Abstract
We screened 533 and 361 methicillin (meticillin)-resistant Staphylococcus aureus strains isolated in a university hospital in 2002 and 2003 and in 2006 and 2007, respectively, and identified 4 (0.8%) of the strains in the first group and 8 (2.2%) of the strains in second group as heterogeneous vancomycin-resistant S. aureus (heterogeneous VISA) strains and 3 (0.8%) of the strains in the second group as VISA strains. This is the first report of VISA strains isolated from patients in Thailand.
The existence of methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) was first reported nearly 50 years ago (20), and MRSA is now an increasingly common pathogen associated with nosocomial and community-acquired infections (1, 2, 29), such as surgical-site infections, bloodstream infections, and pneumonia. Vancomycin, which has been available for more than 50 years, was not widely used initially because of oto- and nephrotoxicities caused by impurities in the early preparations (11). Since the late 1970s, new methods of purification have resulted in improved vancomycin preparations with lower levels of oto- and nephrotoxicities in animal models (3, 7); consequently, with the worldwide emergence of methicillin-resistant staphylococci in the 1970s, interest in vancomycin has been rekindled.
Since the 1980s, vancomycin has been used widely in many countries for the treatment of serious infections caused by gram-positive bacteria. Its frequent use for MRSA infections gradually selected MRSA strains with reduced susceptibilities to glycopeptide antibiotics, which led to the discovery in Japan in 1997 of MRSA strains with intermediate resistance to vancomycin (13). Although the resistant strains are designated intermediate, they are practically resistant to the concentrations of vancomycin used in clinical chemotherapy (4, 9, 28). Most recently, several highly vancomycin-resistant strains of S. aureus—carrying the enterococcal vanA transposon—were recovered from patients coinfected with MRSA and vancomycin-resistant enterococci (21, 26, 27).
Vancomycin-intermediate S. aureus (VISA) is not easily generated from vancomycin-susceptible MRSA, but it is frequently generated from certain MRSA strains which are heterogeneously resistant to vancomycin (10). The vancomycin MICs for heterogeneous vancomycin-resistant S. aureus (hetero-VISA) strains are within the susceptible range (<4 μg/ml), but these strains include subpopulations that exhibit reduced susceptibilities to vancomycin (15). These organisms are clinically important, because they persist latently when they are disseminated in the hospital environment and are able to generate VISA during vancomycin therapy in patients infected with hetero-VISA strains (23, 34).
In Thailand, MRSA is an important pathogen causing serious nosocomial infections. A study in 2001 reported the detection of 3 hetero-VISA strains among 155 MRSA isolates from various hospitals in Thailand (32). Later, a preliminary report announced the identification of 7 hetero-VISA strains among 246 MRSA strains isolated from patients at a university hospital (24). We therefore considered it important to continue surveillance of the vancomycin susceptibility profiles of MRSA clinical strains in hospitals where MRSA is isolated at high frequencies and where relatively generous use of vancomycin is inevitable. This communication thus reports the prevalences of MRSA isolates with reduced susceptibilities to vancomycin among patients at a university hospital in northeast Thailand over two time periods. We present the first evidence of the isolation of VISA in Thailand.
A total of 894 MRSA isolates from patients at the 842-bed Srinagarind Hospital, Khon Kaen University, Khon Kaen, Thailand, were recovered and studied. Between August 2002 and April 2003, 533 isolates were collected, and between November 2006 and December 2007, 361 isolates were obtained. All of the isolates were identified by conventional coagulase tube testing and DNase testing and then were stored in skim milk supplemented with 30% glycerol at −70°C. The references strains used for the vancomycin susceptibility tests were the hetero-VISA strain Mu3 (ATCC 700698) (13), the VISA strain Mu50 (ATCC 700699) (14), and a vancomycin-susceptible S. aureus strain (ATCC 29213).
