from serotyping and MLST were added together, 52 patterns that represent the wide diversity of the S. agalactiae population studied and the major STs and CCs were obtained (Fig. 1).
FIG. 1.
MLST tree based on the unweighted-pair group method using average linkages for 110 S. agalactiae strains used to test MALDI-TOF MS for identification of S. agalactiae strains, with the log(score) obtained for each strain analyzed. The MALDI-TOF MS analysis calculates the log(score), which allows for classification of identification results into four categories: (i) a log(score) of ≥2.3 indicates a highly probable level of species identification, (ii) a log(score) of ≤2.299 and ≥2.0 indicates highly probable genus identification and probable species identification, (iii) a log(score) value of ≤1.999 and ≥1.7 indicates probable genus identification, and (iv) a log(score) of ≤1.699 indicates nonreliable identification. Whatever the phylogenetic origin of the strain tested, no log(score) of ≤1.999 was found. The initial MALDI Biotyper reference library consisted of main spectra of S. agalactiae ATCC 27956 THL and S. agalactiae DSM 2134. The new reference library consisted of the five newly created mass spectra for S. agalactiae strains representative of each major intraspecies lineage (ST1, ST10, ST17, ST19, and ST23).
For MALDI-TOF-MS analysis, cell extracts were prepared, using 10 colonies for each preparation. Samples were prepared according to the microorganism profiling ethanol/formic acid extraction procedure as recommended by the manufacturer, with minor modifications as recently described (1). After drying and addition of a chemical matrix, the samples were analyzed by MALDI-TOF MS on a Bruker Ultraflex TOF/TOF III in positive linear mode (Bruker Daltonique, Wissembourg, France). The spectra were recorded as recently described (1). For each spectrum, 500 laser shots were collected and analyzed. For automated data analysis, raw spectra were processed using MALDI Biotyper 1.1 software (Bruker Daltonique, France) with default settings. To identify bacteria, the peak lists generated were used for matches against the initial MALDI Biotyper reference library (S. agalactiae ATCC 27956 THL and S. agalactiae DSM 2134) by directly using the integrated pattern-matching algorithm of the software (1). In a typical analysis of S. agalactiae strains by MALDI-TOF MS, 70 prominent ion peaks were noted in the spectra in the region between 2,000 and 20,000 Da, with the highest-intensity peaks being consistently in the range of 3,000 to 10,000 Da. The log(score) of the MALDI Biotyper pattern-matching algorithm is calculated according to the log of the product of three factors: the matches of the unknown spectrum against the reference spectrum in the database (main spectrum), the reverse matches of the main spectrum with the unknown spectrum, and the correlation of relative intensities of the unknown spectrum and the main spectrum. The product has a maximum value of 1,000, leading to a maximum log(score) of 3. Differences in the distribution of the strains in log(score) groups were tested by the chi-square test (STAT-ITCF software, Paris, France). A P value of <0.05 was considered to indicate statistical significance.
On this basis, the log(score) values obtained by MALDI-TOF MS correctly identified all 110 S. agalactiae isolates at the species level [log(score), ≥2.0], with 86 of the 110 isolates (78%) being identified with excellent scores [log(score), ≥2.3] (Fig. 1, Table 1). Nevertheless, significant variations in log(score) were observed according to serotypes and major STs. Indeed, excellent scores [log(score), ≥2.3] were obtained significantly less frequently (64%) for serotype III strains than for the other strains (73 to 100%) (Table 1) (P = 0.0027). Similarly, excellent scores [log(score), ≥2.3] were obtained less frequently for ST23 strains (43%) than for the other major STs (77 to 88%) (Table 1) (chi-square test; P = 0.025).
TABLE 1.
Identification scoresa
| S. agalactiae strain(s) (n) | No. (%) of strains with log(score)
|
|||
|---|---|---|---|---|
| Initial reference library
|
New reference library
|
|||
| ≥2.3 | <2.3 and ≥2.0 | ≥2.3 | <2.3 and ≥2.0 | |
| All (110) | 86 (78) | 24 (22) | 109 (99) | 1 (1) |
| Serotype Ia (15) | 11 (73) | 4 (27) | 15 (100) | 0 |
| Serotype Ib (7) | 7 (100) | 0 | 7 (100) | 0 |
| Serotype II (17) | 15 (88) | 2 (12) | 17 (100) | 0 |
| Serotype III (45) | 29 (64) | 16 (36) | 44 (98) | 1 (2) |
| Serotype IV (7) | 7 (100) | 0 | 7 (100) | 0 |
| Serotype V (16) | 15 (94) | 1 (6) | 16 (100) | 0 |
| ST1 (16) | 13 (81) | 3 (19) | 16 (100) | 0 |
| ST10 (8) | 7 (88) | 1 (12) | 8 (100) | 0 |
| ST17 (15) | 12 (80) | 3 (20) | 15 (100) | 0 |
| ST19 (13) | 10 (77) | 3 (23) | 13 (100) | 0 |
| ST23 (7) | 3 (43) | 4 (57) | 7 (100) | 0 |
Scores were produced by the pattern-matching algorithm with the initial MALDI Biotyper reference library (main spectra of S. agalactiae ATCC 27956 THL and S. agalactiae DSM 2134) and with a new reference library (mass spectra of five S. agalactiae strains selected to represent each major intraspecies lineage [ST1, ST10, ST17, ST19, and ST23]) for 110 S. agalactiae strains, according to serotypes and major intraspecies STs.
