Abstract
Previous studies showed that high levels of human immunodeficiency virus type 1 (HIV-1) DNA are associated with a faster progression to AIDS, an increased risk of death, and a higher risk of HIV RNA rebound in patients on highly active antiretroviral therapy. Our objective was to develop and assess a highly sensitive real-time multiplex PCR assay for the quantification of HIV-1 DNA (RTMP-HIV) based on molecular beacons. HIV-1 DNA quantification was carried out by RTMP in a LightCycler 2.0 apparatus. HIV-1 DNA was quantified in parallel with CCR5 as a reference gene, and reported values are numbers of HIV-1 DNA copies/106 peripheral blood mononuclear cells (PBMCs). The clinical sensitivity of the assay was assessed for 115 newly diagnosed HIV-1-infected individuals. The analytical sensitivity was estimated to be 12.5 copies of HIV-1 DNA per 106 PBMCs, while the clinical sensitivity was 100%, with levels ranging from 1.23 to 4.25 log10 HIV-1 DNA copies/106 PBMCs. In conclusion, we developed and assessed a new RTMP-HIV assay based on molecular beacons, using a LightCycler 2.0 instrument. This multiplex assay has comparable sensitivity, reproducibility, and accuracy to single real-time PCR assays.
The rate of human immunodeficiency virus type 1 (HIV-1) disease progression varies greatly among HIV-1-infected individuals (29), depending on several factors, such as baseline HIV RNA level, baseline CD4+ T-cell level, age at seroconversion, gender, etc. (17, 23, 28, 32). Quantification of HIV RNA in plasma has routinely been used for assessment of the efficacy of highly active antiretroviral therapy (HAART), and it provides a strong predictor of HIV-1 disease progression (27, 28). Treatment with HAART allows a drastic reduction of the plasma HIV RNA load, to such an extent that circulating HIV-1 in plasma becomes undetectable (<50 copies/ml) by the most sensitive viral detection assays (27, 28). However, virus eradication is not feasible because viral reservoirs established in body and cell compartments are either not susceptible to the action of antiretroviral drugs or characterized by a long half-life and slow turnover (1, 6, 7, 16, 31).
Replication and immortalization of the HIV genome occur by the reverse transcription of linear double-stranded DNA (dsDNA) and subsequent integration of the proviral HIV-1 DNA genome into human chromosomes. To accomplish successful reverse transcription, HIV-1 undergoes two template switches by using the R and U5 long terminal repeat (LTR) regions and the primer-binding site of its genome. Although the linear dsDNA molecule is the precursor to the provirus, HIV infection of target cells generates a number of nonintegrated DNA species (2, 23, 31). These unintegrated HIV-1 DNA forms, which are linear or circularized, arise when viral dsDNA fails to integrate into the host genome. For example, (i) the ends of the linear DNA may be joined to form a 2-LTR circle, (ii) homologous recombination between the two LTRs in a 2-LTR circle may yield circles with a single LTR (1-LTR), or (iii) linear unintegrated HIV-1 DNA may form. Therefore, aside from proviral DNA (HIV-1 dsDNA integrated into the host genome), several unintegrated forms also persist intracellularly. It is worth mentioning that cell-associated HIV-1 DNA can be found in both actively and latently infected cells (10).
As a result of this multiplicity of forms, HIV-1 DNA has prognostic value as a marker for disease progression (18, 22, 23, 33), in addition to other associated parameters (i.e., plasma HIV RNA or CD4+ T-cell count) (2). In particular, higher levels of HIV-1 DNA in patients not receiving combination therapy are associated with an increased risk of disease progression and a higher rate of death (2, 13, 14, 23, 33, 36). Moreover, HIV-1 DNA was found to be a predictor of a sustained virological response to treatment; patients with lower cellular viral loads showed a more prolonged therapy response (2, 18, 20, 34). The HIV-1 DNA load can also be used as an additional viral marker for treated individuals who have prolonged suppression of HIV-1 replication with HAART. In this case, HIV-1 DNA is the only marker indicative of the residual viral replication in peripheral blood mononuclear cells (PBMCs), lymphoid tissues, and other compartments (10, 35).