All of the isolates were screened for reduced susceptibility to vancomycin by one-point population analysis (13). Overnight cultures in brain heart infusion (BHI) broth were adjusted to an optical density at 578 nm of 0.3 (about 108 CFU/ml). Aliquots of 100 μl of the cell suspensions were spread evenly onto BHI agar plates containing 4 μg/ml of vancomycin. The plates were then incubated at 37°C for 48 h, and cell growth was inspected at 24 and 48 h.
Isolates were suspected of being VISA strains if more than 1,000 colonies grew within 48 h and of being hetero-VISA strains if fewer than 1,000 colonies grew. All of the isolates that grew on the screening agar plates (screening-positive isolates) were subjected to further confirmation tests by population analysis. Briefly, colonies from cultures grown overnight on BHI agar were inoculated into BHI broth. After 24 h of incubation, the cultures were adjusted to an optical density at 578 nm of 0.3 and then 10-fold serial dilutions from 10−1 to 10−6 in normal saline solution were prepared. Aliquots of 100 μl of each dilution were spread evenly onto BHI agar plates containing 0, 1, 2, 3, 4, 5, 6, 7, or 8 μg/ml of vancomycin (or 0, 0.5, 1, 2, 4, 8, 16, 32, 64, or 128 μg/ml of teicoplanin). The plates were then incubated at 37°C for 48 h before the colonies were counted, and the log numbers of CFU per milliliter were plotted against the antibiotic concentrations.
For each screening-positive isolate, MICs of the following antimicrobial agents (all from Sigma Chemical, St. Louis, MO, except for teicoplanin, which was purchased from Sanofi-Aventis, Tokyo, Japan) were determined by testing the indicated ranges of concentrations by the agar dilution method according to the CLSI guidelines (5): vancomycin (0.25 to 8 μg/ml), cefazolin (0.5 to 128 μg/ml), oxacillin (0.5 to 128 μg/ml), ofloxacin (0.5 to 128 μg/ml), tetracycline (0.5 to 128 μg/ml), erythromycin (0.5 to 128 μg/ml), gentamicin (0.5 to 128 μg/ml), and teicoplanin (0.5 to 128 μg/ml).
Isolates from each group of strains, representative of the different antibiotic susceptibility patterns, were analyzed by multiplex PCR to determine staphylococcal cassette chromosome mec (SCCmec) types. Chromosomal DNA was extracted using the Isoplant II kit per the instructions of the manufacturer (Wako Chemical, Tokyo, Japan). Multiplex PCR analyses of mecA, the ccr gene complex type, and the mec gene complex class were performed using primer sets and conditions described previously (22). A 286-bp fragment of the mecA gene was amplified using primers mA1 and mA2. Simultaneously, the ccr gene complex type was identified using primers α1, α2, and α3 with primer βc to amplify the respective 695-, 937-, and 1,791-bp fragments of ccrA1, ccrA2, and ccrA3. Either the 1,287-bp fragment of ccrA4 or the 518-bp fragment of ccrC was amplified using primer α 4.2 with β 4.2 or primer γ R with γ F, respectively. The mec gene complex classes of the isolates were identified by using the primers mI6, IS7, and IS2 with mA7 to amplify the 1,963-, 2,827-, and 804-bp fragments of mec classes A, B, and C, respectively.
Multilocus sequence typing (MLST) of the representative isolates was performed as described previously (6). The alleles of seven loci were evaluated by comparing the sequences to those of the corresponding loci in the S. aureus MLST database (www.mlst.net). Sequence types (STs) were determined according to the combined pattern of the seven alleles, and clonal complexes were identified by the BURST (based upon related STs) program, available at the MLST website. agr typing was performed as described previously (30). Pulsed-field gel electrophoresis (PFGE) analysis of SmaI-digested chromosomal DNA was performed with a contour-clamped homogeneous electric field mapper system according to the instructions of the manufacturer (Bio-Rad), and a 48.5-kb ladder (Bio-Rad) was used as the DNA size marker. After the analysis was run for 22 h, the gel was stained with ethidium bromide and photographed. The band patterns were compared visually and were classified as indistinguishable (clonal), closely related (representative of clonal variants with a difference of three or fewer bands), possibly related (exhibiting a four- to six-band difference), and unrelated, according to previously described criteria (31).
