Abstract
Reverse transcriptase PCR designed to amplify the N1 to N9 neuraminidase (NA) genes of avian influenza viruses detected 118 of the 119 NA genes tested (99.2%) in a subtype-specific manner. This technique successfully subtyped all 167 recent avian influenza viruses isolated from birds. Subtype specificity was confirmed by sequence analyses of all 285 PCR products.
The spread of the H5N1 highly pathogenic avian influenza (AI) virus in Asia, Europe, and Africa raises concerns that a pandemic will occur (1, 15). Migratory ducks that are more resistant to the H5N1 virus can carry the virus long distances, while more-susceptible swans and geese serve as indicators of the infections in wild aquatic bird populations (3, 4). Active surveillance programs have been instituted worldwide to determine the current H5N1 prevalence among wild aquatic birds (1, 14). The AI virus has two glycoprotein spikes, hemagglutinin (HA) and neuraminidase (NA). NA comprises a mushroom-shaped tetramer and is divided serologically into nine subtypes by using an NA inhibition (NI) test (8, 12). NA subtyping using PCR is used for AI global surveillance, since this test can be performed easily in general diagnostic laboratories. In contrast, the NI test can be performed only in reference laboratories.
Comparison of 263 complete nucleotide sequences of NA genes downloaded from the Influenza Sequence Database (http://www.flu.lanl.gov/) (10) revealed that the virus membrane anchor and stalk region have a great deal of nucleotide sequence variability (8). This variability can be exploited to distinguish the different subtypes. For each NA subtype, we thus designed forward primers that were complementary to the membrane anchor (nucleotides ∼53 to 115) and reverse primers complementary to the region carrying the stalk to the 5′ end of the head region (nucleotides ∼209 to 365) ( Table 1). These subtype-specific primers were screened against previously and recently isolated AI viruses and further modified using sequence data for 41 NA genes that were not detected initially by PCR.
TABLE 1.
Primers for subtyping NA genes of AI viruses
| NA subtype | Primer no. | Sequence position | GenBank accession no. for reference sequence | Product size (bp) | Primer sequence (5′ → 3′)a | No. of mixed nucleotides |
|---|---|---|---|---|---|---|
| N1 | 685 | N1-54F | CY003931 | 245 | TCARTCTGYATGRYAAYTGG | 5 |
| 469 | N1-298R | GGRCARAGAGAKGAATTGCC | 3 | |||
| N2 | 470 | N2-59F | CY005306 | 278 | TYTCTMTAACYATTGCRWCARTATG | 6 |
| 695 | N2-336R | GARTT GTCYT TRGAR AAVGG | 5 | |||
| N3 | 687 | N3-79F | AY650272 | 287 | GCCCTTCTYATYGGRRTKGGRAA | 6 |
| 688 | N3-365R | ACTATDRCRTCYTTGTTYTC | 5 | |||
| N4 | 483 | N4-55F | CY003986 | 236 | AGTGYKAGYATTRTAYTRAC | 6 |
| 484 | N4-290R | ARGTCTYTYCCACTRGARTA | 5 | |||
| N5 | 509 | N5-115F | CY005693 | 178 | GARTA ATATCAGYRACVAAAG | 4 |
| 87 | N5-279R | GATACATYRCAGAGAGGTTC | 2 | |||
| N6 | 88 | N6-57F | CY005464 | 264 | AGGAATGACACTATCSGTAGTAAG | 1 |
| 89 | N6-307R | GAYAGRATRTGCCATGAGTTYAC | 4 | |||
| N7 | 472 | N7-53F | CY004347 | 261 | TCWGGAGTGGCMATAGCACT | 2 |
| 473 | N7-313R | CACKACCCAYCCTTCAACWTTG | 3 | |||
| N8 | 107 | N8-93F | CY004056 | 137 | CATRTVGTBAGYATYAYARTAAC | 7 |
| 690 | N8-209R | ACAYTRGYATTGTRCCATTG | 4 | |||
| N9 | 691 | N9-64F | CY004701 | 227 | GTAATAGGCACRATYGCAGT | 2 |
| 692 | N9-290R | CCTTTRGTYARRTTATTGAA | 4 |
Codes for mixed-base positions: B, C/G/T; D, A/G/T; K, G/T; M, A/C; R, A/G; V, A/C/G; W, A/T; Y, C/T.