Given the importance of HIV-1 DNA as a prognostic marker for disease progression and response to therapy, independent of other parameters, it is crucial to use reliable assays for HIV-1 DNA quantification, as well as universal units of quantification. In this study, we report the development and assessment of a multiplex real-time PCR assay using molecular beacons as a detection system to quantify all HIV-1 DNA forms (single-stranded DNA and dsDNA forms and integrated and unintegrated linear or circular forms) (RTMP-HIV) and also the number of PBMCs, in parallel. The newly developed RTMP-HIV assay quantifies all HIV-1 DNA forms in PBMCs, which comprise the major intracellular viral reservoir.
MATERIALS AND METHODS
Preparation of standard DNA.
The external DNA standards used in the RTMP-HIV assay were HIV-1 DNA and CCR5 amplicons with known nucleotide lengths and base compositions. Amplicons were derived from a randomly selected hemophiliac's sample. The standards were used as external standards after quantification in a LightCycler 2.0 instrument (Roche, Molecular Biochemicals, Mannheim, Germany), using a PicoGreen dsDNA quantification kit (Molecular Probes Inc., Invitrogen Detection Technologies). The concentrations of the HIV-1 and CCR5 amplicons were estimated according to standard curves created in the range of 250 to 6.25 ng after 1:2 serial dilutions of the genomic DNA standard. Tenfold serial dilutions of the standards (106 to 10 DNA copies) were used for the generation of standard curves in the multiplex amplification assays, using molecular beacon-based real-time PCR. To mimic the real conditions for multiplex PCR amplification, we included a steady copy number (5 × 105 copies) of CCR5 standard in addition to the HIV-1 DNA standards (23). Similarly, 103 copies of HIV-1 DNA were added to each of the CCR5 standards.
DNA extraction.
Total DNA from PBMCs was isolated by manual extraction using a QIAamp DNA blood mini kit (Qiagen GmbH, Germany) according to the manufacturer's recommendations; extracted DNA was eluted with 100 μl of DNase-free water prior to PCR. PBMCs were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation (Histopaque-1077; Sigma). Aliquots of approximately 106 PBMCs per cryotube were stored at −80°C for 24 h and then placed in a liquid nitrogen tank (about −196°C). PBMC counting was performed using a Neubauer counter, while thawing and washing were performed by standard methods.
RTMP.
PCR was optimized using two sets of primers and molecular beacon probes labeled with two different dyes for the amplification and detection of HIV-1 and CCR5 targets. More specifically, the primers and molecular beacon for HIV-1 amplification and detection were newly designed, targeting the conserved regions of the HIV-1 genome downstream of the LTR (HIV_sense) and gag (HIV_antisense) (in order to quantify all forms of HIV-1 DNA [single-stranded DNA and dsDNA forms and integrated and unintegrated linear or circulating forms, such as 1-LTR and 2-LTR]) (Table 1). Beacon and primer design was based on the alignment of all previously characterized HIV-1 subtypes (A to D, F to H, J, and K) and circulating recombinant forms (http://hiv-web.lanl.gov). In particular, the primers and molecular beacon target very-well-conserved genomic regions of all HIV-1 subtypes and circulating recombinant forms. Note that the target recognition sequence for the CCR5_beacon did not contain the Δ32 deletion of the CCR5 gene and therefore can bind to both the wild-type CCR5 and mutant CCR5Δ32 alleles, as described previously (23). The target recognition sequences for the primers used for HIV-1 DNA and CCR5 quantification are described in Table 1 (MWG-Biotech AG, Germany). The design of the primers and the molecular beacons was performed according to standard requirements, such as the following: (i) one must avoid primer-beacon annealing and primer dimers and (ii) the melting temperatures (Tms) of the primers must be similar and at least 5 to 10°C lower than the Tms of the molecular beacons. The software tool used for the assessment of the Tm calculations was from the Virtual Genome Centre (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For the simultaneous detection of the two PCR targets (in the same capillary tube), we used molecular beacons with two fluorescent dyes excited at overlapping wavelengths but showing no considerable overlap in their emission spectra (Fig. 1). The molecular beacons used in the RTMP-HIV assay were labeled with fluorescein (Biolegio BV, The Netherlands) and Pulsar 650 (Biosearch Technologies, Inc., Novato, CA) fluorescent dyes for the detection of HIV-1 and CCR5 targets (37), respectively (Table 1). Preliminary data showed that the RTMP-HIV assay could be used in both LightCycler 1.0 and LightCycler 2.0 instruments. The data reported in this study reflect the use of the 2.0 platform because of its enhanced testing characteristics.
TABLE 1.
Real-time PCR amplicon sizes and locations of primers and beacons within viral and human reference sequences
FIG. 1.