Among the 533 MRSA strains isolated between August 2002 and April 2003, 19 strains (3.6%) generated colonies on the BHI agar plates with 4 μg/ml of vancomycin, whereas 26 (7.2%) of the 361 strains isolated between November 2006 and December 2007 formed colonies on the screening agar plates. The two groups of strains were isolated from various specimens obtained from patients ranging in age from 1 to 93 years (Table 1).
TABLE 1.
Numbers and characteristics of screening-positive isolates
| Groupa | Total no. of isolates tested | No. of screening-positive isolates | Patient age range (yr) | No. of males | No. of females | No. of isolates from:
|
Range of MICsb (μg/ml) of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sputum samples | Tracheal aspirates | Pus or wound specimens | Blood samples | Body fluids | Van | Cefaz | Oxa | Oflox | Tetra | Erythro | Gen | ||||||
| 1 | 533 | 19 | 1-93 | 10 | 9 | 12 | 2 | 5 | 0.5-2 | 8->128 | 64->128 | 8->128 | 32->128 | 8->128 | >128 | ||
| 2 | 361 | 26 | 1-91 | 18 | 8 | 9 | 8 | 7 | 2 | 2-3 | 128->128 | 64->128 | 64->128 | 32->128 | 64->128 | 1->128 | |
Group 1, isolates recovered in the years 2002 and 2003; group 2, isolates recovered in the years 2006 and 2007.
Van, vancomycin; Cefaz, cefazolin; Oxa, oxacillin; Oflox, ofloxacin; Tetra, tetracycline; Erythro, erythromycin; Gen, gentamicin.
All of the 45 screening-positive strains were tested further by population analyses. Among the 19 screening-positive isolates in the first group, 4 hetero-VISA strains were identified by the confirmatory population analysis profile (PAP) tests. In contrast, 8 hetero-VISA and 3 VISA strains among the 26 strains in the second group were detected. The PAPs of the representative strains in the presence of vancomycin and teicoplanin are shown in Fig. 1 and 2. Statistical analyses using Fisher's exact test showed a significant increase in screening-positive strains (P = 0.02; risk ratio confidence interval, 0.25 to 0.94) and a marginally significant increase in confirmed hetero-VISA strains (P = 0.08; risk ratio confidence interval, 0.07 to 1.29) between the two sampling periods.
FIG. 1.
PAPs of three VISA isolates, one hetero-VISA isolate, and control strains in the presence of vancomycin.
FIG. 2.
PAPs of three VISA isolates and control strains in the presence of teicoplanin.
All 19 screening-positive strains from the first group were susceptible to vancomycin (MICs for the strains, 0.5 to 2 μg/ml), and most were resistant to cefazolin, oxacillin, ofloxacin, tetracycline, erythromycin, and gentamicin. The exceptions were one strain susceptible to cefazolin, two strains susceptible to ofloxacin, and four strains susceptible to erythromycin. All 26 screening-positive isolates from the second group were resistant to cefazolin, oxacillin, ofloxacin, tetracycline, and erythromycin, and all but 4 were resistant gentamicin. The vancomycin MICs for this group of strains were between 2 and 3 μg/ml. The MIC ranges for the screening-positive strains are shown in Table 1. The vancomycin MIC for the 12 PAP-confirmed hetero-VISA strains was 2 μg/ml, and that for the 3 VISA strains was 3 μg/ml (equivalent to 4 μg/ml as determined by the regular twofold dilution system for MIC testing). The teicoplanin MICs for the hetero-VISA strains were between 2 and 8 μg/ml, whereas those for the VISA strains were between 8 and 16 μg/ml (Table 2). By using the paired Wilcoxon rank test, a significant increase in teicoplanin MICs for VISA strains compared to those for hetero-VISA strains was confirmed.
TABLE 2.
