Abstract
HgbA is the sole TonB-dependent receptor for hemoglobin (Hb) acquisition of Haemophilus ducreyi. Binding of Hb to HgbA is the initial step in heme acquisition from Hb. To better understand this step, we mutagenized hgbA by deletion of each of the 11 putative surface-exposed loops and expressed each of the mutant proteins in trans in host strain H. ducreyi FX547 hgbA. All mutant proteins were expressed, exported, and detected on the surface by anti-HgbA immunoglobulin G (IgG). Deletion of sequences in loops 5 and 7 of HgbA abolished Hb binding in two different formats. In contrast, HgbA proteins containing deletions in the other nine loops retained the ability to bind Hb. None of the clones expressing mutant proteins were able to grow on plates containing low concentrations of Hb. Previously we demonstrated in a swine model of chancroid infection that an HgbA vaccine conferred complete protection from a challenge infection. Using anti-HgbA IgG from this study and the above deletion mutants, we show that loops 4, 5, and 7 of HgbA were immunogenic and surface exposed and that IgG directed against loops 4 and 5 blocked Hb binding. Furthermore, loop 6 was cleaved by protease on intact H. ducreyi, suggesting surface exposure. These data implicate a central domain of HgbA (in respect to the primary amino acid sequence) as important in Hb binding and suggest that this region of the molecule might have potential as a subunit vaccine.
Haemophilus ducreyi is the etiologic agent of chancroid, one of the genital ulcer diseases. Genital ulcer diseases are important risk factors for the heterosexual transmission of human immunodeficiency virus (22). Elimination of chancroid, through either vaccination or diagnosis and antibiotic treatment of core groups, could potentially slow human immunodeficiency virus transmission where both diseases are endemic (26, 27). HgbA is a highly effective vaccine in the swine model of chancroid infection (1), and it is conceivable that an HgbA vaccine might be used for the control of chancroid in humans.
H. ducreyi infects only humans under natural conditions and has a small genome relative to those of other organisms that are environmentally diverse (2; http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=nthd01). H. ducreyi lacks the genetic machinery to synthesize heme, and it must obtain heme from its host (3). Heme/iron compounds are transported across the outer membrane in gram-negative bacteria by a class of outer membrane proteins (OMPs) termed TonB-dependent receptors (TB-DRs) (19). During our studies of heme acquisition in H. ducreyi, we identified HgbA (10, 12), one of three TB-DRs in H. ducreyi in type strain 35000HP (17, 29; http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=nthd01). Whereas HgbA expression is required for the establishment of a human experimental infection (4), neither of the other TB-DRs is required for infection (17). Thus, in the human model of experimental chancroid infection, HgbA is the most important TB-DR of H. ducreyi and hemoglobin (Hb) is the most important source of heme/iron.
Since HgbA is an important vaccine candidate for chancroid, understanding its structure and functional domains may greatly impact its effectiveness in vaccine applications. A better understanding of the involvement of surface-exposed loops of HgbA in the initial Hb-binding step might simplify the development of an HgbA vaccine. One objective of this study was to identify surface-exposed loops required for Hb binding. The second objective of this study was to identify which loops of the protective HgbA immunogen, used in the previous vaccine trial, were immunogenic. Finally we sought to determine which loops of HgbA are required for the utilization of heme from Hb.
MATERIALS AND METHODS
Strains and media.
