Abstract
Mycobacterium leprae, the etiological agent of leprosy, is noncultivable on axenic media. Therefore, the viability of M. leprae for clinical or experimental applications is often unknown. To provide new tools for M. leprae viability determination, two quantitative reverse transcriptase PCR (RT-PCR) assays were developed and characterized. M. leprae sodA mRNA and 16S rRNA were used as RNA targets, and M. leprae repetitive element (RLEP) DNA was used to determine relative bacterial numbers in the same purified bacterial preparations or from crude biological specimens. Results demonstrated that both assays were good predictors of M. leprae viability during short-term experiments (48 h) involving rifampin (rifampicin) treatment in axenic medium, within rifampin-treated murine macrophages (MΦ), or within immune-activated MΦ. Moreover, these results strongly correlated those of other M. leprae viability assays, including radiorespirometry-based and Live/Dead BacLight viability assays. The 16S rRNA/RLEP assay consistently identified the presence of M. leprae in eight multibacillary leprosy patient biopsy specimens prior to multidrug therapy (MDT) and demonstrated a decline in viability during the course of MDT. In contrast, the sodA/RLEP assay was able to detect the presence of M. leprae in only 25% of pretreatment biopsy specimens. In conclusion, new tools for M. leprae viability determination were developed. The 16S rRNA/RLEP RT-PCR M. leprae viability assay should be useful both for short-term experimental purposes and for predicting M. leprae viability in biopsy specimens to monitor treatment efficacy, whereas the sodA/RLEP RT-PCR M. leprae viability assay should be limited to short-term experimental research purposes.
Leprosy is a chronic infectious disease of skin and peripheral nerves and is of special concern because it can progress to peripheral neuropathy and permanent progressive deformity. Despite a marked reduction in the prevalence of leprosy since the implementation of multidrug therapy (MDT), the detection rate for new cases has not shown a substantial decline (2). One explanation is that standard immunological and histological approaches for disease assessment are less effective in the diagnosis of early leprosy, and therefore, disease and transmission can progress. In addition, in vitro Mycobacterium leprae viability assays such as those based on radiorespirometry (RR) and the Live/Dead BacLight fluorescent bacterial viability assay require large quantities of bacteria, 107 and 106 bacteria, respectively, for reliable detection and are therefore not applicable for direct detection in clinical specimens (14, 28). The bacterial index (BI) is a long-established method for monitoring the patients' responses to chemotherapy by giving an estimation of the number of acid-fast bacilli present in skin smears of lesions and other specific sites of the skin of leprosy patients. The BI range is 1 to 6, where 1 is the least amount of bacilli detectable and 6 is the most. However, the BI drops very slowly during treatment (1 BI/year of therapy), and in some cases, the BI remains unchanged during and after treatment, making it difficult to determine drug efficacy or relapse of active infection (24).
The fluorescent-based and RR assays are both suitable for viability determinations of nude (nu/nu) mouse footpad (MFP)-derived preparations of M. leprae to serve as an inoculum for in vivo and in vitro experimental infection models because these preparations reproducibly contain very high levels of M. leprae (109 bacteria/ml). However, the ability of these assays to monitor the viability of M. leprae as an indicator of environmental influence or of host cell responses in infected cultures or in animal models, where only low numbers of bacteria are used, is very limited. Therefore, there is great need for a rapid and sensitive viability assay for M. leprae.
To address this, we developed and characterized two quantitative reverse transcriptase PCR (RT-PCR) assays to provide new tools for determinations of Mycobacterium leprae viability for clinical and experimental purposes. M. leprae sodA mRNA and 16S rRNA were used as the RNA targets for these assays, and M. leprae repetitive element (RLEP) DNA was used to determine relative bacterial numbers in the same purified bacterial preparations or from crude biological specimens. The sensitivity and specificity of the assay were examined, and their ability to detect the viability in multibacillary (MB) leprosy patients' biopsy specimens prior to and during MDT was analyzed. Results demonstrated that both assays were good predictors of viability for experimental purposes such as during short-term (48 h) rifampin (rifampicin) treatment in axenic medium or within rifampin-treated murine macrophages (MΦ) or immune-activated MΦ. Analysis of human biopsy specimens from MB leprosy patients monitored for up to 2 years after initiation of leprosy MDT demonstrated that the 16S rRNA/RLEP assay but not the sodA/RLEP assay consistently identified the presence of viable M. leprae in MB biopsy specimens prior to MDT, and 16S rRNA levels declined during MDT treatment.
MATERIALS AND METHODS
M. leprae.
M. leprae Thai-53 bacteria, maintained in continuous serial passage in the hind footpads of athymic nu/nu mice (Hsd, athymic nu/nu; Harlan Inc., Indianapolis, IN), were isolated approximately 6 months postinfection as previously described (28), and acid-fast bacillus counts were then determined by direct counting according to a method described previously by Shepard and McRae (23). The viability of each preparation was determined with axenic medium by the oxidation of [14C]palmitate using the Buddemeyer RR technique described below but using the day 1 cpm as an indicator of viability. The bacterial preparations were held overnight at 4°C pending quality control testing for contamination. Freshly harvested, highly viable bacilli (≥80%) were used within 24 h of harvesting.
RR.
The metabolism of M. leprae was used as an indicator of viability by determining the oxidation of [14C]palmitic acid to 14CO2 using Buddemeyer RR as previously described (5). Briefly, 1 × 107 M. leprae bacteria were suspended in 1.0 ml of Bactec 7H12B medium (Becton Dickinson, San Jose, CA) in a 5-ml glass vial with a loosened cap. This vial was inserted into a wide-mouth liquid scintillation vial lined with filter paper impregnated with NaOH, 2,5-diphenyloxazole (Sigma-Aldrich), and concentrate I (Kodak, Chicago, IL). Daily cpm were recorded, and day 7 cumulative cpm were used as an indicator of viability.
