Abstract
Combination therapy can successfully suppress human immunodeficiency virus (HIV) replication in patients but selects for drug resistance, requiring subsequent resistance-guided therapeutic changes. This report describes the development and validation of a novel assay that offers a uniform method to measure susceptibility to all clinically approved HIV type 1 (HIV-1) drugs targeting reverse transcriptase (RT), protease (PR), integrase (IN), and viral entry. It is an assay in which the antiviral effect on infection within a single replication cycle is measured in triply transfected U87.CD4.CXCR4.CCR5 cells, based on homologous recombination between patient-derived amplicons and molecular proviral clones tagged with the enhanced green fluorescent protein (EGFP) reporter gene and from which certain viral genomic regions are removed. The deletions stretch from p17 codon 7 to PR codon 98 in pNL4.3-ΔgagPR-EGFP, from PR codons 1 to 99 in pNL4.3-ΔPR-EGFP, from RT codons 1 to 560 in pNL4.3-ΔRT-EGFP, from IN codons 1 to 288 in pNL4.3-ΔIN-EGFP, and from gp120 codon 34 to gp41 codon 237 in pNL4.3-Δenv-EGFP. The optimized experimental conditions enable the investigation of patient samples regardless of viral subtype or coreceptor use. The extraction and amplification success rate for a set of clinical samples belonging to a broad range of HIV-1 group M genetic forms (A-J, CRF01-03, CRF05, and CRF12-13) and displaying a viral load range of 200 to >500,000 RNA copies/ml was 97%. The drug susceptibility measurements, based on discrimination between infected and noninfected cells on a single-cell level by flow cytometry, were reproducible, with coefficients of variation for resistance ranging from 7% to 31%, and were consistent with scientific literature in terms of magnitude and specificity.
Despite continued improvements in treatment of human immunodeficiency virus type 1 (HIV-1)-infected patients, therapy failure still occurs, often leading to drug resistance development, which necessitates a change in regimen. At this moment, clinicians have at their disposal 22 drugs, targeting the viral protease (PR), the reverse transcriptase (RT), the integrase (IN), the transmembrane glycoprotein (gp41), or the interaction between the surface glycoprotein (gp120) and the cellular coreceptor CCR5. The choice of which drugs to include in the next regimen is based upon the likelihood of the drugs being active against the mutant virus present in the patient with incomplete suppression of replication (11).
Currently, genotypic drug resistance testing is used more frequently within the clinical setting of patient follow-up due to practical reasons, such as a shorter turnaround time, easier implementation within laboratories, and lower cost, but also due to the fact that no clinical trial has yet provided sufficient evidence for supporting phenotypic over genotypic drug resistance testing. Nevertheless, the interpretation of genotypic drug resistance testing can be very complex, which makes phenotypic drug resistance testing in certain settings very useful and even necessary (48).
Increasing numbers of drug resistance mutations have been identified within gag, pol, and env, reflecting the genetic flexibility of HIV-1. These mutations can directly boost the ability of the virus to specifically replicate in the presence of the drug (major mutations) or indirectly aid the virus by increasing its replication capacity in general, whether in the presence or absence of the drug (minor or compensatory mutations). In general, the major mutations are found at the drug binding sites within the targeted protein (or the viral protein that interacts with the cellular target), whereas the others can be found at distinct sites within the target protein or even other proteins. In this respect, PR inhibitor (PI) mutations have been observed not only within the PR but also at several sites within its Gag substrate (13, 28).
The use of genotypic data to determine if certain drugs are still active against patient-derived virus is further complicated by the presence of natural polymorphisms in non-B strains and by baseline subtype-dependent combinations of mutations that occur during treatment, leading to discordances between different interpretation algorithms (43, 51). Several studies have revealed novel mutations or differences in the prevalence of known mutations in non-B subtypes (1, 3). Others have demonstrated the impact of the genetic background on subtype dependencies in drug susceptibility and resistance development (2, 6, 7). In this respect, phenotypic drug resistance testing is a very useful tool to determine the effects of specific mutations or combinations thereof in their respective backbones and to improve genotypic drug resistance interpretation in the long run.
For newly approved drugs, the relationships between genetic changes in the target region and the clinical response still need to be established. Phenotypic assays can determine whether or not the new drug is active against the patient-derived virus and could be included in the next regimen (23). They also allow the investigation of potential cross-resistance for novel drugs with similar mechanisms of action to those of drugs included in previous regimens (8).
In this paper, we describe a novel assay based on the creation of recombinant replication-competent viruses that enables determination of the impact of mutations and combinations of mutations on susceptibility toward all clinically approved HIV-1 inhibitors. The recombinant viruses are tagged with the enhanced green fluorescent protein (EGFP) reporter gene, which allows for discrimination between infected and noninfected cells on a single-cell level by flow cytometry, which provides a high sensitivity. The assay was designed such that it offers a uniform methodology for all currently approved inhibitors, regardless of subtype or cellular tropism of the viral strains tested.
MATERIALS AND METHODS
Antiviral drugs.
Zidovudine (AZT), abacavir (ABC), didanosine (DDI), efavirenz (EFV), lamivudine (3TC), stavudine (D4T), nelfinavir (NFV), ritonavir (RTV), nevirapine (NVP), and enfuvirtide (ENF) were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH). Raltegravir (RAL), maraviroc (MVC), and AMD3100 were obtained from Merck & Co. (Rahway, NJ), Pfizer (Groton, CT), and AnorMed (Langley, British Columbia, Canada), respectively. Elvitegravir (EVG) and (R)-9-(2-phosphonylmethoxypropyl)adenine (PMPA; also called tenofovir [TDF]) were kindly provided by Gilead Sciences (Foster City, CA).
Cells.
Human embryonic kidney cells (293T cells) were purchased from the ATCC (through LGC Standards, Teddington, United Kingdom) and cultivated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Merelbeke, Belgium) supplemented with 10% fetal calf serum (Perbio Science, Erembodegem, Belgium), 20 μg/ml gentamicin (Invitrogen), and 75 mM NaHCO3 (Invitrogen). U87.CD4.CXCR4, U87.CD4.CCR5, and U87.CD4.CXCR4.CCR5 cells (35) were cultured in DMEM containing 10% fetal calf serum supplemented with 0.2 mg/ml Geneticin (Invitrogen) and 1 μg/ml puromycin (Sigma-Aldrich, Bornem, Belgium). The cell cultures were maintained in a humidified CO2-controlled atmosphere and subcultivated every 2 to 3 days by digestion with trypsin-EDTA (Invitrogen).
Plasmids.
The hemigenomic plasmids p83-2 and p83-10 (16), containing the 5′ half and the 3′ half of the HIV-1 NL4.3 genome, respectively, as well as pT66I (containing the T66I mutation in the NL4.3 integrase [45]), were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH) from Ronald Desrosiers and Vinay K. Pathak, respectively.
The molecular clone pNL4.3-EGFP, containing the gene encoding EGFP between env and nef without affecting expression of any HIV gene, and pNL4.3-Δenv-EGFP, displaying a deletion within the env gene (53), were provided by M. Quiñones-Mateu of The Cleveland Clinic Foundation (Cleveland, OH). The molecular clone pNL4.3-Δenv-EGFP contains a deletion between nucleotides 6404 and 8458.
Viruses. (i) Reference viruses.
For optimization of amplification procedures, a dilution series of the HIV-1 lab strain IIIB (kindly provided by R. C. Gallo, National Institutes of Health, Bethesda, MD) was used.
A CXCR4-tropic (X4) strain was obtained by transfecting pNL4.3-EGFP into 293T cells as described below. The CCR5-tropic (R5) strains Ada and BaL were originally obtained from the NIAID AIDS Research and Reference Reagent Program (Bethesda, MD). The dual-tropic (R5/X4) HIV-1 strain HE was isolated from a Belgian AIDS patient (33).
(ii) Validation samples.