Phenotypic and genotypic characteristics of hetero-VISA and VISA isolates
| Groupa and classification by resistance phenotype | JCSC no. | Source specimen | Date (mo/day/yr) of isolation | Wardb in which patient was hospitalized | Patient sexc | Patient age (yr) | Patient statusd | MICe (μg/ml) of:
|
SCCmec type | ST | agr type | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Van | Cefaz | Oxa | Oflox | Tetra | Erythro | Gen | Teico | |||||||||||
| Group 1 isolates | ||||||||||||||||||
| hVISA | 6902 | Sputum | 8/10/02 | 4C | M | 57 | 2 | >128 | >128 | 32 | >128 | 16 | >128 | 4 | III | 239 | ||
| hVISA | 6903 | Wound sample | 10/15/02 | 5E | F | 53 | 2 | 64 | 64 | 64 | >128 | >128 | >128 | 2 | III | |||
| hVISA | 6904 | Sputum | 1/20/03 | 4B | F | 74 | 2 | >128 | 128 | 32 | 64 | >128 | >128 | 2 | III | |||
| hVISA | 6905 | Sputum | 2/21/03 | 4B | F | 75 | C | 2 | 128 | >128 | 16 | 32 | 16 | >128 | 4 | III | 239 | |
| Group 2 isolates | ||||||||||||||||||
| hVISA | 7192 | Pus | 10/07 | 4C | F | 66 | I | 2 | 128 | 128 | 128 | 128 | >128 | 0.5 | 8 | III | 239 | |
| hVISA | 7194 | Body fluid | 10/15/07 | 3D | F | 12 | I | 2 | >128 | >128 | 64 | 128 | >128 | >128 | 8 | |||
| hVISA | 7196 | Wound sample | 9/22/07 | 3A | F | 71 | C | 2 | 32 | 64 | 64 | 128 | >128 | 0.5 | 8 | III | 239 | |
| hVISA | 7201 | Sputum | 7/13/07 | 4B | M | 47 | C | 2 | >128 | >128 | 64 | >128 | >128 | 1 | NDf | III | 239 | |
| hVISA | 7204 | Blood | 7/26/07 | 3F | M | 52 | I | 2 | >128 | >128 | 128 | 64 | >128 | >128 | ND | III | 239 | |
| hVISA | 7205 | Pus | 7/27/07 | 4B | M | 67 | 2 | >128 | 128 | 64 | >128 | 128 | >128 | ND | ||||
| hVISA | 7206 | Blood | 7/26/07 | 4B | F | 71 | C | 2 | 128 | 64 | 64 | 64 | >128 | >128 | 4 | |||
| hVISA | 7207 | Pus | 12/11/07 | Mk | M | 78 | 2 | >128 | >128 | 128 | >128 | >128 | >128 | ND | ||||
| VISA | 7193 | Pus | 10/14/07 | 3D | M | 12 | I | 3 | >128 | >128 | 128 | 128 | >128 | 128 | 16 | III | 239 | I |
| VISA | 7195 | Body fluid | 9/22/07 | 3B | M | 56 | I | 3 | >128 | >128 | 64 | 64 | >128 | 0.5 | 8 | II | 5 | II |
| VISA | 7203 | Wound sample | 10/31/07 | 3C | F | 9 | C | 3 | 128 | 128 | 64 | 64 | >128 | >128 | 8 | III | 239 | I |
Group 1, isolates recovered in the years 2002 and 2003; group 2, isolates recovered in the years 2006 and 2007; hVISA, hetero-VISA.
3A, surgical ward; 3B, surgical ward; 3C, surgical ward; 3D, pediatric ward; 3F, orthopedic ward; 4B, medical ward; 4C, medical ward; 5E, chemotherapeutic ward; Mk, ward for monks.
M, male; F, female.
I, infected; C, colonized.
Van, vancomycin; Cefaz, cefazolin; Oxa, oxacillin; Oflox, ofloxacin; Tetra, tetracycline; Erythro, erythromycin; Gen, gentamicin; Teico, teicoplanin.
ND, not determined.
