Abstract
Cervical cancer is the second-most prevalent cancer in young women around the world. Infection with human papillomavirus (HPV), especially high-risk HPV types (HR-HPV), is necessary for the development of this cancer. HPV-DNA detection is increasingly being used in cervical cancer screening programs, together with the Papanicolau smear test. We evaluated the usefulness of introducing this new array-based HPV genotyping method (i.e., Clinical Arrays Papillomavirus Humano) in the cervical cancer screening algorithm in our center. The results obtained using this method were compared to those obtained by the hybrid capture II high-risk HPV DNA test (HC-II) and Papanicolau in a selected group of 408 women. The array-based assay was performed in women that were HC-II positive or presented cytological alterations. Among 246 array-positive patients, 123 (50%) presented infection with ≥2 types, and HR-HPV types were detected in 206 (83.7%), mainly HPV-16 (24.0%). Up to 132 (33.2%) specimens were classified as ASCUS (for atypical squamous cells of undetermined significance), and only 48 (36.4%) of them were HPV-DNA positive by either assay; however, 78.7% of these cases were caused by HR-HPV types. The agreement between both HPV-DNA detection techniques was fairly good (n = 367). Screening with Papanicolau smear and HC-II tests, followed by HPV detection and genotyping, provided an optimal identification of women at risk for the development of cervical cancer. Furthermore, with the identification of specific genotypes, either in single or multiple infections, a better prediction of disease progression was achieved. The array method also made allowed us to determine the possible contribution of the available vaccines in our setting.
Cervical cancer is the second most prevalent type of cancer in women worldwide. A total of 500,000 new cases are diagnosed each year and cause more than 270,000 deaths (15). Since the 1940s, screening programs for cervical cancer prevention, mainly based on the Papanicolau smear test, have been implemented in resource-rich countries, resulting in a remarkable decrease in its incidence and related mortality (16). However, this test has a limited sensitivity, especially for detecting precancerous lesions (1, 6).
Genital human papillomavirus (HPV) is a highly common sexually transmitted infection. Although most HPV infections are transient and asymptomatic, epidemiological studies worldwide have demonstrated that persistent infection with certain genotypes is the necessary cause for the development of cervical cancer and its precursor lesions (3, 19, 24). More than 100 HPV types have been described and classified into high-risk types (HR-HPV) and low-risk types (LR-HPV) according to the probability of developing cervical cancer (14). Therefore, in addition to the Papanicolau smear test, HPV detection assays have been implemented in many countries to improve cervical cancer screening. These assays have a higher sensitivity than the Papanicolau smear test for the detection of women at risk of developing precancerous lesions (12).
Since HPV cannot be grown in conventional cell cultures and serological assays are unreliable, molecular techniques constitute the best choice to diagnose HPV infection. Currently, the only assay that has been approved by the U.S. Food and Drug Administration for the detection of HPV-DNA is the Hybrid Capture II system (HC-II; Digene Corp., Gaithersburg, MD). This signal amplification assay was designed to detect LR-HPV and HR-HPV genotypes in two different kits but does not provide genotype information.
The interest of HPV genotyping has increased in light of the recently licensed HPV bivalent and tetravalent vaccines (9, 23). Genotyping also allows clinicians to monitor patients according to the oncogenic risk of the HPV types identified. Several genotyping assays have been developed over the last years with a variety of amplification and detection strategies (reviewed in references 4 and 13). Methods based on consensus PCR and reverse hybridization of PCR products provide high sensitivity and extensive typing information, including identification of multiple infections. Recently, an assay based on amplification and array hybridization has been commercialized for the detection and genotyping of HPV in routine clinical specimens (Clinical Arrays Papillomavirus Humano [CAPH]; Genomica S.A.U., Madrid, Spain). This assay provides the possibility to detect simple or mixed-type infections with 35 HPV types (20 HR-HPV and 15 LR-HPV).
The aim of the present study was to assess the usefulness of introducing this new array-based HPV detection and genotyping method in the cervical cancer screening algorithm in our center, a reference hospital with 600,000-habitant coverage. With this goal, we compared the results obtained using this method with those obtained by HC-II and the cytology findings.
MATERIALS AND METHODS
Study population.