The bacterial strains and plasmids used in this study are shown in Table 1. For routine growth, H. ducreyi was maintained on chocolate agar prepared by following Gonococcal medium base (GCB) instructions (Difco, Detroit, MI) and by incorporation of GGC (0.1% glucose, 0.01% glutamine, 0.026% cysteine) (30) and fetal bovine serum. For maintenance of pLSKS-derivative clones containing hgbA constructs, streptomycin (50 μg/ml) was added to chocolate agar. Agar media containing various heme sources were prepared with GCB and GGC without fetal bovine serum. Human Hb (H7379; Sigma, St. Louis, MO) was dissolved in phosphate-buffered saline (PBS) at a concentration of 10 mg/ml and rocked at room temperature for 2 h or overnight at 4°C prior to filter sterilization. Hb was added to GCB medium to a final nominal concentration of 100 μg/ml, although some loss of Hb occurred during filtration. Heme stock solutions (10 mg/ml) were made by dissolving bovine hemin chloride (Sigma) in 0.1 N NaOH and used without further sterilization. GCB heme plates contained a final concentration of 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| H. ducreyi | ||
| 35000HP | Human-passaged variant of strain 35000 | 3a, 15 |
| FX547 | 35000HP ΔhgbA::CAT; complete deletion of hgbA sequence; unable to grow on Hb | This study |
| E. coli DH5αMCR | recA gyrB; host for cloning | Bethesda Research Laboratories |
| Plasmids | ||
| pBluescript | ColE1 replicon, Ampr | Stratagene |
| pCRII | TA vector | Invitrogen |
| pLSKS | H. ducreyi shuttle vector, Strr | 9 |
| pNC40 | Source of CAT cassette; Ampr Cmr | 12 |
| pUNCH555 | EagI/SpeI fragment of pUNCH579 containing hgbA in pBluescript | 12 |
| pUNCH555loop4 | pUNCH555 lacking 124 aa (A331-K454) in loop 4 | This study |
| pUNCH579 | 8-kb EcoRI fragment containing entire hgbA (strain 35000) in pBluescript; Ampr | 12 |
| pUNCH1261 | EagI/SpeI fragment of pUNCH579 containing hgbA in pLSKS | This study |
| pUNCH1261sx | pUNCH1261 with XhoI, SmaI, HindIII, BamHI, ClaI, and SpeI sites removed | This study |
| pUNCH1261sxplug | pUNCH1261sx lacking 141 aa (V13-I152) in plug region | This study |
| pUNCH1261sxloop1 | pUNCH1261sx lacking 10 aa (Y164-H173) in loop 1 | This study |
| pUNCH1261sxloop2 | pUNCH1261sx lacking 32 aa (N196-S227) in loop 2 | This study |
| pUNCH1261sxloop3 | pUNCH1261sx lacking 31 aa (T256-I286) in loop 3 | This study |
| pUNCH1261loop4 | pUNCH1261 lacking 124 aa (A331-K454) in loop 4 | This study |
| pUNCH1261loop5a | pUNCH1261 lacking 68 aa (G494-G562) in loop 5 | This study |
| pUNCH1261sxloop5n | pUNCH1261 lacking 43 aa (Y509-F551) in loop 5 | This study |
| pUNCH1261sxloop6 | pUNCH1261 lacking 35 aa (R609-F643) in loop 6 | This study |
| pUNCH1261sxloop7a | pUNCH1261 lacking 30 aa (F670-R699) in loop 7 | This study |
| pUNCH1261sxloop7n | pUNCH1261 lacking 16 aa (F670-P685) in loop 7 | This study |
| pUNCH1261sxloop8 | pUNCH1261sx lacking 8 aa (S738-G745) in loop 8 | This study |
| pUNCH1261sxloop9 | pUNCH1261sx lacking 23 aa (R796-N818) in loop 9 | This study |
| pUNCH1261sxloop10 | pUNCH1261sx lacking 11 aa (K863-R873) in loop 10 | This study |
| pUNCH1261sxloop11 | pUNCH1261sx lacking 21 aa (F918-P938) in loop 11 | This study |
| pUNCH1409 | AatII/ClaI fragment deleted from genomic clone pUNCH579 | This study |
| pUNCH1410 | CAT cassette amplified from pNC40 cloned into pCRII | This study |
| pUNCH1411 | CAT cassette from pUNCH1410 replacing deleted hgbA in pUNCH1409 | This study |
Amp, ampicillin; Cm, chloramphenicol; Str, streptomycin; aa, amino acids. The number of deleted amino acids and position coordinates refer to the mature (without a signal peptide) HgbA sequence of H. ducreyi strain 35000HP (GenBank accession number U17281).
Escherichia coli was maintained on Luria-Bertani agar with ampicillin at 100 μg/ml or streptomycin at 30 μg/ml, as appropriate.
Construction of a two-dimensional model of the HgbA receptor.