Fluorescent staining for quantification of bacterial viability.
The membrane integrity of individual M. leprae bacteria (as an indicator of viability in a suspension) was determined using a Live/Dead BacLight viability kit (Molecular Probes, Eugene, OR) as previously described (28). Briefly, M. leprae bacteria were washed twice (10,000 × g for 5 min) in sterile normal saline and incubated for 15 min at room temperature with a final concentration of 6 μM Syto9 and 30 μM propidium iodine. The bacteria were washed twice in normal saline, the pellet was resuspended in 20 μl of 10% (vol/vol) glycerol in normal saline, and 5 μl of the suspension was placed onto a glass slide with a glass coverslip. The total number of bacteria and the number of dead bacteria were enumerated by direct counting of fluorescent green and red bacilli, respectively, using a fluorescence microscope with the appropriate single-band-pass filter sets. The excitation and emission maxima were 480 nm and 500 nm, respectively, for Syto9 and 490 nm and 635 nm, respectively, for propidium iodine. The number of viable bacteria in a preparation was then determined by subtracting the number of dead bacteria (red) from the total number of bacteria (green), and the percentage of viable bacilli in a preparation after treatment was determined by dividing the number of viable bacilli after treatment by that prior to treatment at the same time point.
Rifampin treatment in axenic medium.
M. leprae bacteria were subjected to rifampin treatment in axenic medium using the following procedure. A stock solution of rifampin (Sigma-Aldrich, St. Louis, MO) was made in dimethyl sulfoxide (100 ng/ml) and filter sterilized. Aliquots of 1 × 108 purified nu/nu mouse-derived M. leprae bacteria were added to 5 ml 7H9 medium supplemented with oleic acid-albumin-dextrose-catalase and containing 20 μg/ml (final concentration) rifampin. Cultures were maintained at 33°C, which is the optimum temperature for the maintenance of M. leprae viability (28), and with 5% CO2 for up to 2 weeks. Controls consisted of M. leprae bacteria in 7H9 medium plus the same concentration of dimethyl sulfoxide added to drug-treated M. leprae cells.
Preparation and treatment of macrophage cultures.
RPMI 1640 medium supplemented with 25 mM HEPES, 2 mM glutamine (Sigma-Aldrich), and 10% (vol/vol) fetal calf serum was used throughout these studies. Resident peritoneal cells from Swiss Webster mice (Harlan, Indianapolis, IN) were harvested and allowed to adhere for at least 6 h at 37°C and with 5% (vol/vol) CO2 on Lux plastic coverslips (Miles Laboratory, Elkhart, IN) in 24-well tissue culture plates (Corning Incorporated, Corning, NY) as previously described (19). After washing to remove nonadherent cells, the adherent MΦ were either infected with fresh nu/nu mouse-derived M. leprae bacteria at a multiplicity of infection of 20:1 for 16 h at 33°C or first activated with 500 IU gamma interferon (IFN-γ)/ml (R&D Systems, Minneapolis, MN) and 5 ng/ml lipopolysaccharide (Sigma-Aldrich) for 6 h and then infected with M. leprae. Extracellular M. leprae bacteria were removed by washing the coverslips. Nonactivated MΦ were then treated with rifampin at 20 μg/ml (final concentration) or 50 μg/ml (final concentration) ampicillin (Sigma-Aldrich), and cultures were maintained for up to 48 h. Nontreated infected cells served as controls.
Patient skin biopsy specimens.
A total of 19 skin biopsy specimens (4-mm3 punch) and skin slit smear specimens from eight MB leprosy patients were obtained for leprosy diagnosis and follow-up after initiation of MDT at the National Hansen's Disease Programs Outpatient Clinic at Ochsner's Hospital, Baton Rouge, LA. One-half of each biopsy specimen was formalin fixed and paraffin embedded using standard techniques. The remaining biopsy material was stored frozen in Tissue-Tek optimal cutting temperature (OCT) preservative at −80°C for 3 to 8 years prior to use in this study. Patients were classified by clinical signs, BI of skin smear samples, and histopathology of stained paraffin sections according to the Ridley-Jopling scale (21). All patients in this study were classified as having either borderline lepromatous leprosy or lepromatous leprosy. Before the study was undertaken, it was reviewed by the Louisiana State University Institutional Review Board, Baton Rouge, LA. Since the specimens were unused portions of skin biopsy specimens taken for diagnostic purposes and coded so that names of individual patients were not available to the research staff or for any other purpose of the study, this study was determined to be exempt for the purpose of human subject review.
Extraction of M. leprae RNA and DNA.