As part of the cervical cancer screening program in our hospital, liquid cytology specimens were routinely collected in the following cases: (i) women with cytological alterations referred to hospital, (ii) women with a previously pathological Papanicolau smear, and (iii) women that had been treated or had undergone surgery after developing a cervical intraepithelial lesion. As shown in Fig. 1, in 2005 the screening algorithm was modified to include HPV genotyping with the CAPH, apart from the cytology and HC-II tests, to confirm the presence and to genotype HPV. According to the described algorithm, a total of 408 women were included in the present study.
FIG. 1.
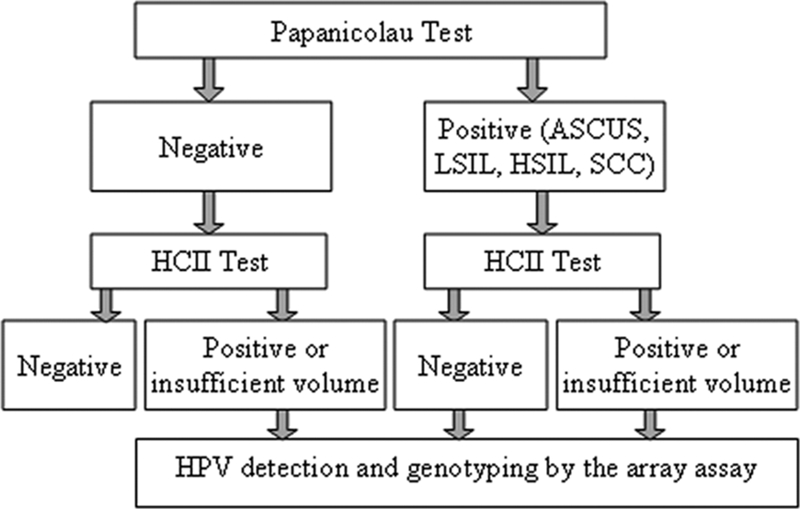
Schematic representation of the screening algorithm used in our center for cervical cancer prevention in patients undergoing liquid cytology.
Sample collection.
Cervical specimens were collected with an Ayre's spatula and an endocervical cytobrush, which was washed in a vial containing PreservCyt Solution (Cytyc Corp., Boxborough, MA) in order to obtain a liquid cytology for HPV-DNA testing.
Cytological study.
Slides were prepared from collected cervical specimens with the automated ThinPrep system (Cytyc Corp.). Slides were Papanicolau stained, observed under a microscope, and classified according to the Bethesda classification as ASCUS (for atypical squamous cells of undetermined significance), LSIL (for low-grade squamous intraepithelial lesion), HSIL (for high-grade squamous intraepithelial lesion), and SCC (for squamous cell carcinoma) (20).
HC-II HR assay.
The HC-II HR HPV DNA test is a sandwich capture molecular hybridization assay that uses a signal amplification detection method based on chemiluminescence. Thirteen HR-HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) can be detected with this assay. The test was performed according to manufacturer's instructions, starting from 4 ml of liquid cytology specimens. The results were considered threshold values when the value of the sample/cutoff ratio was between 1 and 2.5 relative light units.
CAPH genotyping test.
HPV detection and genotyping with the CAPH v.1 assay was performed in three steps according to the manufacturer's protocol from 1 ml of liquid cytology specimens. Briefly, the HPV L1 region was amplified by PCR together with an internal control used to exclude inhibition, and the human β-globin gene used to ensure correct specimen collection. Amplicons were detected by hybridization in a low-density microarray containing triplicate DNA probes specific to 35 genotypes: 20 HR-HPV (types 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, 82, and 85) and 15 LR-HPV (types 6, 11, 40, 42, 43, 44, 54, 61, 62, 71, 72, 81, 83, 84, and 89).
Statistical analysis.
Statistical analysis was performed using SPSS v14.0 software (SPSS, Inc., Chicago, IL), with a significance level of 0.05. Agreement between HPV-DNA detection techniques (HC-II and CAPH) was assessed through determination of Spearman's ρ and kappa coefficient (κ) values, assuming a good agreement for κ ≥ 0.6. Comparisons between groups were performed with the Student t test for quantitative variables and the chi-square or Fisher exact tests for categorical variables.
RESULTS
Study population.