Plasma samples were obtained from patients attending the AIDS Reference Centers at the University Hospitals Leuven (a kind gift of E. Van Wijngaerden).
Plasmids containing the HIV-1 wild-type RT or env sequence were generated by amplifying the RT region from p83-2 or the env region from p83-10, BaL, or HE, using PfuTurbo DNA polymerase (Stratagene, Amsterdam, The Netherlands), and subsequently cloning the sequence by use of a Topo XL PCR cloning kit (Invitrogen). These constructs were used as templates in site-directed mutagenesis experiments to generate the following reference plasmids: p83-10-gp41-D36G (wild type for ENF), p83-10-gp41-D36G-V38M, envBaL-gp41-V38M, envHE-gp41-V38M, RT-Q151M, RT-K70R, RT-K65R, and RT-A62V-S68G-V75I-I77L-F116Y-Q151M. All nucleotide sequences were verified.
Quantification of virus and infected cells. (i) Viral load.
The plasma viral load was determined using Versant HIV-1 RNA 3.0 assay (bDNA assay; Bayer HealthCare, Brussels, Belgium) and Abbott RealTime HIV-1 assay (Abbott Molecular, Louvain-La-Neuve, Belgium).
(ii) Viral core antigen.
HIV-1 core antigen (p24 Ag) in the supernatant was analyzed with an Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (Perkin-Elmer Life and Analytical Sciences, Brussels, Belgium).
(iii) Viral titer.
A viral stock was titrated by plating 20,000 U87.CD4.CCR5.CXCR4 cells per well in a 96-well plate 2 h in advance and infecting them afterwards with 1/2 serial viral dilutions (in triplicate). Twenty-four hours after infection, the supernatant was removed, and cells were washed with phosphate-buffered saline (PBS), trypsinized, and fixed with 2% paraformaldehyde in PBS. Subsequently, EGFP expression was quantified by flow cytometry.
(iv) Cells infected with EGFP-tagged virus.
The percentage of EGFP-expressing cells was determined using a FACSCantoII flow cytometer (BD Biosciences, Erembodegem, Belgium) equipped with a high-throughput sampler. EGFP was excited using a 488-nm-wavelength argon-ion laser, and EGFP expression was detected using a 530/30-nm-band-pass filter. Data were analyzed using FACSDiva v5.0.2 software (BD Biosciences). Forward- versus side-scatter plots were used to exclude dead cells and debris from analysis. Acquisition was stopped when 10,000 gated events were counted. Analytical gates were set in such a manner that fewer than 0.1% of uninfected cells were within the EGFP-positive region.
Extraction. (i) RNA extraction.
For clinical samples and their IIIB reference, 1-ml plasma samples or virus supernatant was ultracentrifuged at 37,100 × g for 1 h to pellet the virus, and samples were extracted subsequently using the extraction procedure from the Viroseq HIV-1 genotyping system (Abbott Molecular, Louvain-La-Neuve, Belgium).
For recombinant viruses, 20 μl of virus supernatant was incubated with 560 μl AVL lysis buffer (Qiagen, Venlo, The Netherlands) for 10 min at room temperature. Subsequently, viral RNA was precipitated with isopropanol, washed with 70% ethanol, and resuspended in 50 μl of RNase-free water (Sigma-Aldrich).
(ii) DNA extraction.
Plasmid DNA was purified using a QIAprep Spin miniprep kit (Qiagen) or an Endofree plasmid maxi kit (Qiagen) when DNA was used for transfection experiments.
Primer design.
The PCR and sequencing primers were designed using sequence alignments for several HIV-1 group M strains (19). Primers were developed and analyzed using Oligo software (Medprobe, Oslo, Norway). The primers were synthesized by Invitrogen.
cDNA synthesis and amplification.
Ten microliters of a 50-μl RNA extract was reverse transcribed and amplified in a one-step RT-PCR, using the SuperScript III one-step RT-PCR system with Platinum Taq high-fidelity polymerase (Invitrogen) in the presence of 10 U of Protector RNase inhibitor (Roche Diagnostics, Vilvoorde, Belgium). Inner PCR products were obtained by adding 1 to 5 μl outer PCR product to an inner PCR mix, using the Expand high-fidelity PCR system (Roche Diagnostics). Primers and cycling conditions are displayed in Tables 1 and 2, respectively. Amplification products were separated in a 1% agarose gel and visualized by ethidium bromide staining. The images were processed on a video imager (ImageMaster VDS; GE Healthcare, Diegem, Belgium).
TABLE 1.
Primers used for amplification of different HIV-1 regions
| Amplified region | Inner or outer region | Primer name | Primer sequence (5′-3′) | Positionb | Sense or antisense | Reference |
|---|---|---|---|---|---|---|
| Gag-PR | Outer | KVL064 | GTTGTGTGACTCTGGTAACTAGAGATCCCTCAGA | 570-603 | Sense | 49 |
| Outer | KVL065 | TCCTAATTGAACYTCCCARAARTCYTGAGTTC | 2797-2828 | Antisense | 49 | |
| Inner | KVL066 | TCTCTAGCAGTGGCGCCCGAACAG | 626-649 | Sense | 49 | |
| Inner | KVL067 | GGCCATTGTTTAACYTTTGGDCCATCC | 2597-2623 | Antisense | 49 | |
| PRa | Outer | KVL064 | GTTGTGTGACTCTGGTAACTAGAGATCCCTCAGA | 570-603 | Sense | 49 |
| Outer | KVL065 | TCCTAATTGAACYTCCCARAARTCYTGAGTTC | 2797-2828 | Antisense | 49 | |
| Inner | AV75 | TGTACTGAGAGACAGGCTAATTTTTTAGGG | 2065-2094 | Sense | ||
| Inner | KVL067 | GGCCATTGTTTAACYTTTGGDCCATCC | 2597-2623 | Antisense | 49 | |
| RT | Outer | AV190-1 | GCTACAYTAGAAGAAATGATGACAGCAT | 1810-1838 | Sense | 42 |
| Outer | CR1 | GATTCTACTACTCCTTGACTTTGGGGATTGTAGGGAA | 4687-4651 | Antisense | 42 | |
| Inner | KVL098 | GGAAGCTCTATTAGAYACAGGAGCAGAT | 2312-2339 | Sense | ||
| Inner | KVL099 | CTGGACTACAGTCTACTTGTCCATG | 4380-4404 | Antisense | ||
| IN | Outer | KVL068 | AGGAGCAGAAACTTWCTATGTAGATGG | 3854-3880 | Sense | 50 |
| Outer | KVL069 | TTCTTCCTGCCATAGGARATGCCTAAG | 5955-5981 | Antisense | 50 | |
| Inner | KVL070 | TTCRGGATYAGAAGTAAAYATAGTAACAG | 4013-4041 | Sense | 50 | |
| Inner | KVL084 | TCCTGTATGCARACCCCAATATG | 5243-5265 | Antisense | 50 | |
| env | Outer | EnvA | GCCTTAGGCATCTCCTATGGCAGGAAGAA | 5953-5981 | Sense | 14 |
| Outer | KVL009 | GCCAATCAGGGAAGWAGCCTTGTGT | 9135-9159 | Antisense | 47 | |
| Inner | EnvB | AGAAAGAGCAGAAGACAGTGGCAATGA | 6198-6224 | Sense | 14 | |
| Inner | HIV-8726-R | TTGTACTACTTCTATAACCCTATCTGT | 8690-8716 | Antisense |
The inner PCR for amplification of PR can also be performed on the RT outer PCR product.
Positions according to pNL4.3 sequence (GenBank accession no. AF324493).
TABLE 2.