M. leprae RNA and DNA were simultaneously extracted from the same sample using TRIzol reagent (Invitrogen) and a modification of the previously described single-tube homogenization/RNA extraction protocol using FastRNA Blue tubes (FastRNA kit-Blue; MP Biomedicals, Santa Ana, CA), and a FastPrep FP 24 instrument (MP Biomedicals) (32). M. leprae cells from axenic media were pelleted and washed two times in cold phosphate-buffered saline (PBS) at 14,000 × g and at 4°C for 10 min and resuspended in 80 μl sterile diethyl pyrocarbonate-treated H2O. For M. leprae in frozen skin biopsy specimens, OCT was partially thawed on ice. The tissue (approximately 30 mg) was removed from OCT, rinsed three times briefly in sterile cold PBS, and minced into small pieces with a sterile scalpel in 200 μl TRIzol reagent. For M. leprae in infected MΦ cultures, MΦ were lysed with 0.1 N NaOH (750 μl) for 3 min and then neutralized with an equal volume of 0.1 N HCl. Bacteria were pelleted (4°C) and washed twice in cold PBS. TRIzol reagent was added to all sample preparations to a total volume of 1 ml prior to adding to FastRNA Blue tubes. Samples were homogenized twice in the FastPrep FP 24 instrument at a speed setting of 6.5 for 45 s. Tubes were allowed to cool for 2 min between homogenizations. After homogenization, tubes were chilled on ice for 5 min, 200 μl chloroform-isoamyl alcohol (24:1) was then added, and tubes were mixed by vortexing for 10 s and then spun at 700 × g and at 4°C for 5 min. The liquid was transferred into a new tube and spun again at 14,000 × g for 10 min. M. leprae RNA was purified from 400 μl of the aqueous phase, and DNA was removed from RNA preparations using the DNA-free kit (Ambion, Inc., Austin, TX) as specified by the manufacturer, precipitated using standard techniques, resuspended in 30 μl diethyl pyrocarbonate-treated H2O, and stored at −80°C until use.
DNA was purified from the remaining aqueous phase and interphase of the FastRNA Blue tubes. Briefly, 100 μl of 10 mM Tris-EDTA (pH 8.0) and 150 μl chloroform-isoamyl alcohol (24:1) were added to the remaining aqueous phase and interphase material (∼500 μl) and homogenized in the FastPrep FP 24 instrument twice. After centrifugation at 14,000 × g for 10 min, the aqueous phase was transferred into another tube and precipitated with 0.3 M sodium acetate and 2 volumes of cold ethanol. The DNA pellet was washed in 70% ethanol, dissolved in 30 μl of sterile distilled water, and stored at −80°C until use.
Reverse transcription of M. leprae RNA.
RNA (500 ng) was converted to cDNA using Advantage cDNA polymerase mix and an Advantage RT-for-PCR kit (BD Biosciences) according to the manufacturer's recommendations by using random hexamer primers, which are a mixture of oligonucleotides representing all possible sequences for a hexamer and which were included in the kit. Controls for DNA contamination consisted of total RNA incubated with the reverse transcription reagents excluding RT and human and mouse cDNAs.
Real-time PCR.
The levels of M. leprae sodA mRNA and 16S rRNA in M. leprae from axenic medium, from cultured MΦ, or in skin biopsy specimens were determined using real-time RT-PCR. These levels were normalized for bacterial numbers using a previously characterized, DNA-based, real-time PCR assay for RLEP (29). Primers and probes were designed using Primer Express 2.0 software (Applied Biosystems) (Table 1). Purified M. leprae DNA or cDNA (5 μl) was added to a total PCR mixture of 25 μl containing 2× TaqMan master mix, 500 nM of each primer, and 100 nM of each probe for RNA-based PCR assays (sodA mRNA or 16S rRNA) or 200 nM of each primer and 100 nM of the probe for the DNA-based PCR assay (RLEP). Reaction mixtures were subjected to 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min using a 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA). A standard curve for each PCR assay was generated by using serial 10-fold dilutions of purified M. leprae DNA ranging from 10 ng to 10 fg. Unknown values were interpolated automatically for each sample using the standard curve method and normalized using the RLEP data for the same sample. The viability of M. leprae in human skin biopsy specimens was determined by comparing normalized 16S rRNA values for patients prior to and up to 2 years after the start of MDT treatment.
TABLE 1.
Primer and probe sequences for M. leprae real-time PCR and RT-PCR TaqMan assays
| Target | Description | Primer | Primer sequence |
|---|---|---|---|
| 16S rRNA | 16S rRNA | ML16S rRNATaq-F | 5′-GCA TGT CTT GTG GTG GAA AGC-3′ |
| ML16S rRNATaq-R | 5′-CAC CCC ACC AAC AAG CTG AT-3′ | ||
| ML16S rRNATaq-Probe | 5′CAT CCT GCA CCG CA-3′ | ||
| sodA mRNA | Superoxide dismutase A | MLsodATaq-F | 5′-ACC ACG CCG CAT ATG TCA-3′ |
| MLsodATaq-R | 5′-CGC GTG CCT CGT CAA GT-3′ | ||
| MLsodATaq-Probe | 5′-TGG CAA GCG CGT CAT TGA CAC CT-3′ | ||
| RLEP | Repetitive element | MLRLEPTaq-F | 5′-GCA GCA GTA TCG TGT TAG TGA A-3′ |
| MLRLEPTaq-R | 5′-CGC TAG AAG GTT GCC GTA T-3′ | ||
| MLRLEPTaq-Probe | 5′-CGC CGA CGG CCG GAT CAT CGA-3′ |
Specificity and sensitivity of assays.
The specificity of each real-time PCR TaqMan assay was determined by analyzing 10 ng of purified DNA from M. leprae; nine other mycobacterial species including Mycobacterium tuberculosis H37Rv ATCC 27294, M. marinum ATCC 927, M. bovis BCG ATCC 35734, M. ulcerans ATCC 19423, M. simiae ATCC 25275, M. avium ATCC 25291, M. intracellulare ATCC 13950, M. kansasii ATCC 35775, and M. smegmatis ATCC 14468; and other bacterial species including Staphylococcus epidermidis ATCC 12228, Streptococcus pyogenes ATCC 12345, and Escherichia coli ATCC 25992. In addition, mouse and human cDNAs were also analyzed. The sensitivity or lower limit of detection of each assay was determined by analyzing 10-fold serial dilutions of M. leprae DNA and identifying the highest dilution, which still gave a positive value (threshold cycle [CT] values of <37).