The mean age of the women included in the study was 38.8 years (range, 15 to 80 years), and 155 (38%) of them were human immunodeficiency virus (HIV) positive. All women (n = 408) were tested by the CAPH test; 246 (60.3%) tested positive, and 206 (83.7%) showed infection with at least one HR-HPV. Among all HPV-positive women, 123 (50%) presented infection with ≥2 HPV types: 75 (61%) with 2, 31 (25%) with 3, 12 (10%) with 4, 3 (2.4%) with 5, and 2 (1.6%) with 6 types. Specimens collected from 41 women had insufficient volume to perform the HC-II assay; among the rest (n = 367), 199 (54.2%) were positive for HR-HPV types according to this test.
The percentage of HPV-DNA positivity was not significantly different in HIV-negative and HIV-positive women (63.2% versus 55.5% according to the CAPH assay, 55.9% versus 51.4% according to the HC-II assay, and 69.2% versus 58.1% according to the number of positives by either assay). Similarly, the percentage of multiple infections according to the CAPH assay was comparable in HIV-negative and HIV-positive women (51.3% versus 47.7%, respectively); the prevalences of a negative cytology result (15.0% versus 17.2%), ASCUS (30.5% versus 37.7%), LSIL (43.9 versus 35.8%), and HSIL (10.2 versus 9.3%) were also similar. The most frequent HR and LR types were type 16 (25.6% in HIV-negative and 20.1% in HIV-positive women) and type 6 (12.5% in HIV-negative and 16.3% in HIV-positive women), respectively (Fig. 2).
FIG. 2.
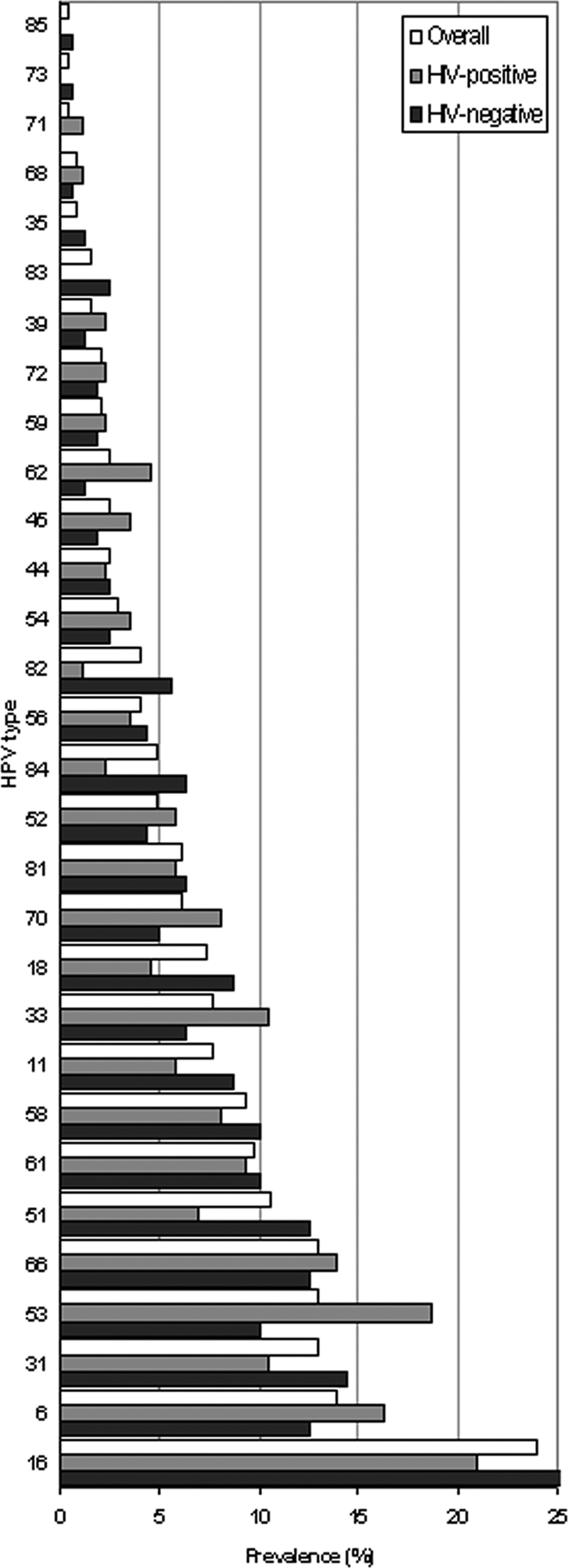
Prevalence of HPV types in the population studied.
Agreement between HPV-DNA detection techniques.