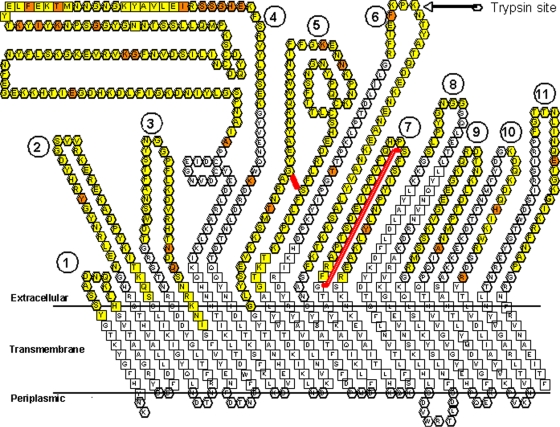
A variety of programs and servers and visual inspection were used to construct Fig. 1. Analyses of the primary predicted amino acid sequence by homology to membrane-spanning regions and beta sheets (strands) of FepA were used in the construction of a two-dimensional barrel model of HgbA. Visual comparison of transmembrane FepA residues with HgbA revealed obvious homologies in the N-terminal and C-terminal barrel domains. Prediction of surface-exposed loop regions was necessary to guide deletion mutagenesis, and we used several methods for this. We used the general principle that surface-exposed loops of OMPs are subjected to immunologic pressure and often vary between strains but functional residues are maintained. We performed amino acid sequence alignments of HgbA homologs within the family Pasteurellaceae (H. ducreyi HgbA; Haemophilus influenzae HgbA, HgbB, and HgbC [8]; and Pasteurella multocida HgbA [5]) to identify variable, potentially surface-exposed, loops (data not shown). Some of these Hb receptors contained deletions of loops present in H. ducreyi HgbA; thus, we hypothesized that they were not necessary for function. Subsequently, we built a three-dimensional atomic model based on the crystallographic structure of FepA with InsightII (www.accelrys.com). We took care to conserve important structural features of the plug region. Using this three-dimensional model, we then generated a two-dimensional model of transmembrane beta sheets. We confirmed our two-dimensional model by using two prediction servers, Proteus and PRED-TMBB. Proteus predicted all 22 of our putative transmembrane strands, with only slight variations in beta-strand endpoints. PRED-TMBB predicted 20 of our 22 strands.
FIG. 1.
Two-dimensional model of the Hb receptor of H. ducreyi HgbA. This model was derived by in silico comparison of the primary sequence of HgbA with that of FepA, whose crystal structure has been solved. The letters represent the amino acid sequence in one-letter code. The 11 putative surface-exposed loops of HgbA are numbered 1 through 11. Amino acids predicted to be outside the membrane shown to vary between H. ducreyi strains are orange. Yellow residues were deleted in first-generation loop mutants, with the exception that the first residue missing in the loop 4 deletion, an A, is orange, since it was variable between strains. For the second-generation loop 5 and 7 deletions, a red line shows the boundaries of the deleted amino acids in loops. The amino acid residues predicted to form β strands are boxed. Loops were formed to fit the page, and no loop structure is proposed or implied. The N-terminal plug (155 amino acids missing), believed to be located in the periplasm and within the pore, is not shown. Bold residues at the bottom of the figure are proposed to be located in the periplasmic space.
Construction of an hgbA plasmid expression system in H. ducreyi. (i) Construction of host strain FX547.
To stabilize complementing plasmids containing hgbA and to lessen the possibility of recombination with the chromosome, we constructed an isogenic mutant lacking the hgbA sequence. Plasmid pUNCH1409 was constructed by deletion of the AatII/ClaI fragment from genomic clone pUNCH579 (12). This first step left 1.3 kb of flanking DNA upstream of hgbA. Plasmid pUNCH1409 was restricted with AflII and HpaI to remove the entire hgbA sequence, and the larger fragment was isolated by agarose gel electrophoresis. This larger AflII-and-HpaI fragment lacked all hgbA sequence but contained sequences flanking hgbA, as well as plasmid vector sequences. HpaI digestion of pUNCH1409 did not disrupt either the open reading frame or the putative promoter region of the upstream gene mutS. A chloramphenicol resistance cassette was amplified from a pNC40 template and cloned into the TA vector pCRII (Invitrogen, Carlsbad, CA) to form pUNCH1410. After digestion of pUNCH1410 with AflII and HpaI, the chloramphenicol acetyltransferase (CAT) cassette (these sites were engineered into DNA by use of PCR primers; see Table S1 in the supplemental material) was isolated and ligated with the larger AflII-to-HpaI fragment of pUNCH1409 to form pUNCH1411. Thus, pUNCH1411 has a CAT cassette replacing the deleted hgbA allele with sufficient flanking DNA to mediate a double-crossover allelic replacement event. Plasmid pUNCH1411 DNA was linearized by restriction with a unique polylinker enzyme and electroporated into strain 35000HP with selection for chloramphenicol resistance as previously described (12) to form H. ducreyi hgbA deletion mutant FX547. FX547 was confirmed by PCR and in Western blot assays with anti-recombinant HgbA (rHgbA) antibodies (data not shown; see Fig. 3). This strain served as the host for hgbA plasmids.