Statistical analysis.
The standard curves for each RT-PCR assay using CT values versus the serial DNA or cDNA concentration were calculated using a linear regression model (GraphPad InStat version 3 software). The means and standard deviations of the results of sodA/RLEP and 16S rRNA/RLEP RT-PCR assays for the detection of M. leprae viability in axenic medium and MΦ cultures under various treatment conditions were calculated using the Student t test. An alpha value of 0.05 was used for all analyses. All statistical comparisons were made using the linear Pearson correlation coefficient (r) (GraphPad InStat version 3 software) as a measure of correlation between assays at a particular time interval.
RESULTS
Identification of suitable targets for viability assays.
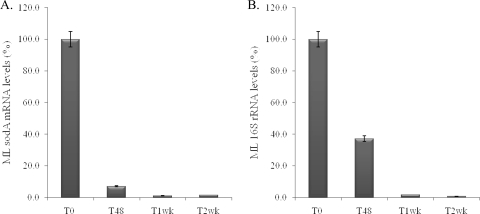
Results of sodA/RLEP and 16S rRNA/RLEP RT-PCR assays demonstrated that levels of sodA gene transcripts were reduced to 5% of those of nontreated controls 48 h after rifampin treatment (Fig. 1A) and that these levels were significantly lower (P < 0.001) than those of the 16S rRNA gene, which were reduced to 38% of those of nontreated controls (Fig. 1B). However, after a week of treatment, both were essentially background levels. In addition, a reduction of sodA mRNA levels could be observed as early as within 12 h of rifampin treatment compared to untreated controls (data not shown).
FIG. 1.
Molecular viability analyses of rifampin-treated M. leprae. (A) M. leprae (ML) sodA/RLEP levels. (B) M. leprae 16S rRNA/RLEP levels. T0, untreated; T48, 48 h after rifampin treatment; T1wk, 1 week after rifampin treatment; T2wk, 2 weeks after rifampin treatment. The data are representative of three replicates for three independent experiments.
Specificity and sensitivity of molecular M. leprae viability assays.
The specificities and lower limits of detection of each assay, the sodA mRNA/RLEP and 16S rRNA/RLEP RT-PCR assays, were analyzed using DNA from nine other mycobacterial species, Staphylococcus epidermidis, Streptococcus pyogenes, E. coli, and mouse and human DNA. Results showed that all three assays resulted in CT values of >39, indicating 100% specificities of these assays for M. leprae (data not shown). The sensitivity of detection of the M. leprae viability assays, defined as the lower limit of detection of M. leprae for each assay, was analyzed using serial 10-fold dilutions of purified M. leprae DNA and the standard curve method. The detection limit of the sodA/RLEP assay was 1 pg (CT = 34.73 ± 0.3), and that of the 16S rRNA gene was 100 fg (CT = 36.42 ± 0.6). Since each M. leprae genome is equivalent to 3 fg, these data suggest that these assays can detect 300 and 30 bacteria, respectively.
M. leprae viability determination in infected MΦ cultures.
The ability of the RT-PCR-based M. leprae viability assays to detect the effects of antileprosy drug treatment of infected MΦ or the effects of immune-activated murine MΦ (the host cell of M. leprae) on M. leprae viability was evaluated. The sodA/RLEP RT-PCR assay results showed that 16% viable M. leprae cells remained in MΦ after 48 h of rifampin treatment and that only 1% viable M. leprae cells remained after 48 h of infection of IFN-γ-activated MΦ (Table 2). Similar trends were noted for the 16S rRNA/RLEP RT-PCR, Live/Dead BacLight bacterial viability, and RR assays. Thus, data from both molecular-based assays, the 16S rRNA/RLEP and sodA/RLEP assays, strongly correlated (P < 0.0001) with RR and Live/Dead BacLight bacterial viability assay data (Table 2). However, the effects of the different treatments on M. leprae viability were lower than those determined by the sodA/RLEP RT-PCR viability assay. Ampicillin treatment led to a 20% decrease in M. leprae viability using the sodA/RLEP RT-PCR assay; however, the Live/Dead BacLight and RR assays both showed that ampicillin did not have an effect on M. leprae viability. The 16S rRNA/RLEP assay showed that ampicillin has less of an effect on M. leprae viability than the sodA/RLEP assay. This result was expected since M. leprae contains a β-lactamase enzyme, which makes it resistant to ampicillin-like drugs (18).
TABLE 2.
Comparison of various methods for M. leprae viability determination using infected murine MΦ
| M. leprae-infected MΦ treatment | Mean % viability ± SD
|
|||
|---|---|---|---|---|
| sodAa | 16S rRNAb | BacLightc | RRd | |
| Rifampin (20 pg/ml) | 16 ± 4.1 | 57 ± 14.6 | 60 ± 5.5 | 49 ± 3.2 |
| Ampicillin (50 μg/ml) | 80 ± 5.5 | 90 ± 5.1 | 100 ± 6.6 | 97 ± 6.8 |
| IFN-γ (100 IU) | 1 ± 0.3 | 13 ± 0.6 | 23 ± 1.4 | 11 ± 1.3 |
sodA/RLEP RT-PCR values for M. leprae in treated murine MΦ divided by that in untreated murine MΦ.
16S rRNA/RLEP RT-PCR values for M. leprae in treated murine MΦ divided by that in untreated murine MΦ.
Live/Dead BacLight bacterial viability assay values for M. leprae in treated murine MΦ divided by that in untreated murine MΦ.