The CAPH assay is able to detect 20 HR and 15 LR-HPV types, while the HC-II can only detect 13 HR-HPV types. Therefore, in order to calculate agreement between both assays, CAPH results were recoded, and only HR-HPV types detected by the HC-II test were considered positive results. As shown in Table 1, agreement was fairly good (Spearman's ρ = 0.6321). Eleven threshold values were obtained with the HC-II test; seven of them were positive by the CAPH assay (three infections with HR types 16 or 18, which should have been detected as positive by the HC-II assay, and four infections with types 53, 58, 54, 68 and 71, which are not included in the HC-II test). Agreement was similar when data were reanalyzed, excluding specimens with threshold values for HC-II (rho = 0.650, κ = 0.625).
TABLE 1.
Agreement between the HC-II and CAPH assays for the detection of the HR-HPV types included in the HC-II test
| CAPH assay resulta | HC-II assay result
|
Total | ||
|---|---|---|---|---|
| No. positive | No. negative | No. at threshold value | ||
| Positive | 140 | 9 | 3 | 152 |
| Negative | 59 | 148 | 8 | 215 |
| Total | 199 | 157 | 11 | 367 |
Array results were recorded, and only HR-HPV types detected by the HC-II test were considered positive results, while infection with any other type was considered as a negative result, together with those where no HPV-DNA was detected.
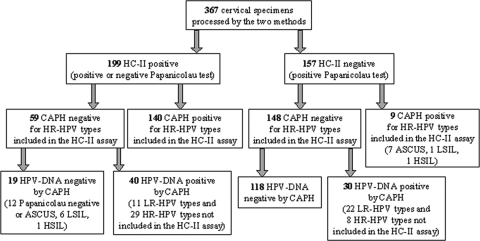
Detailed results are shown in Fig. 3. A positive HC-II result was obtained in 19 specimens that were CAPH negative (12 of them [63.2%] were cytology negative or ASCUS, 6 [31.6%] were LSIL, and 1 was HSIL), and in 40 specimens containing HPV types not included in the HC-II assay (LR-HPV types 6, 11, 61, 72, and 81 and HR-HPV types 53, 66, 70, and 85). Among the later 40 specimens, 11 (33.3%) were cytology negative or ASCUS, 20 (60.6%) were LSIL, and three (9.1%) were HSIL. A negative HC-II result was obtained in nine women with an infection with HR-HPV types included in the HC-II assay (16, 18, 31, 33, 58, and 68); seven of them were ASCUS, one was LSIL and another one was HSIL. The CAPH assay was thus able to detect 38 HPV infections (17 HR and 22 LR-HPV types) not detected by the HC-II assay.
FIG. 3.
Results obtained in the cervical specimens processed by both HC-II and CAPH assays.
Results according to cytology.
Table 2 shows the results of both HPV-DNA detection methods according to cytology. With regard to specimens with a negative Papanicolau test, following the algorithm shown in Fig. 1, only those with an HC-II positive (n = 36) or threshold (n = 5) result were tested by the CAPH assay. The presence of the HR-HPV types included in the HC-II was confirmed by the CAPH assay in 24 cases, while in 6 of them other types were detected, and 11 were negative.
TABLE 2.
Detection of HPV-DNA according to cytology results in specimens processed by both the HC-II and CAPH assays in women with a positive Papanicolau test
| HPV-DNA detection method | No. (%) of positive results obtained with HPV-DNA method for cytology
|
||
|---|---|---|---|
| ASCUS (n = 124) | LSIL (n = 150) | HSILb (n = 36) | |
| HC-II (13 HR-HPV types) | 29 (23.4) | 98 (65.3) | 29 (80.5) |
| CAPH (13 HR-HPV types)a | 26 (21.0) | 70 (46.6) | 26 (72.2) |
| CAPH (35 HPV types) | 45 (36.3) | 110 (73.3) | 32 (88.9) |
| Either method | 48 (38.7) | 116 (77.3) | 33 (91.7) |
Only types detected by the HC-II test were considered positive.
Includes one case of squamous cell carcinoma by HPV-16 positive by both methods.
Up to 132 (33.2%) specimens were classified as ASCUS, and only 48 (36.4%) of them were identified as HPV-DNA positive by either assay. A total of 37 (78.7%) of those that tested positive by the array method (n = 47) had one or more HR-HPV types. Among the patients with an ASCUS result, 22 came back for a second visit and only 4 of them (18.2%) turned negative. Among the rest, 8 (36.4%) remained ASCUS and 10 (45.5%) progressed to LSIL or HSIL.