Viral RNA was extracted from allantoic fluid by using Trizol solution (Invitrogen, Carlsbad, CA) and used for cDNA synthesis with random primers (6-mers) and PrimeScript reverse transcriptase (Takara, Shiga, Japan) (13). The PCR amplification was performed using the GeneAmp PCR System 9700 (ABI, Foster City, CA) and Ex Taq polymerase (Takara, Shiga, Japan) (13). PCR products were sequenced directly using forward and reverse primers (ABI-3100 sequencer; PE ABI, Foster City, CA) (13).
The detection rate of NA genes by the NA subtyping primer sets was determined using previously isolated AI viruses (n = 119; isolated between 1946 and 2006). These 119 viruses consisted of 83 duck strains, 16 chicken strains, 7 turkey strains, and 13 strains from other birds. They included 77 strains from Japan and 42 strains from Korea, Thailand, Italy, The Netherlands, the United States, or Chile; alternatively, the viruses can be viewed as 92 Eurasian and 27 American lineages. The primer sets amplified 118 NA genes: 12 N1, 32 N2, 20 N3, 6 N4, 12 N5, 17 N6, 3 N7, 5 N8, and 11 N9 genes. The NA subtype specificity was confirmed by sequence analyses of all 118 PCR products by using the same primers. Only one gene, an N2 gene, was not amplified by PCR. Full genome analysis showed that the unamplified N2 gene belonged to the American lineage. Serial 10-fold dilutions of allantoic fluids containing nine AI viruses (one strain for each of subtypes N1 to N9) were used to compare PCR sensitivity to infectivity, expressed as 50% egg-infective dose. The detection limits of the PCRs were 102.5 to 104.5 50% egg-infective doses.
The cross-reactivity of the primer sets was tested with 45 AI viruses (5 viruses per subtype for nine NA subtypes). Homologous NA genes were amplified efficiently; faint, smeared, or size-different PCR products that were distinguishable from specific products by their size or intensity on agarose gels were occasionally detected. Sequence analyses showed that some faint bands were confirmed as representing cross-reactivity or nonspecific products, while two clear NA genes in a single sample were indicative of the presence of two AI viruses. Such faint bands might be inevitable in our NA subtyping PCR. The NA subtyping PCR could detect a deletion in the NA stalk region which was present in some AI viruses, such as A/ chicken/Korea/ES/2004 (H5N1) and A/turkey/Italy/4580/99 (H7N1). It is surmised that the deletion might be associated with the adaptation of AI virus strains to terrestrial birds, such as chickens or turkeys.
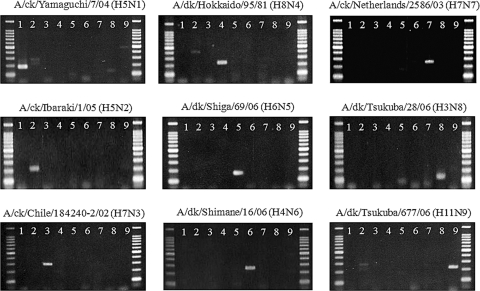
The usefulness of the PCR for subtyping NA genes was assessed using 167 AI viruses: 163 low-pathogenicity (LP) AI viruses isolated from wintering migratory ducks in Japan in 2006/2007 and 2007/2008 (K. Tsukamoto, T. Ashizawa, K. Nakanishi, N. Kaji, R. Kakizawa, M. Shishido, K. Suzuki, T. Tada, and M. Mase, unpublished data) and 4 H5N1 highly pathogenic AI viruses isolated from chickens in Japan in 2007 (M. Mase, Y. Yamamoto, M. Okamatsu, Y. Uchida, T. Saito, K. Nakamura, K. Tsukamoto, and S. Yamaguchi, unpublished data). All 167 AI viruses were successfully subtyped, and nucleotide sequence analysis of the PCR products confirmed the NA subtype specificity. PCRs that determined the NA subtypes of nine AI viruses (one strain per NA subtype) are shown in Fig. 1. PCR was used successfully to subtype 18 N1, 40 N2, 24 N3, 9 N4, 10 N5, 15 N6, 7 N7, 23 N8, and 21 N9 genes. The total rate of detection of NA genes by PCR was 99.7% (285 genes/286 genes).
FIG. 1.
Subtyping PCR for NA genes of nine AI viruses.