Day 7 cumulative cpm using Buddemeyer RR of M. leprae viability in treated murine MΦ divided by that in untreated murine MΦ.
M. leprae viability in paired MB leprosy patient skin biopsy specimens.
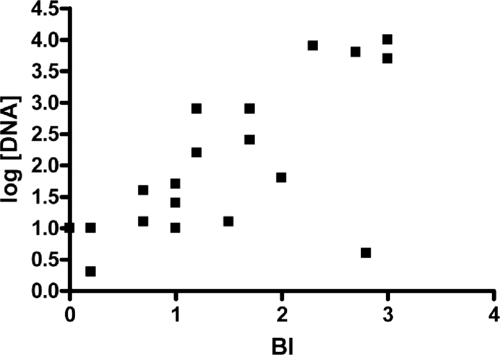
A preliminary experiment was performed to determine the utility of the sodA/RLEP and16S rRNA/RLEP RT-PCR M. leprae viability assays to detect M. leprae viability directly from skin biopsy materials of MB leprosy patients. Results indicated that the sodA/RLEP assay was able to detect sodA cDNA from only two of eight pretreatment biopsy specimens (data not shown), and therefore, the remaining biopsy specimens taken from the same patients after the initiation of MDT were not evaluated. In contrast, the 16S rRNA/RLEP assay showed positive results for all pretreatment specimens, which declined during MDT treatment (Table 3). Moreover, when the BI values of patients' skin slit smear samples were compared to the number of M. leprae cells as a function of M. leprae DNA using real-time RLEP PCR, a significant association was observed (r = 0.6942; P = 0.001). Higher DNA concentrations correlated with higher BI values, and lower DNA concentrations correlated with lower BIs (Fig. 2). However, when 16S rRNA/RLEP viability data were compared to patients' BIs, no correlation was observed (r = 0.4604; P = 0.0842).
TABLE 3.
Analysis of M. leprae viability in MB leprosy patients prior to and during leprosy MDT
| Patient | Biopsy specimen | Leprosy classificationa | MDT treatment | BIb | Mean 16S rRNA/RLEP value ± SD (% viability)c |
|---|---|---|---|---|---|
| 1 | 04-01 | BL | Untreated | 3.5 | 4,314.3 ± 1,395.7 |
| 05-01 | 1 yr | 3.17 | 305.4 ± 112.5 (9) | ||
| 06-01 | 2 yr | 1.5 | 294 ± 76.4 (3) | ||
| 2 | 04-02 | BL | Untreated | 3 | 14.8 ± 3.1 |
| 05-02 | 1 yr | 2 | 8.8 ± 3.2 (59) | ||
| 06-02 | 2 yr | 2.8 | 8.3 ± 3.7 (56) | ||
| 3 | 03-03 | LL | Untreated | 1 | 50.2 ± 11.5 |
| 04-03 | 1 yr | 0.66 | 6.8 ± 4.9 (13) | ||
| 05-03 | 2 yr | 0.17 | 0.1 ± 0.05 (0.2) | ||
| 4 | 00-04 | LL | Untreated | 1 | 337.7 ± 87.8 |
| 00-04 | 6 mo | 1 | 11.5 ± 4.0 (4) | ||
| 5 | 03-05 | LL | Untreated | 1.2 | 4.6 ± 0.6 |
| 04-05 | 1 year | 0.7 | 1.3 ± 0.2 (28) | ||
| 6 | 02-06 | LL | Untreated | 2.3 | 10.5 ± 1.2 |
| 03-06 | 1 year | 1.7 | 1.0 ± 0.1 (9.4) | ||
| 7 | 03-07 | LL | Untreated | 2.3 | 39.5 ± 2.8 |
| 04-07 | 1 year | 1.7 | 0.6 ± 0.3 (1.6) | ||
| 8 | 05-08 | LL | Untreated | 1.7 | 120.8 ± 23.2 |
| 06-08 | 1 year | 1.2 | 0.5 ± 0.2 (0.4) |
Leprosy patient classification according to the Ripley-Jopling scale (21). BL, borderline lepromatous leprosy; LL, lepromatous leprosy.
BI (acid-fast bacillus count) of skin slit smear specimens from a leprosy patient (20).
Means and standard deviations of 16S rRNA/RLEP RT-PCR values derived from cDNA of skin biopsy specimens from untreated and treated leprosy patients and percent M. leprae viability for assays of each treated specimen derived by dividing the number of 16S rRNA/RLEP RT-PCR values for leprosy patients' biopsy specimens after treatment by that of biopsy specimens from untreated patients.
FIG. 2.
Comparison of M. leprae viability using real-time RLEP PCR and BI analyses of patients' biopsy specimens as a function of pretreatment and posttreatment using linear Pearson correlation between BI and DNA concentrations for MB leprosy patients (P = 0.001; r = 0.6942).
DISCUSSION
Determinations of M. leprae viability are extremely difficult due primarily to the inability to cultivate this bacterium on axenic media. The discovery by Shepard and McRae in 1960 (23) of the MFP technique to demonstrate the replication of M. leprae bacteria was a research milestone, permitting the testing of new antileprosy drugs, determination of drug-resistant strains of M. leprae from patient specimens, and initial evaluation of vaccine protection. Variations of the MFP technique involving titration in large numbers of mice have been shown to be helpful for the detection of differences in the relative viabilities of different suspensions of M. leprae (30). However, this labor-intensive, time-consuming, expensive technique is impractical for the study of interactions of M. leprae with its host cell in vitro. Further complicating this is the declining availability of MFP laboratories for M. leprae viability testing around the world.