(A) Absorption spectra for both dyes used as beacons. Red line, fluorescein-labeled beacon; blue line, Pulsar 650-labeled beacon. (B) Emission spectra for these dyes. The two fluorescent dyes absorb in the same wavelength, but the emission spectra have very little overlap.
The reaction mixture for real-time PCR (19) contained 2.5 μl 10× LightCycler FastStart Taq reaction mix (Roche, Molecular Biochemicals, Mannheim, Germany), 6 mM MgCl2, 1.1 μM HIV_sense primer, 1.1 μM HIV_anti primer, 0.08 μM CCR5_sense primer, 0.08 μM CCR5_anti primer, 0.4 μM HIV_beacon, 1.2 μM CCR5_beacon, 1.5 U FastStart Taq DNA polymerase, 1 U of uracil-DNA glycosylase, and 10 μl extracted DNA in a final reaction volume of 25 μl. The amplification conditions were optimized for LightCycler 1.0 and 2.0 as follows: 1 cycle of denaturation at 95°C for 10 min followed by 45 cycles of amplification at 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. Uracil-DNA glycosylase was used to eliminate PCR carryover contaminations from previous PCRs (24, 25). For each run, two standard curves were created in a 6-log range by 1:10 serial dilutions, one with HIV-1 DNA and one with CCR5 external standards. The slope and correlation coefficient of each standard curve were calculated based on the average cycle threshold (CT) value measured for each dilution point, ranging from 106 to 101 copies of standard DNA template. Quantification of HIV-1 DNA and the CCR5 reporter gene in the unknown samples was performed by using these external standards for HIV-1 and CCR5. HIV-1 DNA was quantified by using the CCR5 gene as a reference gene, for which there is a steady number of copies per cell (two copies per cell), as described previously (23). Therefore, the numbers of HIV-1 DNA and CCR5 copies in a sample can easily be converted to HIV-1 DNA copies/106 PBMCs by using the following formula: number of HIV-1 DNA copies/number of CCR5 copies × 2 × 106 = number of HIV-1 DNA copies per 106 PBMCs, given that a single PBMC contains two copies of the CCR5 gene (for example, 15 copies of HIV-1 DNA and 1,500 CCR5 copies in the same sample equals 2 × 104 HIV-1 DNA copies per 106 PBMCs).
Clinical specimens.
HIV-1 DNA was quantified in total DNAs extracted from PBMCs from 115 randomly selected newly diagnosed HIV-1-infected individuals before the initiation of HAART. The clinical specimens (whole blood) were collected anonymously from September 2002 to September 2005. PBMCs were stored in a liquid nitrogen tank (about −196°C) until their use. Plasma HIV-1 RNA was measured with the Versant HIV-1 RNA 3.0 (bDNA) assay (Bayer, Tarrytown, NY), which has a dynamic range of 50 to 500,000 HIV-1 RNA copies/ml, according to the manufacturer's recommendations. CD4+ T-cell counts were measured by flow cytometry by standard procedures.
Statistical analysis.
The variability between replicate tests of HIV-1 DNA and CCR5 standards was described using the percent coefficient of variation (CV) for log10-transformed data. Pearson's correlation coefficient calculation and ordinary least-squares regression (OLR) on log10-transformed values for HIV-1 DNA and CCR5 standards were performed to compare experimentally observed to expected concentrations. Pearson's correlation coefficient, OLR, and Deming regression were used to assess the association of HIV-1 DNA with HIV RNA and the CD4+ T-cell count. The Deming regression is similar to the OLR but takes into account that both y and x are measured with error (8).
RESULTS
The RTMP-HIV assay is the first to implement multiplex methodology using molecular beacons as a detection system with the LightCycler platform. Primary experiments were carried out with a LightCycler 1.0 apparatus; therefore, the analytical sensitivity of the RTMP-HIV assay was primarily estimated with the LightCycler 1.0 platform and thereafter with the LightCycler 2.0 platform. These preliminary data showed that the RTMP-HIV assay could be used on both platforms (LightCycler 1.0 and LightCycler 2.0) and that the latter had superior performance characteristics. The data reported in this study with regard to the reproducibility of the RMTP-HIV assay and testing of clinical samples reflect the use of the LightCycler 2.0 platform because of its enhanced performance characteristics.
Analytical sensitivity of RTMP-HIV assay with LightCycler 1.0 and LightCycler 2.0 platforms.