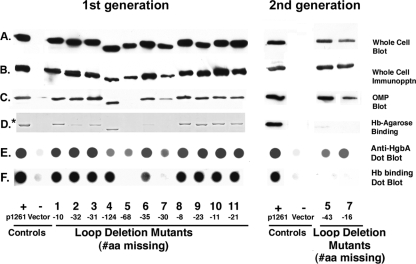
FIG. 3.
Analysis of first-generation (left panel) and second-generation (right panel) HgbA Δ loop mutant proteins and strains. FX547 hgbA containing pUNCH1261 (expressing full-length hgbA), empty vector pLSKS, or each of the indicated Δ loop deletion plasmids was analyzed for expression and cellular localization of HgbA and the ability of each HgbA protein to bind Hb. In panels A to C, Western blot assays with polyclonal rabbit anti-rHgbA antibodies were used to visualize the presence of HgbA from total cellular proteins, whole-cell immunoprecipitation, and Sarkosyl-insoluble OMP, respectively. In panel D, the ability of HgbA mutant proteins to bind Hb was examined. OMPs were solubilized in Zw and bound to Hb-agarose. After washing and elution, OMPs bound to Hb-agarose from first-generation mutants were analyzed by SDS-PAGE and Coomassie staining (*, left panel). Alternatively, proteins bound to Hb from second-generation mutants were analyzed by Western blotting with rabbit anti-rHgbA antibodies. In panels E and F, dot blots were blocked and probed with iodinated pig anti-nHgbA IgG (E) or iodinated human Hb (F) and subjected to autoradiography. Whole cells of loop mutant strains were immobilized and analyzed by using a dot blot assay. #aa, number of amino acids.
(ii) Construction of hgbA plasmids.
The EagI-to-SpeI fragment (3.4 kb) of genomic clone pUNCH579 (12) was used in the construction of plasmid pUNCH1261 (this study, Table 1) or pUNCH555 (12) by excision and ligation into the same sites of pLSKS or pBluescript, respectively. The EagI-to-SpeI fragment contains the entire hgbA structural gene with its putative promoter. Since pLSKS constructs replicate in H. ducreyi, mutant HgbA protein function can be examined in H. ducreyi host strain FX547 hgbA. To eliminate redundant restriction sites and facilitate cloning, plasmid pUNCH1261sx was constructed by excision of the small SpeI-to-XhoI fragment from the polylinker region of clone pUNCH1261. DNA from pUNCH1261 was digested with XhoI and SpeI and religated after filling in with the Klenow fragment of DNA polymerase I and deoxynucleoside triphosphates.
Generation of Δ loop hgbA plasmids.
Thirteen deletions in HgbA were created to test the predictions of our topological model (Fig. 1). The strategy was to delete or partially delete putative surface loops; all 11 loops and the plug region were targeted in this analysis. To introduce a deletion into hgbA, a double-step cloning protocol was used only for loops 4 and 6, first in pUNCH555 and after selection in E. coli, subcloning inserts into pUNCH1261 or pUNCH1261sx. In all subsequent constructs, we cloned PCR products directly into suitably restricted vector pUNCH1261 or pUNCH1261sx. An example of one of the deletion plasmids (loop 4 deletion mutant) that were generated is described in detail in Fig. 2.
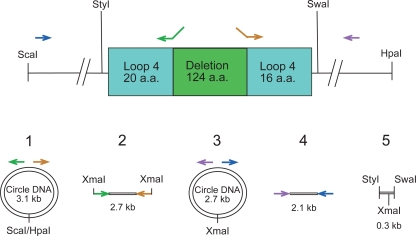
FIG. 2.