Thermal cycling profiles for amplification of different HIV-1 regions
| PCR round | Cycling profile for amplified region
|
||||
|---|---|---|---|---|---|
| gag-PR | PR | RT | IN | env | |
| Outer RT-PCR | 55°C for 30 min | 55°C for 30 min | 55°C for 30 min | 55°C for 30 min | 55°C for 30 min |
| 94°C for 2 min | 94°C for 2 min | 94°C for 2 min | 94°C for 2 min | 94°C for 2 min | |
| 94°C for 15 s | 94°C for 15 s | 94°C for 15 s | 94°C for 15 s | 94°C for 15 s | |
| 57°C for 30 s and 68°C for 2 min (40 times) | 57°C for 30 s and 68°C for 2 min (40 times) | 61°C for 30 s and 68°C for 3 min (40 times) | 53°C for 30 s and 68°C for 2 min 30 s (40 times) | 55°C for 30 s and 68°C for 3 min 30 s (40 times) | |
| 68°C for 5 min | 68°C for 5 min | 68°C for 5 min | 68°C for 5 min | 68°C for 5 min | |
| 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | |
| Inner PCRa | 95°C for 2 min | 94°C for 2 min | 94°C for 2 min | 94°C for 2 min | 94°C for 2 min |
| 95°C for 15 s | 94°C for 15 s | 94°C for 15 s | 94°C for 15 s | 94°C for 15 s | |
| 58°C for 30 s and 68°C for 2 min 30 s (40 times) | 57°C for 30 s and 72°C for 1 min (40 times) | 54°C for 30 s and 72°C for 2 min (40 times) | 53°C for 30 s and 72°C for 1 min 30 s (40 times) | 55°C for 30 s and 68°C for 2 min 30 s (40 times) | |
| 72°C for 10 min | 72°C for 7 min | 72°C for 7 min | 72°C for 7 min | 68°C for 5 min | |
| 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | 4°C (infinite) | |
Starting from cycle 11, the extension time was elongated by 5 s for each cycle, and the annealing temperature for PR amplification was decreased to 55°C.
Sequencing.
PCR products for population sequencing were purified with Microspin S-400 (GE Healthcare). Sequencing was performed using an ABI Prism BigDye Terminator v3.1 ready reaction cycle sequencing kit (42, 47, 49, 50). The sequencing reactions were run on an ABI 3100 genetic analyzer (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands). The sequences were analyzed using Sequence Analysis, version 3.7, and SeqScape, version 2.0 (Applied Biosystems).
Transfection, subcultivation, and harvest of recombinant viruses.
On the day before transfection, 700,000 293T cells were subcultivated in a 5-cm dish with 5 ml of DMEM. Two micrograms of purified PCR product (QIAquick PCR purification kit; Qiagen) was mixed and coprecipitated with 10 μg of XbaI-digested proviral vector. DNA and CaCl2 were gently mixed and added to an equal volume of 2× HEPES-buffered saline. After 20 min of incubation at room temperature, the mixture was added to the 293T cells. The medium was refreshed with 5 ml DMEM after overnight incubation. Transfection was monitored through fluorescence microscopy as described previously (5). Two days after transfection, 5 ml supernatant was transferred to freshly seeded U87.CD4.CCR5.CXCR4 cells. Cell cultures were monitored for EGFP expression by fluorescence microscopy. Virus supernatants were harvested by low-speed centrifugation and stored in 1-ml aliquots at −80°C for further use.
Drug susceptibility testing.
One hundred microliters of medium containing 20,000 U87.CD4.CCR5.CXCR4 cells was seeded within each well of a 96-well tray. Two hours later, triplicate fivefold serial dilutions of the drugs (RT inhibitors, IN inhibitors, and entry inhibitors) were performed within the tray, followed by the addition of 100 μl of virus-containing medium to reach a final volume of 200 μl/well. Virus was prediluted based upon viral titration experiments to obtain 10% EGFP-expressing U87.CD4.CCR5.CXCR4 cells in each well, without any inhibition of antiviral drugs. For MVC and AMD3100, infection was not carried out until half an hour after the addition of the compound to allow for interaction of the compounds with CCR5 and CXCR4, respectively. Twenty-four hours after infection, the percentage of EGFP-expressing cells was determined by flow cytometry. The 50% inhibitory concentration (IC50) was calculated according to the method of Reed and Muench (37).
For PI susceptibility testing, 100 μl medium containing 15,000 U87.CD4.CCR5.CXCR4 cells was seeded within each well of a 96-well tray. Two hours later, 100 μl of virus-containing medium was added. Another 3 h later, supernatant was replaced with 200 μl medium containing fivefold dilutions of the PI. Forty hours after infection, the supernatant was transferred to U87.CD4.CCR5.CXCR4 cells seeded at a density of 20,000 cells/well 2 h earlier. Twenty-four hours after the second infection round, the percentage of EGFP-expressing cells was determined by flow cytometry. The IC50 was calculated according to the method of Reed and Muench (37).
RESULTS
Construction of molecular proviral clones.
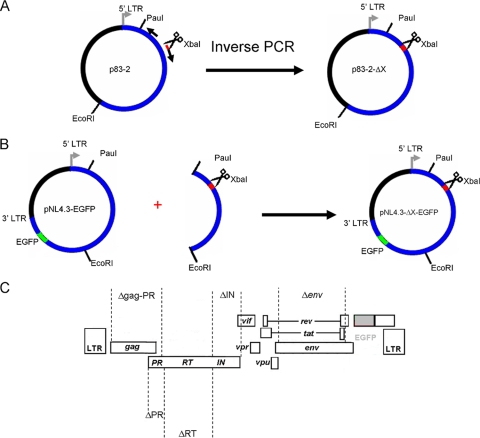
The plasmids pNL4.3-ΔgagPR-EGFP, pNL4.3-ΔPR-EGFP, pNL4.3-ΔRT-EGFP, and pNL4.3-ΔIN-EGFP were generated by deleting specific regions from the p83-2 plasmid by inverse PCR using PfuUltra high-fidelity DNA polymerase (Stratagene) (Fig. 1A). The primers were designed such that each encoding fragment was removed and an XbaI restriction enzyme recognition site was inserted in the deleted region. The following combinations of primers were used to generate the deletions: for the gag-PR region, 5′-TCTAGATTTTCCCATTAGTCCTATTGAGACTGTACCAG-3′ (positions 2546 to 2577 in pNL4.3 [GenBank accession no. AF324493]) and 5′-CGACGCTCTCGCACCCATCTCTG-3′ (positions 785 to 807); for the PR region, 5′-TCTAGACCCATTAGTCCTATTGAGACTGTACCAGTA-3′ (positions 2550 to 2579) and 5′-GAAGCTAAAGGATACAGTTCCTTGTCTATCG-3′ (positions 2222 to 2252); for the RT region, 5′-TCTAGATTTTT AGATGGAATAGATAAGGCCCAAGAAGAA-3′ (positions 4230 to 4262) and 5′-AAAATTTAAAGTGCAGCCAATCTGAGTCAACAG-3′ (positions 2517 to 2549); and for the IN region, 5′-CACATGGAAAAGATTAGTAAAACACCATATGTATATTTC-3′ (positions 5097 to 5135) and 5′-TCTAGATAGTACTTTCCTGATTCCAGCACTGACCA-3′ (positions 4201 to 4229). The XbaI restriction enzyme recognition site is shown in bold, and the first codons upstream and downstream of the gag-PR, PR, RT, and IN coding regions are shown in italics. Parental DNA was digested with DpnI (Fermentas), and the PCR fragments were self-ligated using T4 DNA ligase (Invitrogen). The PauI-EcoRI fragments of the deleted p83-2 plasmids were subcloned into pNL4.3-EGFP (Fig. 1B). The full-length HIV-1 sequences of all molecular proviral clones were verified.
FIG. 1.
Construction of molecular proviral clones containing deletions and the EGFP reporter gene. ΔX indicates a deletion of gag-PR, PR, RT, or IN. The deletions were generated in the 5′-hemigenomic molecular clone p83-2 by inverse PCR and self-ligation (A), followed by subcloning into pNL4.3-EGFP (B). (C) Schematic representation of generated vectors. (Adapted from reference 53 with permission of the publisher.)