When large numbers of nu/nu mouse-derived M. leprae bacteria became available to the leprosy research community, RR, first described by Franzblau in 1988 (5), was utilized to determine the viability of a bacterial preparation based on the rate of oxidation of [14C]palmitate by M. leprae. RR was found to correlate well with growth in MFP (28) and was therefore useful to evaluate the susceptibility of M. leprae to novel antileprosy drugs (6), a variety of environmental conditions (28), as well as UV (27) and gamma (1) radiation. This technique was also useful for characterizations of the role of activated MΦ in host resistance to leprosy (19) and the effects of Schwann cells (the target of M. leprae in peripheral nerves) on M. leprae viability (7). However, the need for large quantities of bacteria (107 bacteria) for RR-based M. leprae viability assays limits the use of these assays for clinical purposes and in experiments where low numbers of bacteria are available (14, 19). In addition, RR requires the use of radioactive substances, which is highly restricted in many areas of the world.
Recently, a fluorescence-based assay for M. leprae viability determination, the Live/Dead BacLight bacterial viability assay, was developed (14). This assay, based on cell membrane integrity, also correlated very well with data from MFP and RR assays for determinations of M. leprae viability; however, it also depends on the use of relatively large numbers of bacteria (106 bacteria) for analysis and therefore limits it usefulness for clinical purposes and experimental conditions for which only low numbers of bacteria are available.
PCR assays based on the amplification of various DNA sequences within genes encoding the 18-kDa, 36-kDa, and 65-kDa proteins; Ag 85; and the multicopy repeat sequence RLEP of M. leprae have been successfully used to detect M. leprae in crude biological specimens even when low numbers of bacteria are present (11, 12, 15, 17, 26, 31). However, an important limitation of these DNA-based PCR assays is their inability to distinguish between viable and dead organisms and thereby provide information for drug efficacy in clinical settings and for short-term experimental procedures in vitro.
The detection of M. leprae RNA has been proposed to be a promising tool for the rapid detection and the measurement of the viability of pathogenic mycobacteria, since the degradation of RNA is relatively rapid upon cell death (3, 13). A previous study of M. tuberculosis measured levels of M. tuberculosis 85B (alpha antigen) mRNA, 16S rRNA, and IS6110 DNA using RT-PCR of patients' sputa to ascertain whether they could serve as potential markers of a response to chemotherapy (4). Results showed a rapid disappearance of M. tuberculosis mRNA from sputum while DNA persisted in sputum from certain patients even after tuberculosis was cured. A preliminary study using the M. leprae 16S rRNA gene as a predictor of viability showed this nucleic acid species to be a suitable target for the detection of M. leprae and its viability in clinical specimens using RT-PCR (8), including skin slit smear samples from treated patients (9, 16). Although these assays were able to detect M. leprae viability in clinical specimens, they were not tested on paired samples from the same patient obtained before and during or after MDT or under short-term experimental conditions where viability determination can be a critical denominator.
In the present study, the utility of two RNA-based RT-PCR assays, one using the 16S rRNA gene as the RNA target and the other using the more labile sodA mRNA as the target, was evaluated for both experimental and clinical usefulnesses. The sodA mRNA transcript, encoding superoxide dismutase A (ML0072c) of M. leprae, was selected as the mRNA target for this assay because of the gene transcripts tested, including hsp18 (ML1795), gyrA (ML0006), and rpoB (ML1891c); the sodA transcript levels remained stable for at least 48 h after harvesting from the MFP tissues but rapidly degraded after the death of M. leprae cells (data not shown). Also, since real-time RLEP DNA-based PCR was previously characterized to be a rapid and objective molecular enumeration tool for detecting and quantifying bacterial numbers in an M. leprae preparation (29), it was chosen to serve as a normalizer for these assays.
Results from in vitro experiments demonstrated that both sodA mRNA/RLEP and 16S rRNA/RLEP RT-PCR assays were very good predictors of M. leprae viability in short-term experiments (up to 48 h) when bacteria were exposed to lethal concentrations of rifampin (the only bactericidal drug in the MDT regimen for leprosy) in axenic medium or within infected mouse MΦ cultures. Assuming that the biological tests like the RR and Live/Dead BacLight assays are “gold standards” for M. leprae viability, the 16S/RLEP assay appeared to be the most sensitive molecular assay for viability determination (r = 0.9817; P = 0.0005), even though the sodA/RLEP assay also presented a significant correlation with data obtained by gold-standard methods (r = 0.9463; P = 0.043).
The ability of these assays to detect the effects of rifampin treatment further demonstrates their ability to differentiate between live and drug-killed M. leprae cells in axenic culture and within their host cells (MΦ). Therefore, these assays may be useful as rapid screening tools to identify effective antileprosy drugs as well as for experiments to study host-parasite interactions. However, because of the inherent instability of sodA mRNA species, resulting in rapid degradation following the death of bacilli, the sodA/RLEP RT-PCR assay may be more useful for experiments that are designed to investigate early effects, <48 h, of drugs or immune factors on the viability of M. leprae. Preliminary results from our laboratory suggest that the sodA/RLEP assay can detect a loss in M. leprae viability as early as 12 h posttreatment (data not shown). These assays were found to be not only highly specific but also more sensitive than either RR (107 bacteria) or Live/Dead BacLight bacterial viability (106 bacteria) analysis for determining the presence and viability of M. leprae bacteria under short-term experimental conditions.