Among women with HSIL, LR-HPV types were found in a greater proportion in HIV-positive women (3 of 13 [23.1%]) than in HIV-negative women (2 of 22 [9.1%]).
As shown in Table 3, according to the prevalence of the genotypes included in the recently approved quadrivalent HPV vaccine (6, 11, 16 and 18) administered in Spain, 19 of 35 (54.3%) women with severe dysplasia (HSIL) included in the present study would have been protected from infection with these types (95% confidence interval, 37.8 to 70.8%). However, only 11 (31.4%, confidence interval 16.1 to 46.8%) of all women with HSIL would have been totally protected from HPV infection, since 8 presented mixed-type infections with other LR or HR-HPV types not included in the vaccine.
TABLE 3.
Estimated vaccine coverage in HPV-DNA positive women with HSIL, according to the genotypes detected by the array-based method
| HPV typea | No. (%) of patients infected with HPV types included in the vaccine
|
||
|---|---|---|---|
| HIV negative (n = 22) | HIV positive (n = 13) | Overall (n = 35) | |
| 6/11 | 4 (18.2) | 2 (15.4) | 6 (17.1) |
| 16/18 | 12 (54.4) | 3 (23.1) | 15 (42.9) |
| 6/11/16/18 | 14 (63.6) | 5 (38.5) | 19 (54.3) |
| 6/11/16/18/31*/45*/52*/58* | 18 (81.8) | 6 (46.2) | 24 (68.6) |
*, HPV types potentially covered by the vaccine due to cross-reactivity.
DISCUSSION
In this study we evaluated the usefulness of introducing a new array-based HPV detection and genotyping method in the cervical cancer screening algorithm in our center. The results obtained using this method were compared to those obtained by the HC-II and the Papanicolau smear tests.
A fairly good agreement was found between the results obtained by the HC-II and the CAPH assays, in concordance with another study comparing the HC-II test with an hybridization-based commercialized assay (18). Since neither of the two methods used can be considered the gold standard, it is difficult to establish sensitivity and specificity. However, in 20.1% of the HC-II-positive specimens, the CAPH assay identified LR- and HR-HPV types that are not included in the first assay. These results could point to unspecific cross-reactivity between the probes used for the 13 HR-HPV types included in the HC-II kit and other types (i.e., types 6, 11, 53, 61, 66, 70, 72, 81, and 85). Similar results were obtained in studies that compared the HC-II assay to other methods based on restriction fragment length polymorphism and dot blot hybridization (5, 17). False-positive results are less likely in the CAPH assay, since it can detect the 35 most common HPV genotypes. Regarding sensitivity, 5.7% of the specimens with some degree of dysplasia in the Papanicolau smear test (ranging from ASCUS to HSIL) and an HC-II-negative result were CAPH positive for one or more of the genotypes included in the first assay, probably reflecting the higher analytical sensitivity of the CAPH assay, which is based on target amplification. On the other hand, 9.5% of HC-II-positive specimens were negative by the CAPH assay, and 44% of them had a negative Papanicolau smear test.
In our study, patients with ASCUS represented approximately one-third of the specimens submitted to the Microbiology Service for HPV genotyping, and 36.4% of them tested positive by either HPV-DNA detection method. The relevance of detecting HPV-DNA in ASCUS specimens is controversial, since most atypical cytology results in young women represent transient infections (21) and may represent public health costs and an unnecessary anxiety for patients. However, HR-HPV types were detected in 81.3% of HPV-positive ASCUS specimens in our study. Some of these patients came for a second visit, and about half of them progressed to LSIL or HSIL, while only 18% became cytology negative, although HPV-DNA was still detectable in all of them. Thus, we believe that testing ASCUS specimens is critical for the detection of women at early stages of infection with HR-HPV types, who are at risk for developing precancerous lesions and who should be closely monitored.
We observed that HPV-DNA might still be positive after the intervention of cervical lesions, even though the cytology becomes negative. In these cases, genotyping is also useful for patient follow-up, since it helps to differentiate persistent infections from newly acquired infections with other HPV types.