No mutations in comparison to the parental DNA were observed, and sequencing confirmed deletions going from p17 amino acid 7 to PR amino acid 98 in pNL4.3-ΔgagPR-EGFP, from PR amino acids 1 to 99 in pNL4.3-ΔPR-EGFP, and from IN amino acids 1 to 288 in pNL4.3-ΔIN-EGFP. Sequencing confirmed the deletion of amino acids 1 to 560 of RT in pNL4.3-ΔRT-EGFP but also revealed the presence of one mutation in comparison to the parental DNA. At position 2528, a G-to-T mutation was present, which would change a glutamine to a histidine at position 94 in PR. Since this mutation was close to the XbaI restriction site (positions 2550 to 2556), it was within the part of the virus that was derived from the PCR amplicon after recombination (as verified by sequencing of recombinant virus [data not shown]) and thus did not interfere with susceptibility testing. pNL4.3-Δenv-EGFP displayed a deletion from gp120 amino acid 34 to gp41 amino acid 237 (53).
Amplification performance.
The primers were designed to enable the amplification of divergent strains from different HIV-1 group M subtypes and the amplification of fragments sufficiently long to enable homologous recombination with the respective molecular proviral clones (at least 100 bp of overlap) (Table 1). Cycling conditions were optimized with a dilution series of HIV-1 IIIB in PBS, ranging from 1,000,000 to 10 RNA copies/ml (Table 2). The optimized nested PCR procedures were specific, generating only a single amplification band, and they were very sensitive (detecting 10 to 100 RNA copies/ml).
Subsequently, they were each validated on a set of clinical samples belonging to a broad range of HIV-1 group M subtypes (A, B, C, D, F, G, H, J, CRF01, CRF02, CRF03, CRF05, CRF12, and CRF13) and displaying a viral load range of 200 to >500,000 RNA copies/ml (Table 3). The success rate varied from 91 to 100% depending on the viral load range and from 95 to 100% depending on the specific assay. The total amplification success rate was 97% (including only samples above the cutoff of 1,000 RNA copies/ml recommended for resistance testing in clinical practice).
TABLE 3.
Sensitivities of amplification assays for gag-PR, PR, RT, IN, and env from clinical HIV-1 samplesa
| Viral load (RNA copies/ml) | Success rate (% [no. of successes/no. of attempts])
|
|||||
|---|---|---|---|---|---|---|
| Gag-PR (1,998 bp) | PR (559 bp) | RT (2,093 bp) | IN (1,253 bp) | Env (2,519 bp) | Total | |
| 200-1,000 | 100 (1/1) | 100 (3/3) | 100 (3/3) | 90 (9/10) | 100 (11/11) | 96 (27/28) |
| 1,000-5,000 | 89 (8/9) | 90 (12/13) | 90 (7/8) | 89 (17/19) | 100 (5/5) | 91 (49/54) |
| 5,000-10,000 | 100 (6/6) | 100 (9/9) | 100 (5/5) | 89 (8/9) | 100 (1/1) | 97 (29/30) |
| 10,000-100,000 | 100 (15/15) | 100 (21/21) | 100 (26/26) | 100 (27/27) | 100 (2/2) | 100 (91/91) |
| >100,000 | 100 (8/8) | 100 (5/5) | 100 (9/9) | 100 (9/9) | 100 (3/3) | 100 (34/34) |
| Total | 97 (38/39) | 98 (50/51) | 98 (50/51) | 95 (70/74) | 100 (22/22) | 97 (230/237) |
Samples belonged to the genetic forms A (12%), B (17%), C (9%), D, (8%) F (6%), G (8%), H (1%), J (4%), CRF01_AE (8%), CRF02_AG (11%), CRF03_AB (3%), CRF05_DF (1%), CRF12_BF (5%), and CRF13_cpx (5%) or were a unique recombinant form (2%), as determined by the Rega subtyping tool (9; http://www.bioafrica.net/subtypetool/html/). The failed amplifications of the PR and RT regions and one in the IN region were performed on the same sample, belonging to CRF03_AB, and we believe that this can be attributed to poor quality of the extracted RNA (50). Since no more sample was available, we could not reextract the RNA. The other failed amplifications belonged to subtype B (gag-PR), subtype C, CRF01_AE, and CRF02_AG (IN).
Production and characterization of replication-competent recombinant viruses from reference strains and clinical samples.
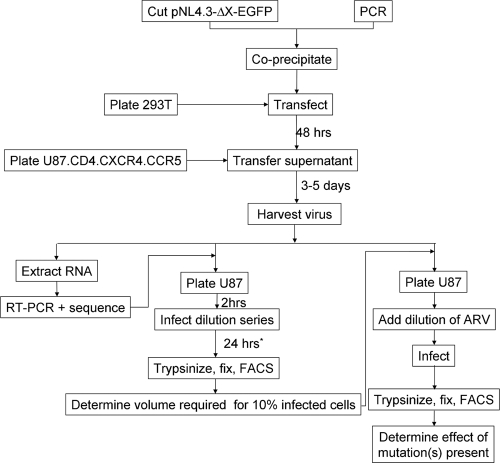
At first, the transfection, homologous recombination, and subcultivation procedures were fine-tuned for the X4 strain NL4.3, starting from amplicons covering the gag-PR, PR, RT, IN, and env regions of the NL4.3 template and the respective XbaI-cut molecular proviral clones, i.e., pNL4.3-ΔgagPR-EGFP, pNL4.3-ΔPR-EGFP, pNL4.3-ΔRT-EGFP, pNL4.3-ΔIN-EGFP, and pNL4.3-Δenv-EGFP. Using optimized conditions (as mentioned in Materials and Methods), virus supernatant could typically be harvested 3 to 5 days after infection of U87.CD4.CCR5.CXCR4 cells (see Fig. 4).
FIG. 4.
Schematic overview of recombinant virus assay. *, for testing of susceptibility to PIs, the titration procedure as well as the IC50 determination should be modified as indicated in the text. FACS, fluorescence-activated cell sorting; ARV, antiretroviral.
Next, replication-competent viruses containing env genes of different R5 subtype B strains (BaL and Ada) and a dual-tropic (R5/X4) HIV-1 strain (HE) were generated by the same methodology. Potential changes in the tropism of the virus (due to growth on cells expressing high levels of both CXCR4 and CCR5) were controlled by infecting U87.CD4.CXCR4 cells and U87.CD4.CCR5 cells with the respective recombinant viruses, and no changes in tropism were observed (as monitored by fluorescence microscopy), even after cultivation on U87.CD4.CCR5.CXCR4 cells for more than 2 weeks (data not shown).
Finally, 22 recombinant viruses with recombination within the different regions were generated starting from several clinical HIV-1 samples. Irrespective of the viral load (range, 2,000 to 120,000 RNA copies/ml), subtype (A, B, C, D, G, J, and CRF02_AG included), and drug resistance profile, the procedure was successful in generating replication-competent recombinant viruses within a similar time frame to that for the reference strains. For the samples that were tested, no major selection biases occurred, as observed by population sequencing of the harvested recombinant strains. Sequences from all viruses that were generated using the PCR product from a single cloned sequence were identical to the original clone from which they were derived. For a few recombinant viruses generated directly from amplified plasma samples, population mixtures present in the plasma sample were no longer present in the recombinant virus stock. All major resistance-associated mutations were unaltered, and the observed changes in mixtures at positions other than major resistance positions did not change the resistance level according to the Rega interpretation system (46; data not shown). Differences in gag between the original isolate and the recombinant virus were E62KE, A81TA, QL108L, KR411K, and NS441S for a subtype G sample, KR28R, IM31I, ND47N, and IML104IM for a CRF02_AG sample, and T239TA and F463FV for a subtype J sample. In PR, the following changes were observed: PS39PAS in a subtype D sample and VL33V in a CRF02_AG sample. For IN recombinants, the following changes were observed: IM50I and M281MV for a subtype A sample and T124TA for a subtype C strain.