Strategy used to construct loop deletions by PCR. Steps are numbered 1 through 5 in this example of a loop 4 deletion mutant. In step 1, pUNCH555 was restricted with ScaI and HpaI to generate a 3.1-kb hgbA fragment with blunt ends as shown at the top. This product was self-ligated under dilute conditions to form the circle shown in step 1. This circle contains the entire hgbA open reading frame and is the template for the generation of all 11 loop mutants. In step 2, inverse PCR was done with primers containing XmaI sites at their ends (Loop4F and Loop4R) to generate a 2.7-kb fragment in the example for the loop 4 mutant construction shown here. These XmaI sites are at the desired point of deletion in loop 4 (Pro330 to Gly455 of the parent protein). In step 3, the 2.7-kb PCR product was restricted with XmaI, self-ligated under dilute conditions, and then PCR amplified with hgbA primers flanking the deletion (Hgb1.05 and Hgb4) to form the product shown in step 4. In step 5, the PCR product was digested with StyI and SwaI, which cut at sites that flank the deletion, generating a 293-bp product that was cloned into the same sites in pUNCH555 to create pUNCH555 Δloop4. The arrows indicate the primers used for PCR. The SwaI-to-StyI insert of pUNCH555 Δloop4 was moved into the same sites of pUNCH1261 to create pUNCH1261Δloop4. a.a., amino acids.
We introduced translationally silent restriction sites into pUNCH1261 by PCR. The sites introduced were ClaI in pUNCH1261sxΔplug; BamHI in pUNCH1261sxΔloop1, pUNCH1261sxΔloop2, pUNCH1261sxΔloop3, pUNCH1261sxΔloop5n, pUNCH1261sxΔloop6, pUNCH1261sxΔloop7a, pUNCH1261sxΔloop7n, pUNCH1261sxΔloop8, and pUNCH1261sxΔloop10; XmaI in pUNCH1261Δloop4 and pUNCH1261sxΔloop11; and KasI in pUNCH1261Δloop5a and pUNCH1261sxΔloop9. By using convenient hgbA restriction sites (HindIII for Δloop1, Δloop2, and Δloop3; StyI and SwaI for Δloop4; SwaI and BsiWI for Δloop5a, Δloop5n, Δloop6, Δloop7a, Δloop7n, and Δloop8; BsiWI and HpaI for Δloop9, Δloop10, and Δloop11; and AflII and SwaI for Δplug), receptor genes were constructed with the smallest possible PCR-generated fragment containing the desired deletion in the pUNCH1261 or pUNCH1261sx vector. These incorporated silent restriction sites are unique. In order to confirm that each plasmid expressed the appropriate-size protein, mutants were examined in Western blot assays of total cellular proteins. Plasmid DNA was confirmed by sequencing of the nongenomic, PCR-generated regions. In some cases, a single-base PCR error was inadvertently introduced, resulting in an in-frame point mutation. If that mutant protein bound Hb-agarose, it was concluded that neither the intended deletion nor the unintended mutation affected Hb binding. If a mutant protein did not bind Hb-agarose and contained inadvertent PCR artifacts, a new mutant clone was selected and verified. In the cases of loop mutants 5 and 7, which did not bind Hb-agarose, additional “second-generation” mutants (designated by the letter n for new) were made with smaller deletions to confirm the original phenotype and to narrow down residues required for Hb binding.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and dot blotting.
Western blots were probed with rabbit anti-rHgbA antibodies. Alkaline phosphatase-conjugated anti-rabbit goat immunoglobulin G (IgG) was used as a secondary antibody (Sigma) and visualized with Lumi-Phos (Pierce, Rockford, IL) as previously described (31). To examine the ability of whole cells of H. ducreyi to bind Hb or anti-native HgbA (nHgbA) IgG, dot blot assays were performed as previously described (10). Briefly, H. ducreyi (2 × 107 CFU/well) was immobilized on nitrocellulose and the filter was blocked and probed with either iodinated Hb or iodinated anti-nHgbA IgG. After washing unbound probe, the filters were subjected to autoradiography at −80°C with an intensifying screen.
Whole-cell immunoprecipitation.
Whole-cell immunoprecipitation was performed as previously described (11), with pig anti-nHgbA antibodies that recognize surface-exposed epitopes of nHgbA (1). H. ducreyi strains were grown on chocolate agar plates with streptomycin (50 μg/ml). A 1-ml suspension of H. ducreyi (optical density at 600 nm [OD600] = 1.0) was mixed with antisera (25 μl) for 30 min to allow antibodies to bind to surface-exposed epitopes. After washing to remove unbound antibody, H. ducreyi was solubilized with 2% Zwittergent 3-14 (Zw) in TEN buffer (5 mM Tris/HCl, 0.1 M NaCl, 1 mM EDTA, pH 8.0) at 37°C for 1 h with agitation. Zw-soluble proteins were added to 30 μl of protein G-agarose (50% slurry) to bind antigen-antibody complexes and incubated for 2 h to overnight. Protein G-agarose was pelleted, washed extensively with 0.2% Zw/TEN buffer, and subjected to Western blotting as described above, with anti-rHgbA primary antibody (31).