To summarize, NA-subtyping PCR for the N1 to N9 genes of AI viruses is both sensitive (99.7%; 285/296) and highly specific (100%; 285/285) and useful for amplifying widely diverse NA genes from 59 reference strains and 227 wild-duck strains. The strains from wild ducks may be representative of Eurasian strains since they were isolated in five prefectures over a 3-year period in Japan; these strains comprise a variety of subtypes and represent molecularly diverse viruses. Alvarez et al. (2) developed a universal primer set for detecting all nine NA subtypes with 97% (32/33) sensitivity; however, sequencing of the PCR products was essential for NA subtyping (2). Qiu et al. (11) utilized nine primer sets for NA subtyping using 101 reference strains or isolates; the reported sensitivity was 97.3%, and the specificity was 91.1% (11). Fereidouni et al. (5) developed an NA subtyping PCR to analyze a total of 162 NA genes from 43 reference strains and 119 diagnostic samples (5). We designed the PCR to be subtype specific by targeting the most subtype-specific regions of NA genes. The subtype specificity was confirmed by using 45 isolates and sequence analysis of all 285 PCR products. Although direct detection of NA genes in diagnostic samples was not performed in this study, PCR is a viable option if the cDNA is prepared appropriately. Our NA subtyping PCR could be useful in determining the adaptation of AI viruses to terrestrial birds, such as chickens or turkeys. It can also be used to detect mixed viruses (data not shown) and is rapid (∼4 h) compared to the conventional serological NI test (2 days). In contrast to the use of microarray technology, PCR does not require expensive equipment or training (6, 7, 9). Our NA-subtyping PCR, together with HA-subtyping PCR (13), holds great potential for use in general laboratories for simultaneous subtyping of HA and NA genes of AI viruses.
Nucleotide sequence accession numbers.
The sequences of the 41 NA genes were registered in GenBank under accession numbers AB472013 to AB472040 and AB472051 to AB472063.
Acknowledgments
This study was supported by a grant-in-aid from the Zoonoses Control Project from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Alexander, D. J. 2007. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002-2006. Avian Dis. 51161-166. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, A. C., M. E. Brunck, V. Boyd, R. Lai, E. Virtue, W. Chen, C. Bletchly, H. G. Heine, and R. Barnard. 2008. A broad spectrum, one-step reverse-transcription PCR amplification of the neuraminidase gene from multiple subtypes of influenza A virus. Virol. J. 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. D., D. E. Stallknecht, J. R. Beck, D. L. Suarez, and D. E. Swayne. 2006. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 121663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. D., D. E. Stallknecht, and D. E. Swayne. 2008. Experimental infection of swans and geese with highly pathogenic avian influenza virus (H5N1) of Asian lineage. Emerg. Infect. Dis. 14136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fereidouni, S. R., E. Starick, C. Grund, A. Globig, T. C. Mettenleiter, M. Beer, and T. Harder. 2009. Rapid molecular subtyping by reverse transcription polymerase chain reaction of the neuraminidase gene of avian influenza A viruses. Vet. Microbiol. 135253-260. [DOI] [PubMed] [Google Scholar]
- 6.Gyarmati, P., T. Conze, S. Zohari, N. LeBlanc, M. Nilsson, U. Landegren, J. Baner, and S. Belak. 2008. Simultaneous genotyping of all hemagglutinin and neuraminidase subtypes of avian influenza viruses by use of padlock probes. J. Clin. Microbiol. 461747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kessler, N., O. Ferraris, K. Palmer, W. Marsh, and A. Steel. 2004. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 422173-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamp, R. A., and R. M. Krug. 2001. Otrhomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe and R. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 9.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection. Elsevier Science, Amsterdam, The Netherlands.
- 11.Qiu, B. F., W. J. Liu, D. X. Peng, S. L. Hu, Y. H. Tang, and X. F. Liu. 2009. A reverse transcription-PCR for subtyping of the neuraminidase of avian influenza viruses. J. Virol. Methods 155193-198. [DOI] [PubMed] [Google Scholar]
- 12.Swayne, D. E., and D. A. Halvorso. 2003. Influenza, p. 135-160. In Y. M. Saif, J. H. Barnes, J. R. Glisson, A. M. Fadly, L. R. McGougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames, IA.
- 13.Tsukamoto, K., H. Ashizawa, K. Nakanishi, N. Kaji, K. Suzuki, M. Okamatsu, S. Yamaguchi, and M. Mase. 2008. Subtyping of avian influenza viruses H1 to H15 on the basis of hemagglutinin genes by PCR assay and molecular determination of pathogenic potential. J. Clin. Microbiol. 463048-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallensten, A., V. J. Munster, N. Latorre-Margalef, M. Brytting, J. Elmberg, R. A. Fouchier, T. Fransson, P. D. Haemig, M. Karlsson, A. Lundkvist, A. D. Osterhaus, M. Stervander, J. Waldenstrom, and O. Bjorn. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster, R. G., M. Peiris, H. Chen, and Y. Guan. 2006. H5N1 outbreaks and enzootic influenza. Emerg. Infect. Dis. 123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]