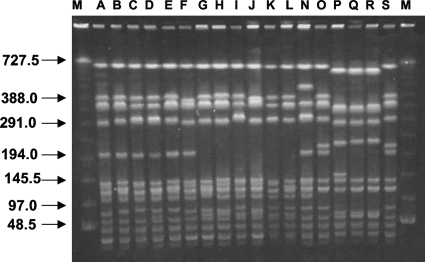
It was found that the genotypes of 6 hetero-VISA strains (arbitrarily chosen from among the 12 confirmed hetero-VISA strains) classified the strains as ST239 and SCCmec type III. Two of the three VISA strains, JCSC 7193 and JCSC 7203, had the ST239-SCCmec type III genotype, whereas the third one, JCSC 7195, was ST5 and SCCmec type II, as were strains Mu3 and Mu50. Strains JCSC 7193 and JCSC 7203 were agr type I, whereas JCSC 7195 was agr type II (Table 2). PFGE analyses of these isolates revealed that 1 of the 12 hetero-VISA strains (JCSC 6905) was identical to a hospital-associated MRSA (HA-MRSA) strain from the same hospital. The remaining 11 hetero-VISA and 2 VISA strains, JCSC 7193 and JCSC 7203, were closely related to the HA-MRSA strain. In contrast, the VISA strain JCSC 7195 had a banding pattern similar to those of Mu3 and Mu50 (Fig. 3).
FIG. 3.
PFGE banding patterns of SmaI-digested chromosomal DNA samples. Lanes contained samples from the following strains: A, JCSC 6903; B, JCSC 6904; C, JCSC 7194; D, JCSC 7204; E, JCSC 7206; F, JCSC 7207; G, JCSC 7192; H, JCSC 7196; I, JCSC 7201; J, JCSC 7205; K, JCSC 7193; L, JCSC 7203; N, JCSC 6902; O, JCSC 6905; P, JCSC 7195; Q, Mu3; R, Mu50; and S, JCSC 6890 (a HA-MRSA isolate from the same hospital). Lanes M, molecular size markers.
We found three VISA strains in samples from three patients in different wards during the second period of the surveillance. The first strain (JCSC 7193) was isolated in October 2007 from a 12-year-old boy with underlying acute leukemia who had a fractured leg and a ruptured spleen after a motorcycle accident. After being hospitalized and undergoing open reduction and internal fixation in a provincial hospital, he developed chronic osteomyelitis. He was referred to our tertiary-care hospital in September 2007 with septic shock caused by extended-spectrum β-lactamase-producing Escherichia coli and chronic osteomyelitis caused by MRSA. Between October 2007 and March 2008, he was treated periodically with vancomycin combined with either an aminoglycoside, a fluoroquinolone, or a carbapenem. The second strain (JCSC 7195) was found in a 56-year-old man with an underlying triple-vessel coronary artery anomaly and chronic renal failure. After receiving a coronary artery bypass graft, he underwent continuous ambulatory peritoneal dialysis and became infected with MRSA. Vancomycin was used to treat him. The third strain (JCSC 7203) was found in a 9-year-old girl with underlying osteofibrous dysplasia and persistent abscesses due to reconstructive surgery. The patient achieved good recovery after receiving cefazolin for 5 days.
The results of our screening test indicated that 3.6% of MRSA isolates from the years 2002 and 2003 and 7.2% of MRSA isolates from the years 2006 and 2007 could be suspected of having reduced vancomycin susceptibilities. After confirmation by PAPs, the prevalence of hetero-VISA during the first period was determined to be 0.8%, compared to 2.2% during the second period. In addition, 0.8% of the strains from the second period exhibited VISA phenotypes, implying that the trend toward the reduction of vancomycin susceptibility in our hospital had gradually increased over the previous half decade. The strains with reduced vancomycin susceptibilities were associated with infection as well as colonization of the patients. Two of the three VISA strains caused serious infections refractory to vancomycin therapy and the prolonged hospitalization of two patients. These two VISA strains were genetically different, suggesting that reduced vancomycin susceptibility in our hospital occurred among different clones of MRSA. This finding coincides with the data in a previous report of the emergence of reduced susceptibility to vancomycin in various lineages of MRSA (17).