Trypsin digestion of HgbA.
H. ducreyi strain 35000HP was grown under heme-depleted conditions (1% GGC and 2.5% fetal bovine serum in gonococcal broth) for 14 to 16 h at 34.5°C. The bacterial cells were harvested and washed twice in PBS to remove proteins from the growth medium. The final suspension was adjusted to an OD600 of 0.4 (approximately 2 × 108 CFU/ml). Trypsin (20 μg/ml; Sigma) was added to this bacterial suspension, it was incubated for 10 min at 37°C, and protease inhibitor cocktail (P8465; Sigma) was added to stop the reaction. The trypsin-treated sample was centrifuged at 14,000 rpm for 3 min, the supernatant was removed, the cells were resuspended in 10 mM HEPES buffer with lysozyme (10 mg/ml; Sigma), and the cells were frozen overnight at −20°C. OMPs prepared from this sample (12) were subjected to SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, and HgbA polypeptides were detected with anti-rHgbA antibodies (31). Bands of interest were cut out of the membrane for N-terminal sequencing at the University of North Carolina Microsequencing facility.
Affinity purification of Hb-binding proteins.
The ability of HgbA proteins with deletions in individual loops to qualitatively bind Hb-agarose was assessed as previously described, by an analytical-scale method (1, 10), which is the most reliable method for determination of Hb binding in H. ducreyi without a high background level (10, 21, 28). Hb-agarose was prepared by covalently attaching bovine Hb to Affigel 15 (Bio-Rad Laboratories, Hercules, CA) by following the manufacturer's directions. Sarkosyl-insoluble OMPs were solubilized in 1% Zw/TEN buffer for 2 h with end-over-end tumbling at 37°C. After centrifugation at 14,000 × g for 10 min, each mutant HgbA protein present in the original OMP preparation was found in the Zw-soluble fraction. Hb-agarose beads (40 μl of a 50% slurry) were prewashed with TEN buffer containing 0.5% Zw and incubated with Zw-soluble OMP (diluted to 0.5% Zw) for 2 h. To remove nonspecifically bound proteins, the protein-loaded agarose beads were repeatedly washed with 0.2% Zw/TEN buffer; this was followed by boiling of the Hb-agarose in Laemmli sample buffer to release bound proteins. Samples were analyzed by SDS-PAGE, and the proteins were either stained with Coomassie brilliant blue (Fig. 3, first-generation mutants) or transferred to nitrocellulose and visualized in Western blot assays with anti-rHgbA antibodies (Fig. 3, second-generation mutants).
For Hb affinity chromatography with total cellular proteins, H. ducreyi cultures grown on chocolate agar plates were harvested, washed in gonococcal broth, and stored at −80°C. After thawing on ice, cells were suspended in PBS with 4% Zw and disrupted by passage through an 18-gauge needle 5 to 10 times, until the cell suspension cleared. Following centrifugation (8,000 × g, 10 min, room temperature [RT]) to pellet unlysed cells and debris, the supernatant was diluted in PBS to reduce the concentration of Zw to 0.5% and affinity purification was performed as described above.
For detection of mobility and expression differences in HgbA, we used OMP fractions or total cellular protein fractions of FX547 expressing hgbA from Δ loop plasmids.
Absorption of polyclonal pig anti-HgbA to produce loop-specific antiserum.
Each loop mutant shown in Fig. 3, along with controls, was used to extensively absorb pooled IgG from four pigs vaccinated with nHgbA (1). Protein G-purified IgG (35 μg in 120 μl PBS) was mixed with each individual loop mutant (the cell pellet from 1 ml of a suspension with an OD of 1.0) and incubated for 60 min on ice. Bacteria were centrifuged, and the absorption was repeated twice for 30 min each time. Absorbed IgG was sterilized by passage through a 0.45-μm filter and stored at −80°C in aliquots.
Hb-binding ELISA.