The analytical sensitivity of the RTMP-HIV assay was estimated by repetitively testing (20 replicates) different concentrations of HIV-1 DNA standards, ranging from 106 to 3.125 copies/reaction (3.125, 6.25, 10, 12.5, 25, 102, 103, 104, 105, and 106) (Table 2). The seven highest concentrations of the HIV-1 DNA proviral standards (12.5 to 106 copies per reaction) were examined using the LightCycler 1.0 platform, while the three lowest concentrations (3.125 to 10 copies per reaction) were examined with the LightCycler 2.0 platform. The lowest concentration of HIV-1 DNA standard that could be quantified at a frequency of 100% by RTMP-HIV with the LightCycler 1.0 platform was 12.5 copies per reaction (in a multiplex setting with 5 × 105 copies of the CCR5 standard), which equals 50 copies of HIV-1 DNA per 106 PBMCs. The analytical sensitivity of the RTMP-HIV assay was found to be higher with the LightCycler 2.0 platform (3.125 copies per reaction, which equals 12.5 copies of HIV-1 DNA per 106 PBMCs). The latter was set as the cutoff for the quantification of HIV-1 DNA for the LightCycler 2.0 platform.
TABLE 2.
Experimental results for repetitively quantified (20 times) serial dilutions of HIV-1 DNA standards in LightCycler 1.0 and LightCycler 2.0 platformsa
| Exptl platform | No. of HIV-1 DNA copies per reaction | No. of HIV-1 DNA copies/106 PBMCs | Mean ± SD of exptl valuesb | CV (%) of exptl valuesb |
|---|---|---|---|---|
| LightCycler 1.0 | 106 | 4 × 106 | 5.99 ± 0.08 | 1.36 |
| 105 | 4 × 105 | 5.08 ± 0.20 | 3.86 | |
| 104 | 4 × 104 | 3.95 ± 0.09 | 2.16 | |
| 103 | 4 × 103 | 2.99 ± 0.15 | 5.02 | |
| 102 | 400 | 2.21 ± 0.27 | 12.37 | |
| 25 | 100 | 1.44 ± 0.27 | 18.87 | |
| 12.5 | 50 | 0.79 ± 0.21 | 25.93 | |
| LightCycler 2.0 | 10 | 40 | 1.03 ± 0.13 | 12.99 |
| 6.25 | 25 | 0.87 ± 0.31 | 35.32 | |
| 3.125 | 12.5 | 0.17 ± 0.40 | 232.26 |
Each reaction was performed in a multiplex setting with 5 × 105 copies of the CCR5 gene in a final reaction volume of 25 μl. The experimental results were 100% positive for all concentrations with both platforms.
Log10-transformed values.
RTMP-HIV assay performance with LightCycler 2.0 platform.
To examine the reproducibility and linearity of the RTMP-HIV assay, we quantified different concentrations of HIV-1 DNA standards, ranging from 10 to 106 copies per reaction (10, 102, 103, 104, 105, and 106), in a multiplex setting with CCR5 standards (5 × 105 copies of CCR5 were present in each sample) in 28 replicates and estimated the CV for each concentration (Table 3). According to the estimated values, the %CV was low (2.0% and 3.2%) for high concentrations of HIV-1 DNA (for 106 and 105 copies per reaction, respectively), while more variation (11.6% and 27.3%) was observed for lower HIV-1 DNA values (102 and 10 copies/reaction, respectively) (Table 3). The standard deviation (SD) of experimental values ranged from 0.12 (for 106 copies per reaction) to 0.27 (for 10 copies per reaction). Furthermore, the theoretically calculated values showed a very high correlation versus the experimentally estimated concentrations (Pearson's r = 0.999; P < 0.001) (Fig. 2).
TABLE 3.