Determination of optimal time point for quantification of EGFP-expressing cells as a marker for viral titer.
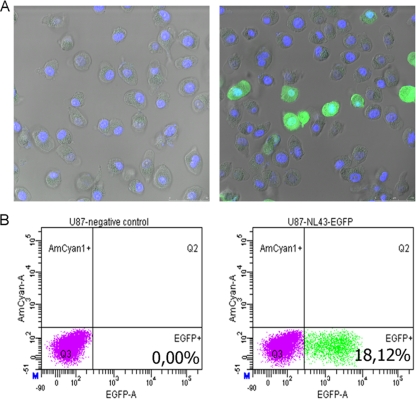
Taking into account practical considerations and the aim to develop a single-cycle assay, it was necessary to investigate whether infection of U87.CD4.CCR5.CXCR4 cells could accurately be quantified at 24 h postinfection based upon EGFP expression. Both fluorescence microscopy and flow cytometry allowed for detection of EGFP expression at this time point (Fig. 2). Moreover, a linear relationship was observed between volume of viral input and the percentage of EGFP-expressing cells (as determined by flow cytometry).
FIG. 2.
Discrimination between infected and noninfected cells. U87.CD4.CCR5.CXCR4 cells were infected with HIV-1-NL4.3-EGFP (right) or were mock infected (left). Twenty-four hours later, cells were washed with PBS, trypsinized, and fixed in 2% paraformaldehyde for 10 min. (A) Afterwards, nuclei were stained for 5 min with DAPI (4′,6-diamidino-2-phenylindole). Subsequently, cells were washed three times with PBS, resuspended in PBS, and visualized by fluorescence microscopy. (B) After fixation, cells were analyzed by flow cytometry.
Susceptibility testing with PIs.
In using expression of recombinant virus-encoded EGFP as a marker for viral production (5), drug susceptibility testing needs to be adjusted to determine PI susceptibility profiles. This is because PIs act by inactivating the viral PR produced in the first replication cycle, rendering the produced virus noninfectious, and thus the infectivity can be measured only in a second replication cycle. Therefore, the viral production after a first round of infection in the presence of a dilution series of PI was quantified in a subsequent viral titration experiment. The optimal time point for transferring the supernatant from the first round to the second round was determined based upon several experiments in which p24 Ag production and virus production were determined for different reference strains and clinical samples (data not shown). Typically, production of virus did not start before 30 to 32 h postinfection, ensuring that transfer of supernatant at 40 to 42 h to the subsequent viral titration procedure generated sufficient virus for reproducible measurements and that only infectious particles produced by cells that were initially infected were detected. Additional experiments revealed that correction for the percentage of infected cells during the first cycle was not necessary to measure the IC50 for PIs (data not shown).
Reproducibility and accuracy of drug resistance testing.
In order to assess the ability of the novel assay to determine susceptibility, the susceptibility of wild-type NL4.3 virus toward different inhibitors was first determined (Table 4). Since NL4.3 is CXCR4 tropic, it is not inhibited by MVC. Therefore, MVC was tested on a recombinant virus containing the envelope of the R5 strain BaL, and inhibition was indeed observed. For ENF, susceptibility was tested for three different recombinant viruses that harbored the envelope of either an X4 (NL4.3), an R5 (BaL), or an R5/X4 (HE) strain. The coefficients of variation ranged from 7 to 27%, with an outlier of 48% for testing of susceptibility of BaL against ENF.
TABLE 4.
Intra- and interassay reproducibility of HIV-1 susceptibility testing with several inhibitors from different drug classes
| Recombinant virus | Inhibitor | IC50 (ng/ml)a | SD (ng/ml)b | CV (%)c | FCIC50d | SD of FCIC50e | CV of FCIC50f |
|---|---|---|---|---|---|---|---|
| RT-NL4.3 | DDI | 565.4 | 54 | 9.6 | |||
| AZT | 1.29 | 0.35 | 27 | ||||
| ABC | 392 | 43 | 11 | ||||
| D4T | 4.00 | 0.58 | 14 | ||||
| TDF | 390 | 27 | 7 | ||||
| 3TC | 84.8 | 15 | 18 | ||||
| EFV | 1.77 | 0.25 | 14 | ||||
| NVP | 18.5 | 3.15 | 17 | ||||
| RT-NL4.3-Q151 M | AZT | 1.48 | 0.11 | 7.4 | |||
| ABC | 2.68 | 0.46 | 17 | ||||
| RT-NL4.3-K70R | D4T | 0.95 | 0.15 | 15 | |||
| PR-NL4.3 | RTV | 75.2 | 18.3 | 24 | |||
| NFV | 1.66 | 0.29 | 17 | ||||
| PR-CSg | NFV | 9.52 | 1.15 | 12 | |||
| IN-NL4.3 | RAL | 6.3 | 1.1 | 17 | |||
| EVG | 0.40 | 0.06 | 15 | ||||
| IN-NL4.3-T66I | EVG | 5.22 | 1.63 | 31 | |||
| Env-NL4.3 | ENF | 85 | 11 | 13 | |||
| AMD3100 | 32 | 4 | 13 | ||||
| MVC | No inhibition | ||||||
| Env-NL4.3-V38 M | ENF | 5.26 | 1.57 | 30 | |||
| Env-He | ENF | 110 | 16 | 15 | |||
| Env-BaL | ENF | 62 | 30 | 48 | |||
| MVC | 0.92 | 0.14 | 15 |
Means for one triplicate experiment.
Standard deviations of IC50s for one triplicate experiment.
Coefficients of variation of IC50s for one triplicate experiment.
Mean fold changes in IC50 (FCIC50) versus wild-type NL4.3 (env-gp41 D36G in the case of ENF) for three independent experiments.
Standard deviations of FCIC50 for three independent experiments.
Coefficients of variation of FCIC50 for three independent experiments.
Recombinant virus generated from a cloned PR of a clinical sample belonging to subtype B that contains I13V, I15V, K20R, E35D, M36I, I54V, L63P, I64V, A71V, N88D and L90M mutations in PR.
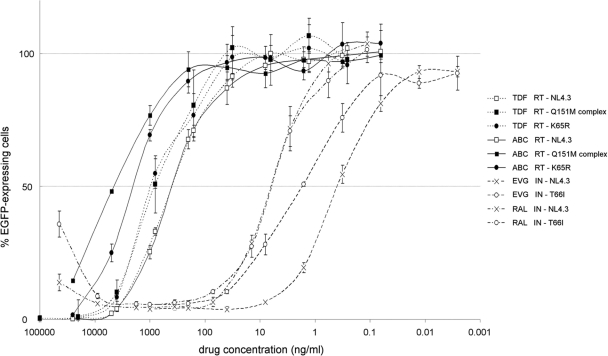
Next, recombinant viruses containing specific mutations were generated using plasmids created by site-directed mutagenesis or cloned patient samples with known resistance mutations as templates for PCR. The change in drug susceptibility relative to that of the wild type was determined (Table 5). Representative examples of susceptibility curves are shown in Fig. 3. All measurements of altered susceptibility were consistent with the scientific literature in terms of magnitude and specificity. For example, the T66I mutation in the IN gene is known to confer resistance to EVG but not to RAL (18). When several resistance-associated mutations were included in the recombinant virus, an increase in the degree of change in susceptibility was observed (e.g., Q151M complex mutation versus single Q151M mutation). Lastly, inclusion of the V38M mutation in gp41 of X4, R5, or R5/X4 strains resulted in a decreased susceptibility relative to that of recombinant viruses carrying the respective wild-type envelopes, and no evidence of cross-resistance to the CXCR4 receptor antagonist AMD3100 was observed.
TABLE 5.