However, the sodA/RLEP assay was able to detect the presence of M. leprae in only 25% of the pretreatment biopsy specimens tested. In contrast, the 16S rRNA/RLEP assay was able to detect the presence of M. leprae in all pretreatment biopsy specimens analyzed. Several parameters may have contributed to these observed results. The first parameter is the inherent labile nature of the mycobacterial mRNA compared to that of rRNA (22), which may be further affected by the low degree of viability of M. leprae within the skin biopsy specimens of leprosy patients in general (10). The second parameter is the relatively lower sensitivity in detecting sodA than that in detecting the 16S rRNA gene due to the increased copy number of rRNA levels compared to that of mRNA.
In addition, results for the 16S rRNA/RLEP assay suggested a strong correlation between the length of therapy and decline of M. leprae viability. Even though the case numbers were low, these results confirm the potential utility of this assay for monitoring antileprosy MDT therapy and thereby potentially identifying leprosy cases that are not responding to MDT due to drug resistance, noncompliance, or potential bacterial growth during or after treatment (relapse). In contrast, the lack of detectable M. leprae sodA gene transcripts in the majority of human biopsy specimens demonstrated that this assay was not suitable for monitoring of M. leprae viability in crude biological specimens.
In the present study, M. leprae viability was determined using skin biopsy specimens based on 16S rRNA levels normalized by RLEP DNA levels. When RLEP data were used as an indicator of M. leprae numbers in an MB patient's biopsy specimen, there was a strong correlation between these results and BI results obtained by microscopic examination of skin slit smear samples from the same patients. Thus, the RLEP PCR assay alone may be useful for defining the clinical form of the disease and the potential up- or downgrading of disease status. However, no correlation between BI and 16S rRNA/RLEP levels was found, indicating that an assessment of bacterial load per se does not reflect viability in most instances. This was anticipated because DNA-based PCR-positive signals and the presence of acid-fast bacilli by microscopic examination persist in some cases for years after treatment but do not necessarily reflect the real impact of treatment on bacterial viability (25).
In conclusion, this study has identified additional tools for leprosy diagnosis and monitoring of antileprosy drug efficacy for clinical purposes and for M. leprae viability in short-term experimental studies that include the study of host cell-M. leprae interactions. This has been made possible by the development of a procedure for the simultaneous isolation of M. leprae RNA and DNA from the same sample and by the incorporation of the previously described real-time RLEP PCR as a good predictor of M. leprae numbers. Due to the stability and copy number of the 16S rRNA gene as well the degradation of this molecule over time, the 16S rRNA/RLEP assay should be useful for determinations of viable bacterial loads in skin biopsy specimens from MB leprosy patients and therefore may be important in determining MDT efficacy and the ability of patients to still be infectious posttherapy. Hence, because of its high sensitivity and specificity, this assay may also constitute a very sensitive and specific assay for the early detection of M. leprae in skin biopsy specimens and may therefore potentially be a predictor of the clinical form of leprosy.
Acknowledgments
We thank J. P. Pasqua for his excellent technical contributions to this work.
This research was partially funded by the HRSA, BPHC, Division of the National Hansen's Disease Programs, NIH/NIAID contract number Y1-AI-2646-01; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Adams, L. B., N. A. Soileau, J. R. Battista, and J. L. Krahenbuhl. 2000. Inhibition of metabolism and growth of Mycobacterium leprae by gamma irradiation. Int. J. Lepr. Other Mycobact. Dis. 681-10. [PubMed] [Google Scholar]
- 2.Britton, W. J., and D. N. Lockwood. 2004. Leprosy. Lancet 3631209-1219. [DOI] [PubMed] [Google Scholar]
- 3.Chae, G. T., M. J. Kim, T. J. Kang, S. E. Lee, Y. Shin, J. P. Kim, Y. H. Ko, S. H. Kim, and N. H. Kim. 2002. DNA-PCR and RT-PCR for the 18-kDa gene of Mycobacterium leprae to assess the efficacy of multi-drug therapy for leprosy. J. Med. Microbiol. 51417-422. [DOI] [PubMed] [Google Scholar]
- 4.Desjardin, L. E., M. D. Perkins, K. Wolski, S. Haun, L. Teixeira, Y. Chen, J. L. Johnson, J. J. Ellner, R. Dietze, J. Bates, M. D. Cave, and K. D. Eisenach. 1999. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am. J. Respir. Crit. Care Med. 160203-210. [DOI] [PubMed] [Google Scholar]
- 5.Franzblau, S. G. 1988. Oxidation of palmitic acid by Mycobacterium leprae in an axenic medium. J. Clin. Microbiol. 2618-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzblau, S. G., A. N. Biswas, P. Jenner, and M. J. Colston. 1992. Double-blind evaluation of BACTEC and Buddemeyer-type radiorespirometric assays for in vitro screening of antileprosy agents. Lepr. Rev. 63125-133. [DOI] [PubMed] [Google Scholar]
- 7.Hagge, D. A., S. Oby-Robinson, D. Scollard, G. McCormick, and D. L. Williams. 2002. A new model for studying the effects of Mycobacterium leprae on Schwann cell and neuron interactions. J. Infect. Dis. 1861283-1296. [DOI] [PubMed] [Google Scholar]