Quantified values for serial dilutions of standards with LightCycler 2.0 platforma
| Target DNA | No. of copies of DNA per reaction | No. (%) of measurements above the limit of detection for 28 replicates (HIV-1) or 12 replicates (CCR5) | Mean ± SD of exptl valuesb | CV (%) of exptl valuesb |
|---|---|---|---|---|
| HIV-1 | 106 | 28 (100) | 5.89 ± 0.12 | 2.0 |
| 105 | 28 (100) | 4.93 ± 0.16 | 3.2 | |
| 104 | 28 (100) | 3.83 ± 0.16 | 4.0 | |
| 103 | 28 (100) | 2.97 ± 0.12 | 4.0 | |
| 102 | 28 (100) | 2.03 ± 0.24 | 11.6 | |
| 10 | 9 (32.1) | 1.00 ± 0.27 | 27.3 | |
| CCR5 | 106 | 12 (100) | 5.92 ± 0.29 | 4.9 |
| 105 | 12 (100) | 5.03 ± 0.21 | 4.2 | |
| 104 | 12 (100) | 3,94 ± 0.12 | 2.9 | |
| 103 | 12 (100) | 3.06 ± 0.20 | 6.6 | |
| 102 | 12 (100) | 2.05 ± 0.19 | 9.3 | |
| 10 | 12 (100) | 1.05 ± 0.29 | 27.6 |
Each reaction was performed in a multiplex setting, with each reaction mix including, in addition to the standards, 103 copies of HIV-1 DNA and 5 × 105 CCR5 copies for CCR5 and HIV-1 DNA quantifications, respectively, in a final volume of 25 μl.
Log10-transformed data.
FIG. 2.
Mean experimentally estimated concentrations (dots) for six serial dilutions of HIV-1 DNA standards versus theoretically calculated concentrations (log10-transformed data). Error bars indicate the SD for the six 10-fold serially diluted standards.
CCR5 RTMP assay performance with LightCycler 2.0 platform.
The reproducibility and linearity of the newly developed RTMP-HIV assay were estimated similarly for the CCR5 external standard. In particular, the amount of CCR5 was estimated repeatedly (12 replicates) for DNA concentrations ranging between 10 and 106 copies coamplified with HIV-1 DNA (103 copies of HIV-1 DNA were included in each reaction). The CV was calculated for each different serially diluted concentration (Table 3). According to the estimated values, the %CV was low (4.9% and 4.2%) for high concentrations (for 106 and 105 copies of the CCR5 gene per reaction, respectively) and was higher for 102 and 10 copies per reaction (9.3% and 27.6%, respectively) (Table 3). Interestingly, the SD of experimental values ranged from 0.12 (for 104 copies per reaction) to 0.29 (for 10 and 106 copies per reaction). Furthermore, the theoretically calculated values showed a very high correlation versus experimentally estimated concentrations (Pearson's r = 0.999; P < 0.001) (Fig. 3).
FIG. 3.
Mean experimentally estimated concentrations (dots) for six serial dilutions of CCR5 gene standards versus theoretically calculated concentrations (log10-transformed data). Error bars indicate the SD for the six 10-fold serially diluted standards.
Assessment of RTMP-HIV assay with clinical samples.
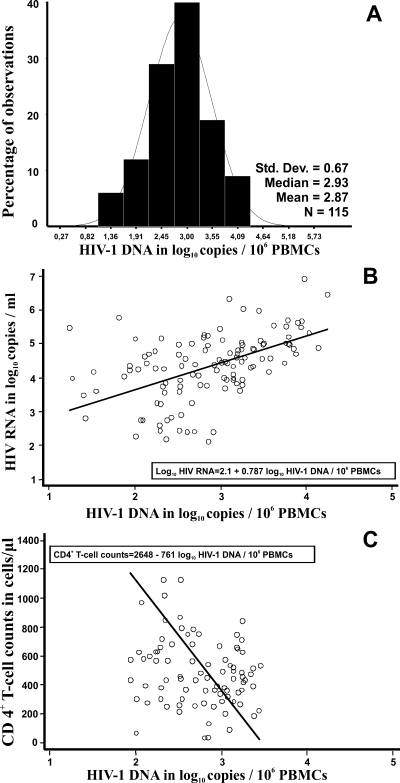
The RTMP-HIV assay was used for the quantification of HIV DNA in 115 newly diagnosed HIV-1-infected individuals prior to administration of therapy. HIV-1 DNA was detectable in 115 samples (100%). The log10 HIV-1 DNA copies/106 PBMCs ranged from 1.23 to 4.25, with a mean ± SD of 2.87 ± 0.67 (Fig. 4A). Importantly, HIV DNA/106 PBMC values fell within a 3-log dynamic range. The log10 HIV-1 DNA copies/106 PBMCs and the number of CD4+ T cells per μl were negatively correlated (Pearson's r = −0.461; P < 0.001), and the strength of this linear relationship was similar to that of HIV RNA levels and CD4+ T-cell counts (Pearson's r = −0.353; P < 0.001) (data not shown). CD4+ T-cell counts ranged from 32 to 1,328 cells/μl, with a median value (25th, 75th percentiles) of 444 cells/μl (279, 600 cells/μl) (Fig. 4C). Finally, we found that the number of HIV-1 DNA copies/106 PBMCs was positively correlated with the HIV-1 RNA level (Pearson's r = 0.488; P < 0. 001). Log10 HIV-1 RNA levels ranged from 2.13 to 6.92, with a mean ± SD of 4.35 ± 0.98 (Fig. 4B).