HPV-16 and HPV-18 confer a greater risk for developing high-grade cervical lesions than other types and are present in up to 70% of cervix carcinoma cases around the world (2, 10). Introducing the genotyping assay in our setting allowed us to identify patients infected by these types for a closer follow-up, as well as to establish that HPV-16 was the most prevalent type. Implementing HPV genotyping also enabled us to describe circulating genotypes, as well as to estimate the proportion of the population at risk theoretically covered by the quadrivalent vaccine administered in our country (54.3% in women with HSIL). Nevertheless, it should be taken into account that some of them had multiple infections with HR-HPV types other than those included in the vaccine and that these types may also cause cervical cancer. Cervical infection with multiple HPV types has also been observed as a frequent event in other studies (8, 11) and has an impact on the efficacy of current vaccines.
It has been observed that in cases of deficient cellular immunity HPV types other than types 16 and 18 have an increased probability of causing dysplasia in comparison with immunocompetent patients (22). In our study, only LR-HPV types were detected by the CAPH assay in 23.1% of the HIV-positive women with HSIL. In HSIL lesions, HR-HPV types are predominantly in latent infection with low copy numbers per cell (7), which could have caused a negative result for these types, even though they could have been present. However, this result was observed in a higher proportion in HIV-positive than in HIV-negative women and could reflect a lower vaccine coverage in the first population.
In conclusion, screening with Papanicolau smear and HC-II tests, followed by HPV detection and genotyping with the CAPH assay, allowed us to optimally identify women at risk for the development of cervical cancer. Testing HC-II-negative and Papanicolau smear-positive specimens by the CAPH assay led to the detection of an additional 10% of HPV infections in the studied women, who were infected by HR- and LR-HPV types. Furthermore, with the identification of specific genotypes, either in single or multiple infections, a better prediction of disease progression was achieved. Finally, the CAP assay also made possible to determine the possible contribution of the available vaccines in our setting.
Acknowledgments
This study was partially supported by grant 2008FI_B01050 (N.G.-S.) from the Comissionat per a Universitats i Recerca del Departament d'Innovació, Universitats i Empresa de la Generalitat de Catalunya i del Fons Social Europeu (Spain), and by grants CD05/00258 (E.M.) (Contratos Postdoctorales de Perfeccionamiento) and CA060193 (M.C.) (Contratos de Apoyo a la Investigación) from the Ministerio de Sanidad y Consumo, within the Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I+D+I).
We thank Marisol Benito for her help with specimen processing.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Belinson, J., Y. L. Qiao, R. Pretorius, W. H. Zhang, P. Elson, L. Li, Q. J. Pan, C. Fischer, A. Lorincz, and D. Zahniser. 2001. Shanxi Province Cervical Cancer Screening Study: a cross-sectional comparative trial of multiple techniques to detect cervical neoplasia. Gynecol. Oncol. 83439-444. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, F. X., X. Castellsague, and S. de Sanjosé. 2008. HPV and cervical cancer: screening or vaccination? Br. J. Cancer 9815-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brink, A. A., P. J. Snijders, and C. J. Meijer. 2007. HPV detection methods. Dis. Markers 23273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castle, P. E., M. Schiffman, R. D. Burk, S. Wacholder, A. Hildesheim, R. Herrero, M. C. Bratti, M. E. Sherman, and A. Lorincz. 2002. Restricted cross-reactivity of hybrid capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol. Biomarkers Prev. 111394-1399. [PubMed] [Google Scholar]
- 6.Chacon, J., I. Sanz, M. D. Rubio, M. L. de la Morena, E. Diaz, M. L. Mateos, and F. Baquero. 2007. Detection and genotyping of high-risk human papillomavirus in cervical specimens. Enferm. Infecc. Microbiol. Clin. 25311-316. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 7.Dehn, D., K. C. Torkko, and K. R. Shroyer. 2007. Human papillomavirus testing and molecular markers of cervical dysplasia and carcinoma. Cancer 1111-14. [DOI] [PubMed] [Google Scholar]