Accuracy of susceptibility testing of mutant HIV-1 recombinant viruses toward PR, RT, IN, and entry inhibitors
| Recombined region | Mutation(s)a | Inhibitor | Fold change in IC50b | Referencec |
|---|---|---|---|---|
| gag-PR | I13V, I15V, K20R, E35D, M36I, I54V, L63P, I64V, A71V, N88D, L90M* | NFV | 7.75 | 25 |
| PR | I13V, I15V, K20R, E35D, M36I, I54V, L63P, I64V, A71V, N88D, L90M* | NFV | 9.46 | 25 |
| RT | Q151M | AZT | 1.60 | 34 |
| ABC | 2.76 | 34 | ||
| TDF | 0.85 | 44 | ||
| K70R | AZT | 1.71 | 30 | |
| ABC | 0.95 | 30 | ||
| TDF | 0.85 | 55 | ||
| K65R | AZT | 1.13 | 40 | |
| ABC | 5.17 | 40 | ||
| TDF | 2.41 | 44 | ||
| A62V, S68G, V75I, F77L, F116Y, Q151M | AZT | 79.07 | 29 | |
| ABC | 11.66 | 29 | ||
| TDF | 2.11 | 29 | ||
| M41L, D67N, K70R, V179I, M184V, Y188L, T215Y** | AZT | 10.10 | 20 | |
| ABC | 23.26 | 20 | ||
| TDF | 2.02 | 31 | ||
| 3TC | >1,100 | 20 | ||
| EFV | 111.80 | 25 | ||
| NVP | >100 | 25 | ||
| IN | T66I | RAL | 0.98 | 18 |
| EVG | 4.29 | 18 | ||
| env | gp41, D36G, V38M | ENF | 5.88 | 22 |
| AMD3100 | 1.15 | 39 | ||
| Env-He | AMD3100 | No inhibition | 35 | |
| MVC | No inhibition | 35 | ||
| Env-He-gp41 V38M | ENF | 2.56 | 39 | |
| Env-BaL | AMD3100 | No inhibition | 35 | |
| Env-BaL-gp41 V38M | ENF | 4.19 | 39 |
Mutations were generated by site-directed mutagenesis, except for the two strains that originated from a clonal sequence from a patient sample (subtype B) (*) and one strain that originated from a clonal sequence from another patient sample (subtype C) (**).
Relative to the wild-type recombinant virus recombined in the respective region (gag-PR-p83-2, PR-p83-2, IN-p83-2, env-p83-10-gp41-D36G, env-He, or env-BaL) (triplicate experiment).
Published reference in which comparable results were obtained.
FIG. 3.
Representative examples of susceptibility curves for different inhibitors and recombinant viruses. Means and standard deviations of percentages of infection relative to the positive control are shown (data for triplicate experiments are shown). Susceptibility curves are shown for wild-type virus (open squares with dashed line) and RT recombinant virus with A62V, S68G, V75I, F77L, F116Y, and Q151M mutations (filled squares with dashed line) or K65R mutation (filled circles with dashed line) against TDF; for wild-type virus (open squares with solid line) and RT recombinant virus with A62V, S68G, V75I, F77L, F116Y, and Q151M mutations (filled squares with solid line) or K65R mutation (filled circles with sold line) against abacavir; for wild-type virus (× with dashed line) and IN recombinant virus with T66I mutation (open circles with dashed line) against EVG; and for wild-type virus (× with dotted and dashed line) and IN recombinant virus with T66I mutation (filled circles with dotted and dashed line) against RAL.
Assay reproducibility for at least one inhibitor of each drug class was evaluated by three independent determinations of the change in IC50 for mutant viruses relative to the wild type (Table 4). The coefficients of variation ranged from 7 to 31%, with the highest variability for EVG and ENF.
DISCUSSION
The use of highly active antiretroviral therapy in the management of HIV disease has resulted in a decrease in HIV-related morbidity and mortality (27). Nevertheless, therapy failure does occur often, with drug resistance as a consequence. The composition of subsequent regimens is guided mostly by genotypic drug resistance testing in the current clinical setting. In cases of multiple failures with broad-drug-class cross-resistance or of failures with new drugs for which genotype-clinical response correlations have not yet been established or are limited, phenotypic drug resistance testing can help in clinical decision-making. Apart from this, phenotypic testing also provides a means to investigate qualitative and quantitative effects of novel mutations or combinations of mutations on drug susceptibilities and is thus a useful tool to improve genotypic interpretation systems.
For practical reasons, such as hands-on time and reproducibility issues, phenotyping is done mostly by recombinant virus assays. Despite the use of different testing strategies, the available assays show good correlations, although care should be taken in extrapolating values near the cutoffs of the different formats (36, 52). The addition of ENF and MVC to the existing range of drugs in clinical practice prompted the need for assays applicable to viruses capable of using either coreceptor. In this respect, the assay described here is a single-cycle format, using U87.CD4.CXCR4.CCR5 cells, based on homologous recombination between patient-derived amplicons and molecular proviral clones (Fig. 4). The proviral clones were tagged with the EGFP reporter gene, and relevant viral genomic regions were removed (p17 codon 7 to PR codon 98 in pNL4.3-ΔgagPR-EGFP, PR codons 1 to 99 in pNL4.3-ΔPR-EGFP, RT codons 1 to 560 in pNL4.3-ΔRT-EGFP, IN codons 1 to 288 in pNL4.3-ΔIN-EGFP, and gp120 codon 34 to gp41 codon 237 in pNL4.3-Δenv-EGFP). Only minor modifications to the described technology would be required to enable susceptibility testing for drugs addressing novel interaction points within the viral replication cycle.
Our assay was optimized for low viral loads and all group M subtypes. Although limited information is available on potential artifacts due to forced intersubtype recombination, intersubtype recombination does occur often in vivo, and recombinant viruses obtained in vitro are clearly viable. Additionally, one in vitro study suggested that using a subtype C backbone as opposed to a subtype B backbone did not significantly alter drug susceptibility measurements (4). The optimized experimental conditions enabled the investigation of patient samples regardless of viral load or subtype. The overall extraction and amplification success rate for a set of clinical samples that belonged to the genetic forms A-J, CRF01-03, CRF05, and CRF12-13 within group M was 97%, even if taking only samples above the clinically useful cutoff of 1,000 RNA copies/ml into account. One concern is how well the initial viral population within the patient is represented within the recombinant virus population, as RNA extraction, amplification, and in vitro cultivation represent potential bottlenecks. To address these issues as much as possible, primers were carefully chosen to target the most conserved sites surrounding the viral regions of interest, and cultures were limited in time. No major selection bias occurred, as observed by sequencing comparison between original plasma samples and recombinant virus stocks.
Drug susceptibility values are based on discrimination between infected and noninfected cells by measuring EGFP expression on a single-cell level through flow cytometry. Standard deviations and coefficients of variation were within the ranges found for other recombinant virus assays, and changes were consistent with the scientific literature in terms of magnitude and specificity (15, 17, 31, 34). The single-cycle format ensures very little sequence evolution and selection bias against less replication-competent variants and ensures that potential effects of mutations present in the recombinant virus on interaction with calmodulin or other cellular proteins involved in apoptosis will have no influence on the assay readout (26). An additional value lies in the potential to study recently described resistance mechanisms. Since almost the complete envelope sequence is recombined (starting from codon 34 and stretching to amino acid 237 of gp41), the impacts of mutations within different sections of gp120 on ENF and MVC susceptibilities can be investigated (10, 21, 38, 54; J. Heera, M. Saag, P. Ive, J. Whitcomb, M. Lewis, L. McFadyen, J. Goodrich, H. Mayer, E. van der Ryst, and M. Westby, oral presentation 40LB, 15th Conference on Retroviruses and Opportunistic Infections, Boston, MA, 2008). The recombination of the entire RT sequence enables the measurement of the influence of mutations within the connection and RNase H domains of RT on RT inhibitor susceptibilities (41, 56). Lastly, the potential to make both PR and gag-PR recombinants allows for investigation of the impacts of both cleavage site and non-cleavage-site mutations within gag on sensitivity to PIs (12, 24, 28, 32).