- 8.Hirawati, K. Katoch, D. S. Chauhan, H. B. Singh, V. D. Sharma, M. Singh, M. Kashyap, and V. M. Katoch. 2006. Detection of M. leprae by reverse transcription-PCR in biopsy specimens from leprosy cases: a preliminary study. J. Commun. Dis. 38280-287. [PubMed] [Google Scholar]
- 9.Jadhav, R. S., R. R. Kamble, V. S. Shinde, S. Edward, and V. K. Edward. 2005. Use of reverse transcription polymerase chain reaction for the detection of Mycobacterium leprae in the slit-skin smears of leprosy patients. Indian J. Lepr. 77116-127. [PubMed] [Google Scholar]
- 10.Job, C. K., J. Jayakumar, M. Aschhoff, and M. M. Mathan. 1996. Viability of Mycobacterium leprae in skin and peripheral nerves and persistence of nerve destruction in multibacillary patients after 2 years of multidrug therapy. Int. J. Lepr. Other Mycobact. Dis. 6444-50. [PubMed] [Google Scholar]
- 11.Kang, T. J., S. K. Kim, S. B. Lee, G. T. Chae, and J. P. Kim. 2003. Comparison of two different PCR amplification products (the 18-kDa protein gene vs. RLEP repetitive sequence) in the diagnosis of Mycobacterium leprae. Clin. Exp. Dermatol. 28420-424. [DOI] [PubMed] [Google Scholar]
- 12.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Böttger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 312882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurabachew, M., A. Wondimu, and J. J. Ryon. 1998. Reverse transcription-PCR detection of Mycobacterium leprae in clinical specimens. J. Clin. Microbiol. 361352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahiri, R., B. Randhawa, and J. L. Krahenbuhl. 2005. Application of a viability-staining method for Mycobacterium leprae derived from the athymic (nu/nu) mouse foot pad. J. Med. Microbiol. 54235-242. [DOI] [PubMed] [Google Scholar]
- 15.Martinez, A. N., C. F. Britto, J. A. Nery, E. P. Sampaio, M. R. Jardim, E. N. Sarno, and M. O. Moraes. 2006. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J. Clin. Microbiol. 443154-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phetsuksiri, B., J. Rudeeaneksin, P. Supapkul, S. Wachapong, K. Mahotarn, and P. J. Brennan. 2006. A simplified reverse transcriptase PCR for rapid detection of Mycobacterium leprae in skin specimens. FEMS Immunol. Med. Microbiol. 3319-328. [DOI] [PubMed] [Google Scholar]
- 17.Plikaytis, B. B., R. H. Gelber, and T. M. Shinnick. 1990. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J. Clin. Microbiol. 281913-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prabhakaran, K., E. B. Harris, B. Randhawa, L. B. Adams, D. L. Williams, and R. C. Hastings. 1993. Use of beta-lactam/beta-lactamase-inhibitor combinations as antimycobacterial agents. Microbios 76251-261. [PubMed] [Google Scholar]
- 19.Ramasesh, N., L. B. Adams, S. G. Franzblau, and J. L. Krahenbuhl. 1991. Effects of activated macrophages on Mycobacterium leprae. Infect. Immun. 592864-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rees, R. J. W. 1985. The microbiology of leprosy, p. 46. In R. C. Hastings (ed.), Leprosy, 2nd ed. Churchill Livingstone, New York, NY.
- 21.Ridley, D. S., and W. H. Jopling. 1966. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 34255-273. [PubMed] [Google Scholar]
- 22.Sela, S., J. E. Clark-Curtis, and H. Bercovier. 1989. Characterization and taxonomic implication of the rRNA genes of Mycobacterium leprae. J. Bacteriol. 17170-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shepard, C. C., and D. H. McRae. 1968. A method for counting acid-fast bacteria. Int. J. Lepr. Other Mycobact. Dis. 3678-82. [PubMed] [Google Scholar]
- 24.Srinivas, D., P. N. Rao, T. S. Lakshmi, and S. Suneetha. 2002. Bacterial index of granuloma and its relevance compared to BI of skin smears. Lepr. Rev. 7379-80. [PubMed] [Google Scholar]
- 25.Torres, P., J. J. Camarena, J. R. Gomez, J. M. Nogueira, V. Gimeno, J. C. Navarro, and A. Olmos. 2003. Comparison of PCR mediated amplification of DNA and the classical methods for detection of Mycobacterium leprae in different types of clinical samples in leprosy patients and contacts. Lepr. Rev. 7418-30. [PubMed] [Google Scholar]
- 26.Truman, R., A. B. Fontes, A. B. De Miranda, P. Suffys, and T. P. Gillis. 2004. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 422558-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truman, R. W., and T. P. Gillis. 2000. The effect of ultraviolet light radiation on Mycobacterium leprae. Int. J. Lepr. Other Mycobact. Dis. 6811-17. [PubMed] [Google Scholar]
- 28.Truman, R. W., and J. L. Krahenbuhl. 2001. Viable M. leprae as a research reagent. Int. J. Lepr. Other Mycobact. Dis. 691-12. [PubMed] [Google Scholar]
- 29.Truman, R. W., P. K. Andrews, N. Y. Robbins, L. B. Adams, J. L. Krahenbuhl, and T. P. Gillis. 2008. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl. Trop. Dis. 2e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch, T. M., R. H. Gelber, L. P. Murray, H. Ng, S. M. O'Neill, and L. Levy. 1980. Viability of Mycobacterium leprae after multiplication in mice. Infect. Immun. 30325-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, D. L., T. P. Gillis, R. J. Booth, D. Looker, and J. D. Watson. 1990. The use of a specific DNA probe and polymerase chain reaction for the detection of Mycobacterium leprae. J. Infect. Dis. 162193-200. [DOI] [PubMed] [Google Scholar]
- 32.Williams, D. L., S. Oby-Robinson, T. L. Pittman, and D. M. Scollard. 2003. Purification of Mycobacterium leprae RNA for gene expression analysis from leprosy biopsy specimens. BioTechniques 35534-536. [DOI] [PubMed] [Google Scholar]