- 8.Gravitt, P. E., L. J. van Doorn, W. Quint, M. Schiffman, A. Hildesheim, A. G. Glass, B. B. Rush, J. Hellman, M. E. Sherman, R. D. Burk, and S. S. Wang. 2007. Human papillomavirus (HPV) genotyping using paired exfoliated cervicovaginal cells and paraffin-embedded tissues to highlight difficulties in attributing HPV types to specific lesions. J. Clin. Microbiol. 453245-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper, D. M., E. L. Franco, C. Wheeler, D. G. Ferris, D. Jenkins, A. Schuind, T. Zahaf, B. Innis, P. Naud, N. S. De Carvalho, C. M. Roteli-Martins, J. Teixeira, M. M. Blatter, A. P. Korn, W. Quint, and G. Dubin. 2004. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 3641757-1765. [DOI] [PubMed] [Google Scholar]
- 10.Khan, M. J., P. E. Castle, A. T. Lorincz, S. Wacholder, M. Sherman, D. R. Scott, B. B. Rush, A. G. Glass, and M. Schiffman. 2005. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 971072-1079. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs, K., A. D. Varnai, M. Bollmann, A. Bankfalvi, M. Szendy, N. Speich, C. Schmitt, L. Pajor, and R. Bollmann. 2008. Prevalence and genotype distribution of multiple human papillomavirus infection in the uterine cervix: a 7.5-year longitudinal study in a routine cytology-based screening population in West Germany. J. Med. Virol. 801814-1823. [DOI] [PubMed] [Google Scholar]
- 12.Lorincz, A. T., and R. M. Richart. 2003. Human papillomavirus DNA testing as an adjunct to cytology in cervical screening programs. Arch. Pathol. Lab. Med. 127959-968. [DOI] [PubMed] [Google Scholar]
- 13.Molijn, A., B. Kleter, W. Quint, and L. J. van Doorn. 2005. Molecular diagnosis of human papillomavirus (HPV) infections. J. Clin. Virol. 32(Suppl. 1)S43-S51. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz, N., F. X. Bosch, S. S. de, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 15.Parkin, D. M., F. Bray, J. Ferlay, and P. Pisani. 2005. Global cancer statistics, 2002. CA Cancer J. Clin. 5574-108. [DOI] [PubMed] [Google Scholar]
- 16.Parkin, D. M., F. I. Bray, and S. S. Devesa. 2001. Cancer burden in the year 2000: the global picture. Eur. J. Cancer 37(Suppl. 8)S4-S66. [DOI] [PubMed] [Google Scholar]
- 17.Poljak, M., I. J. Marin, K. Seme, and A. Vince. 2002. Hybrid Capture II HPV Test detects at least 15 human papillomavirus genotypes not included in its current high-risk probe cocktail. J. Clin. Virol. 25(Suppl. 3)S89-S97. [DOI] [PubMed] [Google Scholar]
- 18.Sandri, M. T., P. Lentati, E. Benini, P. Dell'Orto, L. Zorzino, F. M. Carozzi, P. Maisonneuve, R. Passerini, M. Salvatici, C. Casadio, S. Boveri, and M. Sideri. 2006. Comparison of the Digene HC2 assay and the Roche AMPLICOR human papillomavirus (HPV) test for detection of high-risk HPV genotypes in cervical samples. J. Clin. Microbiol. 442141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiffman, M. H., H. M. Bauer, R. N. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. E. Sherman, R. J. Kurman, S. Wacholder, C. K. Stanton, and M. M. Manos. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85958-964. [DOI] [PubMed] [Google Scholar]
- 20.Solomon, D., D. Davey, R. Kurman, A. Moriarty, D. O'Connor, M. Prey, S. Raab, M. Sherman, D. Wilbur, T. Wright, Jr., and N. Young. 2002. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA 2872114-2119. [DOI] [PubMed] [Google Scholar]
- 21.Stanley, M. 2006. Immune responses to human papillomavirus. Vaccine 24(Suppl. 1)S16-S22. [DOI] [PubMed] [Google Scholar]
- 22.Strickler, H. D., J. M. Palefsky, K. V. Shah, K. Anastos, R. S. Klein, H. Minkoff, A. Duerr, L. S. Massad, D. D. Celentano, C. Hall, M. Fazzari, S. Cu-Uvin, M. Bacon, P. Schuman, A. M. Levine, A. J. Durante, S. Gange, S. Melnick, and R. D. Burk. 2003. Human papillomavirus type 16 and immune status in human immunodeficiency virus-seropositive women. J. Natl. Cancer Inst. 951062-1071. [DOI] [PubMed] [Google Scholar]
- 23.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6271-278. [DOI] [PubMed] [Google Scholar]
- 24.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 18912-19. [DOI] [PubMed] [Google Scholar]