FIG. 4.
Distribution of log10 HIV-1 DNA copies/106 PBMCs (A) and correlation of log10 HIV-1 DNA copies/106 PBMCs with plasma RNA levels (B) and with CD4+ T-cell counts (C) for 115 newly diagnosed HIV-1-infected individuals. The solid lines in panels B and C are fitted regression lines obtained by Deming regression.
DISCUSSION
Our study aimed to develop an RTMP-HIV assay for the quantification of total HIV-1 DNA per PBMC in a single-tube capillary reaction by using a LightCycler instrument. For the accomplishment of this task, we quantified HIV-1 DNA by using CCR5 as a reference gene, as there is a steady number of copies of this gene per cell, as described previously (23). Therefore, HIV-1 DNA quantifications can easily be converted to copies of HIV-1 DNA/106 PBMCs.
The analytical sensitivity of the RTMP-HIV assay was higher with the LightCycler 2.0 platform than with the LightCycler 1.0 platform because of the improved optical and hardware settings installed in the latest version of LightCycler instruments. The analytical sensitivity of the RTMP-HIV assay (12.5 HIV-1 DNA copies/106 PBMCs) is very similar to those of the most sensitive methods (5 to 10 copies of HIV-1 DNA per reaction) based on single real-time PCRs (2), thus suggesting that the RTMP-HIV assay can be used reliably for HIV-1 DNA quantification.
Prior to this study, several PCR-based methodologies had been developed for the quantification of cell-associated HIV-1 DNA, based on end-point and real-time PCR methodology (2). Implementation of real-time PCR methodology considerably improved the quantification of HIV-1 DNA compared to end-point PCR methodology. The former shows improved accuracy and reproducibility and a wider range of linear quantification because the concentration of the target is estimated during the linear phase rather than at the end point (plateau phase) of the amplification step. Most home-brewed real-time PCR methods have implemented TaqMan methodology (2), while three others have used SYBR green for the detection and quantification of HIV-1 DNA (3, 12, 21). Moreover, Christopherson et al. reported a modification of the Amplicor HIV-1 Monitor test (Roche Molecular Systems, Inc., Branchburg, NJ) for total HIV-1 DNA quantification. This assay, based on end-point PCR measurement, is almost identical to the Amplicor HIV-1 Monitor assay for RNA (Roche Molecular Systems, Inc., Branchburg, NJ) but differs in (i) the sample preparation method, (ii) the use of plasmid DNA rather than an RNA transcript as the quantitation standard, and (iii) the normalization of DNA load to total cellular input (4).
In contrast with all previous methods, our assay combines the advantages of real-time PCR (high dynamic range of quantification, less intra- and interassay variability in measurements, and improved sensitivity) with the direct quantification of cell copy numbers by using the CCR5 gene as a reference gene in a multiplex format. Despite the multiplex setting, our assay performed very well with clinical samples, showing a clinical sensitivity of 100% and a linear range of values within 3 log10 copies. Most in-house assays use two single PCRs for the quantification of the amount of HIV-1 DNA per cell. Importantly, the current method comprises the first assay with a multiplex design using the LightCycler 2.0 platform for quantification of the HIV-1 DNA concentration in PBMCs.
We should note that until now there has been no universal way to report HIV-1 DNA values. In particular, some investigators quantify the amount of HIV-1 DNA per 106 PBMCs (2, 5, 9, 11, 23) by estimating the cell copy number directly or indirectly or per number of resting CD4+ T cells (21). In the latter method, the amount of detected HIV-1 DNA depends on the subsets of PBMCs as well as on the methodology used to estimate the number of cell equivalents.
In conclusion, we developed and assessed a novel RTMP-HIV assay for HIV-1 DNA and CCR5 quantification based on molecular beacons and the use of a LightCycler instrument (the assay applies to both LightCycler 1.0 and LightCycler 2.0 platforms). Although fluorescent molecular beacons have been implemented in real-time PCR methodology for several years (15, 26, 30), the RTMP-HIV assay is the first RTMP assay for the LightCycler apparatus that uses molecular beacons as a detection system. The assay with the LightCycler 2.0 platform has comparable performance characteristics to those of several previously described real-time PCR assays, as well as a multiplex design for high throughput and accurate detection of the HIV-1 DNA concentration in PBMCs.