To test the reproducibility of the method, five strains representative of the five main STs (ST1, ST10, ST17, ST19, and ST23) were grown in quintuplicate and analyzed. The mass spectrum was measured five times for each replicate of each of the five representative strains of main STs (125 measurements). The log(score) calculated using the mass spectrum data allowed for determination of the coefficient of variation (CV), calculated by δ/μ, in which δ is the standard deviation and μ is the average. The CV for intraruns (five measurements for each replicate) varied from 0.006 to 0.025, and the highest CV for interruns (five different replicas) was 0.027. Moreover, reproducibility tests were also performed by cultivation of bacteria on different culture media (5% sheep blood agar, CPS ID 2, chocolate PVX agar [bioMérieux], Granada [Biolys, Taluyers, France], and Mueller Hinton agar). A strain of GBS ST17 was grown in triplicate on each medium. All the log(score) values were above 2.3. The low CVs for the intraruns and interruns and the low variations in the scores obtained using various culture media indicate that the reproducibility of the method was high. Thus, the variations found between populations of various serotypes or STs could not be related to the procedure used for preparing the samples.
This study confirmed variations in expression of proteins according to the distribution of strains in serotypes and in various S. agalactiae phylogenetic lineages. Strains of serotype III and of the phylogenetic lineage ST23 expressed greater variation in molecular weight of putative “species-identifying” biomarker ions than strains of other serotypes and STs. The ability to produce an intense peak in the protein pattern correlates with ionization efficiency, combined with the protein quantity used. The variations observed for serotype III strains may be explained by the greater genetic diversity for this serotype population than for the other serotype populations, as shown in Fig. 1 and as previously described (7, 12, 13). As shown previously, CC23 (which mainly contains ST23 strains) is genetically a highly divergent clone in the species (Fig. 1) (5, 7, 16). This characteristic may explain why ST23 strains express differences in the nature or quantities of proteins in comparison to other major ST populations that are genetically more closely related. These variations did not affect the ability of MALDI-TOF MS combined with the MALDI Biotyper software method to identify S. agalactiae at the species level; the log(score) values were ≥2.0 for all strains. Nevertheless, to optimize the ability of MALDI-TOF MS to identify S. agalactiae strains whatever their phylogenetic origins, we selected five strains to constitute a new reference database on the basis of the variations observed for each lineage. These strains were retained because they generated the best representative spectra for each major intraspecies lineage (ST1, ST10, ST17, ST19, and ST23). The log(score) values obtained by using the new reference database were higher (≥2.3) for 109 of the 110 S. agalactiae isolates (>99%) (Fig. 1 and Table 1).
In conclusion, genetic diversity between bacterial species affects the diversity of MALDI patterns observed with MALDI-TOF MS. Consequently, the most effective performance for identification of bacterial strains at the species level was obtained by using a reference database designed not by selecting one or two strains randomly but by choosing them from the major phylogenetic lineages that represent the species studied.
Acknowledgments
M.-F.L. and R.Q. conceived and designed the experiments. M.-F.L., G.H.-A., E.H., A.-S.D., N.V.D.M.-M., P.L., and L.M. performed the serotyping and MLST experiments. M.-F.L. and P.-O.S. performed the MALDI-TOF MS experiments. M.-F.L., M.K., and R.Q. analyzed the data. M.-F.L. and R.Q. wrote the paper.
This study was supported by the Ministère de l'Enseignement Supérieur et de la Recherche and by the Centre Hospitalier Universitaire de Tours.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid Identification and Typing of Listeria species using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 745402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatellier, S., H. Huet, S. Kenzi, A. Rosenau, P. Geslin, and R. Quentin. 1996. Genetic diversity of rRNA operons of unrelated Streptococcus agalactiae strains isolated from cerebrospinal fluid of neonates suffering from meningitis. J. Clin. Microbiol. 342741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domelier, A. S., N. van der Mee-Marquet, A. Grandet, L. Mereghetti, A. Rosenau, and R. Quentin. 2006. Loss of catabolic function in Streptococcus agalactiae strains and its association with neonatal meningitis. J. Clin. Microbiol. 443245-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20157-171. [DOI] [PubMed] [Google Scholar]
- 5.Héry-Arnaud, G., G. Bruant, P. Lanotte, S. Brun, B. Picard, A. Rosenau, N. van der Mee-Marquet, P. Rainard, R. Quentin, and L. Mereghetti. 2007. Mobile genetic elements provide evidence for a bovine origin of clonal complex 17 of Streptococcus agalactiae. Appl. Environ. Microbiol. 734668-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland, R. D., J. G. Wilkes, F. Rafii, J. B. Sutherland, C. C. Persons, K. J. Voorhees, and J. O. J. Lay. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 101227-1232. [DOI] [PubMed] [Google Scholar]
- 7.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 412530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnamurthy, T., P. L. Ross, and U. Rajamani. 1996. Detection of pathogenic and non-pathogenic bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10883-888. [DOI] [PubMed] [Google Scholar]
- 10.Loulergue, J., C. Couhé, C. Grasmick, P. Laudat, and R. Quentin. 2004. Sensibilité aux antibiotiques des souches de streptocoque du groupe B de portage vaginal isolées en France, 2003. Bull. Epidemiol. Hebdomadaire 1869-70. [Google Scholar]
- 11.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 461946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 864731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 332576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenau, A., K. Martins, S. Amor, F. Gannier, P. Lanotte, N. van der Mee-Marquet, L. Mereghetti, and R. Quentin. 2007. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect. Immun. 751310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Mee-Marquet, N., L. Fourny, L. Arnault, A. S. Domelier, M. Salloum, M. F. Lartigue, and R. Quentin. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 462906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]