In conclusion, we have optimized a single-cycle recombinant drug susceptibility assay that allows an accurate and reproducible measurement of susceptibility to all currently approved HIV-1 drugs for patient samples belonging to the major genetic forms within group M. This assay could help to decipher the different pathways leading to drug resistance and could thus improve existing genotypic interpretation systems.
Acknowledgments
We are thankful to Kristien Erven and Liesbet De Dier for performing p24 Ag enzyme-linked immunosorbent assays.
This work was supported by the AIDS Reference Laboratory of Leuven, which receives support from the Belgian Ministry of Social Affairs through a fund within the Health Insurance System, by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (grants G.0266.04 and G.485.08), by the Centers of Excellence of the Katholieke Universiteit Leuven (krediet EF/05/015), and by IUAP grant P6/41. Kris Covens was funded by a Ph.D. grant of the Institute for the Promotion of Innovation through Sciences and Technology in Flanders (IWT).
The molecular clones pNL4.3-EGFP and pNL4.3-Δenv-EGFP were provided by M. Quiñones-Mateu of The Cleveland Clinic Foundation (Cleveland, OH). All rights, title, and interest in these materials are owned by The Cleveland Clinic Foundation.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Abecasis, A. B., K. Deforche, J. Snoeck, L. T. Bacheler, P. McKenna, A. P. Carvalho, P. Gomes, R. J. Camacho, and A. M. Vandamme. 2005. Protease mutation M89I/V is linked to therapy failure in patients infected with the HIV-1 non-B subtypes C, F or G. AIDS 191799-1806. [DOI] [PubMed] [Google Scholar]
- 2.Abecasis, A. B., K. Deforche, L. T. Bacheler, P. McKenna, A. P. Carvalho, P. Gomes, A. M. Vandamme, and R. J. Camacho. 2006. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02_AG. Antivir. Ther. 11581-589. [PubMed] [Google Scholar]
- 3.Brenner, B., D. Turner, M. Oliveira, D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17F1-F5. [DOI] [PubMed] [Google Scholar]
- 4.Choe, S. S., E. Stawiski, and N. T. Parkin. 2007. Interpretation of drug susceptibility and replication capacity results from subtype C HIV-1 protease/RT is not influenced by the subtype of the resistance test vector, poster 1f07. Abstr. XV Int. HIV Drug Resistance Workshop 2007.
- 5.Daelemans, D., C. Pannecoucque, G. N. Pavlakis, O. Tabarrini, and E. De Clercq. 2005. A novel and efficient approach to discriminate between pre- and post transcription inhibitors. Mol. Pharmacol. 671574-1580. [DOI] [PubMed] [Google Scholar]
- 6.Deforche, K., T. Silander, R. Camacho, Z. Grossman, M. A. Soares, K. Van Laethem, R. Kantor, Y. Moreau, A. M. Vandamme, and Non-B Workgroup. 2006. Analysis of HIV-1 pol sequences using Bayesian networks: implications for drug resistance. Bioinformatics 222975-2979. [DOI] [PubMed] [Google Scholar]
- 7.Deforche, K., R. Camacho, Z. Grossman, T. Silander, M. A. Soares, Y. Moreau, R. W. Shafer, K. Van Laethem, A. P. Carvalho, B. Wynhoven, P. Cane, J. Snoeck, J. Clarke, S. Sirivichayakul, K. Ariyoshi, A. Holguin, H. Rudich, R. Rodrigues, M. B. Bouzas, P. Cahn, L. F. Brigido, V. Soriano, W. Sugiura, P. Phanuphak, L. Morris, J. Weber, D. Pillay, A. Tanuri, P. R. Harrigan, J. M. Shapiro, D. A. Katzenstein, R. Kantor, and A. M. Vandamme. 2007. Bayesian network analysis of resistance pathways against HIV-1 protease inhibitors. Infect. Genet. Evol. 7382-390. [DOI] [PubMed] [Google Scholar]
- 8.De Meyer, S., T. Vangeneugden, B. Van Baelen, E. De Paepe, H. Van Marck, G. Picchio, E. Lefebvre, and M. P. De Béthune. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retrovir. 24379-388. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira, T., K. Deforche, S. Cassol, M. Salminen, D. Paraskevis, C. Seebregts, J. Snoeck, E. J. van Rensburg, A. M. J. Wensing, D. A. van de Vijver, C. A. Boucher, R. Camacho, and A. M. Vandamme. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 213797-3800. [DOI] [PubMed] [Google Scholar]
- 10.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor ENF is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 748358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. 29 January 2008, posting date. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. AIDSinfo, Rockville, MD. http://AIDSinfo.nih.gov.
- 12.Dierynk, I., S. De Meyer, K. Cao-Van, S. Van Marck, E. Lathouwers, K. Thys, B. Maes, and M. P. de Béthune. 2007. Impact of gag cleavage site mutation on the virological response to darunavir/ritonavir in treatment-experienced patients in POWER 1, 2 and 3. Antivir. Ther. 12S23. [Google Scholar]
- 13.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 703763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, B. H. Hahn, et al. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res. Hum. Retrovir. 101359-1368. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Perez, J., S. Sanchez-Palomino, M. Perez-Olmeda, B. Fernandez, and J. Alcami. 2007. A new strategy based on recombinant viruses as a tool for assessing drug susceptibility of human immunodeficiency virus type 1. J. Med. Virol. 79127-137. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10343-350. [DOI] [PubMed] [Google Scholar]
- 17.Hertogs, K., M. P. De Béthune, V. Miller, T. Ivens, P. Schel, A. V. Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, G., R. Ledford, F. Yu, M. Miller, M. Tsiang, and D. McColl. 2007. Resistance profile of HIV-1 mutants in vitro selected by the HIV-1 integrase inhibitor, GS-9137 (JTK-303), poster 627. Abstr. 14th Conf. Retrovir. Opportun. Infect., Los Angeles, CA.
- 19.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). 2005. HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 20.Lennerstrand, J., D. K. Stammers, and B. A. Larder. 2001. Biochemical mechanism of human immunodeficiency virus type 1 reverse transcriptase resistance to stavudine. Antimicrob. Agents Chemother. 452144-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, M., J. Mori, P. Simpson, J. Whitcomb, X. Li, D. Robertson, and M. Westby. 2008. Changes in the V3 loop sequence associated with failure of maraviroc treatment in patients enrolled in the Motivate 1 and 2 trials, poster 871. Abstr. 15th Conf. Retrovir. Opportun. Infect., Boston, MA.