The present prevalences of hetero-VISA and VISA strains in our hospital are 2.2 and 0.8%, respectively, which are comparable to those reported in several previous studies, 2.7% in Hong Kong (35, 36), 2.6% in Ireland (8), and 1.1% in Italy (25), but lower than those in university hospitals in Japan in 1996 (9.3 to 20%) (13).
In patients who are colonized by a hetero-VISA strain, subsequent infection with this organism may occur. It has been reported previously that patients with MRSA colonization have a 35% risk of developing invasive disease within a year (19). Moreover, the family members of the patients carrying MRSA may potentially be at risk of becoming colonized as well (12, 16). Although the resistance phenotypes of the strains used in the previous studies were not characterized by laboratory testing, these strains may nevertheless cause infections with increased risks of clinical failure (4).
The strains with reduced susceptibilities to vancomycin could not be detected by a routine susceptibility test. Various screening methods for strains with reduced vancomycin susceptibilities have been described previously (33, 34); however, confirmation of the resistance phenotypes of screening-positive isolates by population analyses is recommended (8, 18). This study revealed a slight increase in strains with reduced vancomycin susceptibilities over time at a university hospital in northeast Thailand. This is the first report of VISA strains in Thailand. The appropriate use of antimicrobial agents and effective control of transmission will help retard the emergence of these organisms.
Acknowledgments
This work was supported by a grant-in-aid for 21st Century COE Research and by the Japan Society for the Promotion of Science.
We thank the CMDL, Faculty of Associated Medical Sciences, Khon Kaen University, for support; Natthanan Hongsrichan, Boualay Norchaleun, and the staff of the Clinical Microbiology Laboratory at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University, for collecting the clinical isolates; and Bryan Roderick Hamman for assistance with the English language presentation of the manuscript.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Appelbaum, P. C. 2006. MRSA—the tip of the iceberg. Clin. Microbiol. Infect. 12(Suppl. 2)3-10. [DOI] [PubMed] [Google Scholar]
- 2.Beam, J. W., and B. Buckley. 2006. Community-acquired methicillin-resistant Staphylococcus aureus: prevalence and risk factors. J. Athl. Train. 41337-340. [PMC free article] [PubMed] [Google Scholar]
- 3.Brummett, R. E. 1981. Effects of antibiotic-diuretic interactions in the guinea pig model of ototoxicity. Rev. Infect. Dis. 3(Suppl.)S216-S223. [PubMed] [Google Scholar]
- 4.Charles, P. G. P., P. B. Ward, D. P. R. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38448-451. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement, M100-S16. CLSI, Wayne, PA.
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farber, B. F., and R. C. Moellering, Jr. 1983. Retrospective study of the toxicity of preparations of vancomycin from 1974 to 1981. Antimicrob. Agents Chemother. 23138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgibbon, M. M., A. S. Rossney, and B. O'Connell. 2007. Investigation of reduced susceptibility to glycopeptides among methicillin-resistant Staphylococcus aureus isolates from patients in Ireland and evaluation of agar screening methods for detection of heterogeneously glycopeptide-intermediate S. aureus. J. Clin. Microbiol. 453263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarwis, T. M. Perl, and F. C. Tenover. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36429-439. [DOI] [PubMed] [Google Scholar]
- 10.Gemmel, C. G. 2004. Glycopeptide resistance in Staphylococcus aureus: is it a real threat? J. Infect. Chemother. 1069-75. [DOI] [PubMed] [Google Scholar]
- 11.Griffith, R. S. 1981. Introduction to vancomycin. Rev. Infect. Dis. 3(Suppl.)S200-S204. [PubMed] [Google Scholar]