Footnotes
Published ahead of print on 6 May 2009.
REFERENCES
- 1.Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 806441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beloukas, A., D. Paraskevis, M. Psichogiou, and A. Hatzakis. 2009. The role of HIV-1 DNA as an additional marker of HIV-1 infection. Curr. HIV Res. 7255-265. [DOI] [PubMed] [Google Scholar]
- 3.Casabianca, A., C. Gori, C. Orlandi, F. Forbici, C. F. Perno, and M. Magnani. 2007. Fast and sensitive quantitative detection of HIV DNA in whole blood leucocytes by SYBR green I real-time PCR assay. Mol. Cell. Probes 21368-378. [DOI] [PubMed] [Google Scholar]
- 4.Christopherson, C., Y. Kidane, B. Conway, J. Krowka, H. Sheppard, and S. Kwok. 2000. PCR-based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387183-188. [DOI] [PubMed] [Google Scholar]
- 6.Chun, T. W., D. C. Nickle, J. S. Justement, D. Large, A. Semerjian, M. E. Curlin, M. A. O'Shea, C. W. Hallahan, M. Daucher, D. J. Ward, S. Moir, J. I. Mullins, C. Kovacs, and A. S. Fauci. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Investig. 1153250-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 9413193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornbleet, P. J., and N. Gochman. 1979. Incorrect least-squares regression coefficients in method-comparison analysis. Clin. Chem. 25432-438. [PubMed] [Google Scholar]
- 9.Eriksson, L. E., T. Leitner, B. Wahren, A. C. Bostrom, and K. I. Falk. 2003. A multiplex real-time PCR for quantification of HIV-1 DNA and the human albumin gene in CD4+ cells. APMIS 111625-633. [DOI] [PubMed] [Google Scholar]
- 10.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 3401614-1622. [DOI] [PubMed] [Google Scholar]
- 11.Gibellini, D., F. Vitone, E. Gori, M. La Placa, and M. C. Re. 2004. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) viral load by SYBR green real-time RT-PCR technique in HIV-1 seropositive patients. J. Virol. Methods 115183-189. [DOI] [PubMed] [Google Scholar]
- 12.Gibellini, D., F. Vitone, P. Schiavone, C. Ponti, M. La Placa, and M. C. Re. 2004. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. J. Clin. Virol. 29282-289. [DOI] [PubMed] [Google Scholar]
- 13.Goedert, J. J., T. R. O'Brien, A. Hatzakis, and L. G. Kostrikis. 2001. T cell receptor excision circles and HIV-1 2-LTR episomal DNA to predict AIDS in patients not receiving effective therapy. AIDS 152245-2250. [DOI] [PubMed] [Google Scholar]
- 14.Goujard, C., M. Bonarek, L. Meyer, F. Bonnet, M. L. Chaix, C. Deveau, M. Sinet, J. Galimand, J. F. Delfraissy, A. Venet, C. Rouzioux, and P. Morlat. 2006. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin. Infect. Dis. 42709-715. [DOI] [PubMed] [Google Scholar]
- 15.Gullsby, K., M. Storm, and K. Bondeson. 2008. Simultaneous detection of Chlamydophila pneumoniae and Mycoplasma pneumoniae by use of molecular beacons in a duplex real-time PCR. J. Clin. Microbiol. 46727-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, and P. A. Volberding. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—USA panel. Top. HIV Med. 14827-843. [PubMed] [Google Scholar]
- 17.Hatzakis, A., G. Touloumi, R. Karanicolas, A. Karafoulidou, T. Mandalaki, C. Anastassopoulou, L. Zhang, J. J. Goedert, D. D. Ho, and L. G. Kostrikis. 2000. Effect of recent thymic emigrants on progression of HIV-1 disease. Lancet 355599-604. [DOI] [PubMed] [Google Scholar]
- 18.Hatzakis, A. E., G. Touloumi, N. Pantazis, C. G. Anastassopoulou, O. Katsarou, A. Karafoulidou, J. J. Goedert, and L. G. Kostrikis. 2004. Cellular HIV-1 DNA load predicts HIV-RNA rebound and the outcome of highly active antiretroviral therapy. AIDS 182261-2267. [DOI] [PubMed] [Google Scholar]
- 19.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6986-994. [DOI] [PubMed] [Google Scholar]