- 22.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 784628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacArthur, R. D., and R. M. Novak. 2008. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin. Infect. Dis. 47236-241. [DOI] [PubMed] [Google Scholar]
- 24.Maguire, M. F., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J. P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 767398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margot, N. A., E. Isaacson, I. McGowan, A. K. Cheng, R. T. Schooley, and M. D. Miller. 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 161227-1235. [DOI] [PubMed] [Google Scholar]
- 26.Micoli, K. J., O. Mamaeva, S. C. Piller, J. L. Barker, G. Pan, E. Hunter, and J. M. McDonald. 2006. Point mutations in the C-terminus of HIV-1 gp160 reduce apoptosis and calmodulin binding without affecting viral replication. Virology 344468-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, E. L., A. C. Collier, K. A. Kalish, S. F. Assmann, M. F. Para, T. P. Flanigan, P. N. Kumar, L. Mintz, F. R. Wallach, and G. H. Nemo. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 13517-26. [DOI] [PubMed] [Google Scholar]
- 28.Nijhuis, M., N. M. Van Maarseveen, S. Lastere, P. Schipper, E. Coakley, B. Glass, M. Rovenska, D. de Jong, C. Chappey, I. W. Goedegebuure, G. Heilek-Snyder, D. Dulude, N. Cammack, L. Brakier-Gingras, J. Konvalinka, N. Parkin, H. G. Kräusslich, F. Brun-Vezinet, and C. A. B. Boucher. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 4152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, S., R. W. Shafer, and T. C. Merigan. 1999. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS 13661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkin, N. T., S. G. Deeks, M. T. Wrin, J. Yap, R. M. Grant, K. H. Lee, D. Heeren, N. S. Hellmanna, and C. J. Petropoulos. 2000. Loss of antiretroviral drug susceptibility at low viral load during early virological failure in treatment-experienced patients. AIDS 142877-2887. [DOI] [PubMed] [Google Scholar]
- 31.Parkin, N. T., N. S. Hellmann, J. M. Whitcomb, L. Kiss, C. Chappey, and C. J. Petropoulos. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkin, N., C. Chappey, E. Lam, and C. Petropoulos. 2005. Reduced susceptibility to protease inhibitors (PI) in the absence of primary resistance-associated mutations. Antivir. Ther. 10S116. [Google Scholar]
- 33.Pauwels, R., K. Andries, J. Desmyter, D. Schols, M. J. Kukla, H. J. Breslin, A. Raeymaekers, J. Van Gelder, R. Woestenborghs, J. Heykants, K. Schelkens, M. A. C. Janssen, E. De Clercq, and P. A. J. Janssen. 1990. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Lett. Nat. 343470-474. [DOI] [PubMed] [Google Scholar]
- 34.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, T. Huan, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Princen, K., S. Hatse, K. Vermeire, E. De Clercq, and D. Schols. 2004. Establishment of a novel CCR5 and CXCR4 expressing CD4+ cell line which is highly sensitive to HIV and suitable for high-throughput evaluation of CCR5 and CXCR4 antagonists. Retrovirology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qari, S. H., R. Respess, H. Weinstock, E. M. Beltrami, K. Hertogs, B. A. Larder, C. J. Petropoulos, N. Helmann, and W. Heneine. 2002. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J. Clin. Microbiol. 4031-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 38.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, M. L. Sharon, P. Harvey, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 9916249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 794991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Røge, B. T., T. L. Katzenstein, N. Obel, H. Nielsen, O. Kirk, C. Pedersen, L. Mathiesen, J. Lundgren, and J. Gerstoft. 2003. K65R with and without S68: a new resistance profile in vivo detected in most patients failing abacavir, didanosine and stavudine. Antivir. Ther. 8173-182. [PubMed] [Google Scholar]
- 41.Santos, A. F., R. B. Lengruber, E. A. Soares, A. Jere, E. Sprinz, A. M. Martinez, J. Silveira, F. S. Sion, V. K. Pathak, and M. A. Soares. 2008. Conservation patterns of HIV-1 RT connection and RNase H domains: identification of new mutations in NRTI-treated patients. PLoS ONE 3e1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snoeck, J., C. Riva, K. Steegen, Y. Schrooten, B. Maes, L. Vergne, K. Van Laethem, M. Peeters, and A. M. Vandamme. 2005. Optimization of a genotypic assay applicable to all human immunodeficiency virus type 1 protease and reverse transcriptase subtypes. J. Virol. Methods 12847-53. [DOI] [PubMed] [Google Scholar]
- 43.Snoeck, J., R. Kantor, R. W. Shafer, K. Van Laethem, K. Deforche, A. P. Carvalho, B. Wynhoven, M. A. Soares, P. Cane, J. Clarke, C. Pillay, S. Sirivichayakul, K. Ariyoshi, A. Holguin, H. Rudich, R. Rodrigues, M. B. Bouzas, F. Brun-Vézinet, C. Reid, P. Cahn, L. F. Brigido, Z. Grossman, V. Soriano, W. Sugiura, P. Phanuphak, L. Morris, J. Weber, D. Pillay, A. Tanuri, R. P. Harrigan, R. Camacho, J. M. Schapiro, D. Katzenstein, and A. M. Vandamme. 2006. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob. Agents Chemother. 50694-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivas, R. V., and A. Fridland. 1998. Antiviral activities of 9-R-2-phosphonomethoxypropyl adenine (PMPA) and bis(isopropyloxymethylcarbonyl)PMPA against various drug-resistant human immunodeficiency virus strains. Antimicrob. Agents Chemother. 421484-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svarovskaia, E., R. Barr, X. Zhang, G. Pais, C. Marchand, Y. Pommier, T. R. Burke, and V. K. Pathak. 2004. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 783210-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Laethem, K., A. De Luca, A. Antinori, A. Cingolani, C. F. Perno, and A. M. Vandamme. 2002. A genotypic drug resistance interpretation algorithm that significantly predicts therapy response in HIV-1 infected patients. Antivir. Ther. 7123-129. [PubMed] [Google Scholar]
- 47.Van Laethem, K., Y. Schrooten, P. Lemey, E. Van Wijngaerden, S. De Wit, M. Van Ranst, and A. M. Vandamme. 2005. A genotypic resistance assay for the detection of drug resistance in the human immunodeficiency virus type 1 envelope gene. J. Virol. Methods 12325-34. [DOI] [PubMed] [Google Scholar]
- 48.Van Laethem, K., and A. M. Vandamme. 2006. Interpreting resistance data for HIV-1 therapy management—know the limitations. AIDS Rev. 837-43. [PubMed] [Google Scholar]
- 49.Van Laethem, K., Y. Schrooten, S. Dedecker, L. Van Heeswijck, K. Deforche, E. Van Wijngaerden, M. Van Ranst, and A. M. Vandamme. 2006. A genotypic assay for the amplification and sequencing of Gag and protease from diverse human immunodeficiency virus type 1 group M subtypes. J. Virol. Methods 132181-186. [DOI] [PubMed] [Google Scholar]
- 50.Van Laethem, K., Y. Schrooten, K. Covens, N. Dekeersmaeker, P. De Munter, E. Van Wijngaerden, M. Van Ranst, and A. M. Vandamme. 2008. A genotypic assay for the amplification and sequencing of integrase from diverse HIV-1 group M subtypes. J. Virol. Methods 153176-181. [DOI] [PubMed] [Google Scholar]
- 51.Vergne, L., J. Snoeck, A. Aghokeng, B. Maes, D. Valea, E. Delaporte, A. M. Vandamme, M. Peeters, and K. Van Laethem. 2006. Genotypic drug resistance interpretation algorithms display high levels of discordance when applied to non-B strains from HIV-1 naive and treated patients. FEMS Immunol. Med. Microbiol. 4653-62. [DOI] [PubMed] [Google Scholar]
- 52.Wang, K., R. Samudrala, and J. E. Mittler. 2004. Antivirogram or phenosense: a comparison of their reproducibility and an analysis of their correlation. Antivir. Ther. 9703-712. [PubMed] [Google Scholar]
- 53.Weber, J., J. Weberova, M. Carobene, M. Mirza, J. Martinez-Picado, P. Kazanjian, and M. E. Quinones-Mateu. 2006. Use of a novel assay based on intact recombinant viruses expressing green (EGFP) or red (DsRed2) fluorescent proteins to examine the contribution of pol and env genes to overall HIV-1 replicative fitness. J. Virol. Methods 136102-117. [DOI] [PubMed] [Google Scholar]
- 54.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 812359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf, K., H. Walter, N. Beerenwinkel, W. Keulen, R. Kaiser, D. Hoffmann, T. Lengauer, J. Selbig, A. M. Vandamme, K. Korn, and B. Schmidt. 2003. Tenofovir resistance and resensitization. Antimicrob. Agents Chemother. 473478-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yap, S. H., C. W. Sheen, J. Fahey, M. Zanin, D. Tyssen, V. D. Lima, B. Wynhoven, M. Kuiper, N. Sluis-Cremer, P. R. Harrigan, and G. Tachedjian. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4e335. [DOI] [PMC free article] [PubMed] [Google Scholar]