- 12.Hicks, N. R., E. P. Moore, and E. W. Williams. 1991. Carriage and community treatment of methicillin-resistant Staphylococcus aureus: what happens to colonized patients after discharge? J. Hosp. Infect. 1917-24. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 3501670-1673. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40135-136. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1147-155. [DOI] [PubMed] [Google Scholar]
- 16.Hollis, R. J., J. L. Barr, B. N. Doebbeling, M. A. Pfaller, and R. P. Wenzel. 1995. Familial carriage of methicillin-resistant Staphylococcus aureus and subsequent infection in a premature neonate. Clin. Infect. Dis. 21328-332. [DOI] [PubMed] [Google Scholar]
- 17.Howe, R. A., A. Monk, M. Wootton, T. R. Walsh, and M. C. Enright. 2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe, R. A., M. Wootton, T. R. Walsh, P. M. Bennett, and A. P. Macgowan. 2000. Heterogeneous resistance to vancomycin in Staphylococcus aureus. J. Antimicrob. Chemother. 45130-131. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S. S., and R. Platt. 2003. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin. Infect. Dis. 36281-285. [DOI] [PubMed] [Google Scholar]
- 20.Jevons, M. P. 1961. ‘Celbenin’-resistant staphylococci. Br. Med. J. 1124-125. [Google Scholar]
- 21.Kaccia, M., and L. C. McDonald. 2004. Vancomycin-resistant Staphylococcus aureus—New York. MMWR Morb. Mortal. Wkly. Rep. 53322-323. [PubMed] [Google Scholar]
- 22.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 473040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lulitanond, A., A. Chanawong, P. Sribenjalux, W. Kaewkes, N. Charoensri, D. Leumsai, and P. Monpou. 2005. Occurrence of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Srinagarind Hospital. J. Infect. Dis. Antimicrob. Agents 229-14. [Google Scholar]
- 25.Marchese, A., G. Balistreri, E. Tonoli, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D., V. Urdaneta, and A. Weltman. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania. MMWR Morb. Mortal. Wkly. Rep. 51902. [PubMed] [Google Scholar]
- 27.Saha, B., A. K. Singh, A. Ghosh, and M. Bal. 2008. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 5772-79. [DOI] [PubMed] [Google Scholar]
- 28.Schwaber, M. J., S. B. Wright, Y. Carmeli, L. Venkataraman, P. C. DeGirolami, A. Gramatikova, T. M. Perl, G. Sakoulas, and H. S. Gold. 2003. Clinical implications of varying degrees of vancomycin susceptibility in methicillin-resistant Staphylococcus aureus bacteremia. Emerg. Infect. Dis. 9657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw, B. E., T. Boswell, J. L. Byrne, C. Yates, and N. H. Russell. 2007. Clinical impact of MRSA in a stem cell transplant unit: analysis before, during and after an MRSA outbreak. Bone Marrow Transplant. 39623-629. [DOI] [PubMed] [Google Scholar]
- 30.Shopsin, B., B. Mathema, P. Alcabes, B. Said-Salim, G. Lina, A. Matsuka, J. Martinez, and B. N. Kreiswirth. 2003. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 41456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murry, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voss, A., J. W. Mouton, E. P. van Elzakker, R. G. Hendrix, W. Goessens, J. A. Kluytmans, P. F. Krabbe, H. J. de Neeling, J. H. Sloos, N. Oztoprak, R. A. Howe, and T. R. Walsh. 2007. A multi-center blinded study on the efficiency of phenotypic screening methods to detect glycopeptide intermediately susceptible Staphylococcus aureus (GISA) and heterogeneous GISA (h-GISA). Ann. Clin. Microbiol. Antimicrob. 69-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 392439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong, S. S., P. L. Ho, P. C. Woo, and K. Y. Yuen. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 29760-767. [DOI] [PubMed] [Google Scholar]
- 36.Wong, S. S., T. K. Ng, W. C. Yam, D. N. Tsang, P. C. Woo, S. K. Fung, and K. Y. Yuen. 2000. Bacteremia due to Staphylococcus aureus with reduced susceptibility to vancomycin. Diagn. Microbiol. Infect. Dis. 36261-268. [DOI] [PubMed] [Google Scholar]