- 20.Hoen, B., D. A. Cooper, F. C. Lampe, L. Perrin, N. Clumeck, A. N. Phillips, L. E. Goh, S. Lindback, D. Sereni, B. Gazzard, J. Montaner, H. J. Stellbrink, A. Lazzarin, D. Ponscarme, S. Staszewski, L. Mathiesen, D. Smith, R. Finlayson, R. Weber, L. Wegmann, G. Janossy, and S. Kinloch-de Loes. 2007. Predictors of virological outcome and safety in primary HIV type 1-infected patients initiating quadruple antiretroviral therapy: QUEST GW PROB3005. Clin. Infect. Dis. 45381-390. [DOI] [PubMed] [Google Scholar]
- 21.Kabamba-Mukadi, B., P. Henrivaux, J. Ruelle, N. Delferriere, M. Bodeus, and P. Goubau. 2005. Human immunodeficiency virus type 1 (HIV-1) proviral DNA load in purified CD4+ cells by LightCycler real-time PCR. BMC Infect. Dis. 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzenstein, T. L., R. S. Oliveri, T. Benfield, J. Eugen-Olsen, C. Nielsen, and J. Gerstoft. 2002. Cell-associated HIV DNA measured early during infection has prognostic value independent of serum HIV RNA measured concomitantly. Scand. J. Infect. Dis. 34529-533. [DOI] [PubMed] [Google Scholar]
- 23.Kostrikis, L. G., G. Touloumi, R. Karanicolas, N. Pantazis, C. Anastassopoulou, A. Karafoulidou, J. J. Goedert, and A. Hatzakis. 2002. Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load. J. Virol. 7610099-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339237-238. [DOI] [PubMed] [Google Scholar]
- 25.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93125-128. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Lopez, J., J. J. Lahuerta, P. Salama, R. Ayala, and J. M. Bautista. 2004. The use of fluorescent molecular beacons in real time PCR of IgH gene rearrangements for quantitative evaluation of multiple myeloma. Clin. Lab. Haematol. 2631-35. [DOI] [PubMed] [Google Scholar]
- 27.Mellors, J. W., L. A. Kingsley, C. R. Rinaldo, Jr., J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122573-579. [DOI] [PubMed] [Google Scholar]
- 28.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 2721167-1170. [DOI] [PubMed] [Google Scholar]
- 29.Pantaleo, G., C. Graziosi, and A. S. Fauci. 1993. New concepts in the immunopathogenesis of human immunodeficiency virus infection. N. Engl. J. Med. 328327-335. [DOI] [PubMed] [Google Scholar]
- 30.Pattyn, F., F. Speleman, A. De Paepe, and J. Vandesompele. 2003. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 31122-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18665-708. [DOI] [PubMed] [Google Scholar]
- 32.Powderly, W. G., M. S. Saag, S. Chapman, G. Yu, B. Quart, and N. J. Clendeninn. 1999. Predictors of optimal virological response to potent antiretroviral therapy. AIDS 131873-1880. [DOI] [PubMed] [Google Scholar]
- 33.Rouzioux, C., J. B. Hubert, M. Burgard, C. Deveau, C. Goujard, M. Bary, D. Sereni, J. P. Viard, J. F. Delfraissy, and L. Meyer. 2005. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J. Infect. Dis. 19246-55. [DOI] [PubMed] [Google Scholar]
- 34.Sarmati, L., S. G. Parisi, E. Nicastri, G. d'Ettorre, C. Andreoni, L. Dori, F. Gatti, M. Montano, A. R. Buonomini, C. Boldrin, G. Palu, V. Vullo, and M. Andreoni. 2007. Cellular HIV-1 DNA quantitation in patients during simplification therapy with protease inhibitor-sparing regimens. J. Med. Virol. 79880-886. [DOI] [PubMed] [Google Scholar]
- 35.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 676-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tierney, C., J. L. Lathey, C. Christopherson, D. M. Bettendorf, R. T. D'Aquila, S. M. Hammer, and D. A. Katzenstein. 2003. Prognostic value of baseline human immunodeficiency virus type 1 DNA measurement for disease progression in patients receiving nucleoside therapy. J. Infect. Dis. 187144-148. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14303-308. [DOI] [PubMed] [Google Scholar]