The ability of anti-nHgbA IgG to block Hb binding to nHgbA was evaluated by enzyme-linked immunosorbent assay (ELISA) as previously described (1) with the following modifications. After immobilization of nHgbA, blocking, and washing, purified anti-nHgbA IgG (previously absorbed or unabsorbed in a volume of 100 μl) was added to each well, and incubation for 20 min at RT was followed by three washes with PBS-Tween (0.05%). Human Hb was labeled with digoxigenin according to the manufacturer's instructions (DIG protein labeling kit; Roche Diagnostic, Indianapolis, IN). Digoxigenin-labeled human Hb (1:100 dilution) was added to the wells, and plates were incubated at RT for 1 h. After three washes, alkaline phosphatase-conjugated anti-digoxigenin antibody (catalog no. 11327720; Roche Diagnostic) was added at a dilution of 1:5,000 in PBS and the mixture was incubated for 1 h at RT. Color development was achieved by adding 50 μl of 1-Step p-nitrophenylphosphate substrate (Pierce) to each well. The plates were incubated for 15 min at RT, and the reaction was stopped by the addition of 50 μl 2 M NaOH. The OD405 was measured on a 1420 VICTOR 2 multilabel spectrophotometer (Wallac Oy, Turku, Finland).
RESULTS
Construction of a stable plasmid expression system in H. ducreyi host strain FX547.
Most forms of rHgbA expressed in E. coli are toxic and, when overexpressed, are found as aggregated protein in inclusion bodies (14). Plasmids expressing hgbA with a leader sequence are unstable in E. coli (data not shown). Moreover, these inclusion body-derived (IBD) rHgbA proteins are not recognized by conformation-dependent anti-nHgbA monoclonal antibodies (20), IBD rHgbA does not bind Hb, and antibodies elicited to IBD rHgbA do not recognize whole cells of H. ducreyi (data not shown). To avoid these issues, we decided to express hgbA from plasmids in H. ducreyi. We first constructed an H. ducreyi isogenic host strain, FX547, containing a complete chromosomal deletion of all hgbA sequence. We reasoned that hgbA plasmid constructs would be more stable in this strain since homologous recombination with the chromosome would be less likely. FX547 containing the empty vector pLSKS did not express HgbA as detected by Western blot assays (Fig. 3, second lane) and was unable to grow on low concentrations of Hb (100 μg/ml) as the sole source of heme (12; data not shown).
Complementation of FX547 with full-length hgbA.
When FX547 was complemented in trans with plasmid pUNCH1261 or pUNCH1261sx (pUNCH1261sx is pUNCH1261 lacking several polylinker sites) containing full-length hgbA, HgbA expression was stable and constitutive, since heme stress (10) resulted in little, if any, heme regulation (data not shown). As expected, HgbA expressed from plasmid pUNCH1261 in FX547 bound Hb-agarose (Fig. 3, first lane) and bacterial growth was indistinguishable from that of wild-type (WT) 35000HP when plated on Hb agar (100 μg/ml) (data not shown). The amount of HgbA expressed from the pUNCH1261 multicopy plasmid approximated the amount of the receptor expressed in H. ducreyi 35000HP cells grown on chocolate agar plates. FX547/pUNCH1261 (and pUNCH1261sx) served as the positive control, and FX547/pLSKS (empty vector) (9) served as the negative control in subsequent experiments.
Generation and characterization of first-generation Δ loop hgbA plasmids.
To test the predictions of our topological model (Fig. 1), we constructed a panel of first-generation deletion mutants lacking each of the 11 putative surface-exposed loops as described in Materials and Methods. We did this in order to identify HgbA loops required to bind Hb and loops necessary for growth on Hb. We assayed strains containing each deletion loop plasmid for expression of HgbA, proper assembly, and export to the surface, since it is known that the conformation of HgbA is critical for function. The results obtained with the first-generation loop mutants are presented in Fig. 3. Western blot assays of total cellular proteins with anti-rHgbA antibodies indicated that most of the 11 mutant clones synthesized HgbA proteins with faster mobility than WT protein expressed from plasmid pUNCH1261 (Fig. 3A), consistent with their deletions. There appeared to be some differences in expression levels in that loop mutants 5 and 7 expressed less HgbA than some of the other strains, as assayed by Western blotting. However, loss of immunogenic loops (detailed in Fig. 4), electrotransfer differences, and other factors may have affected these results (Fig. 3).
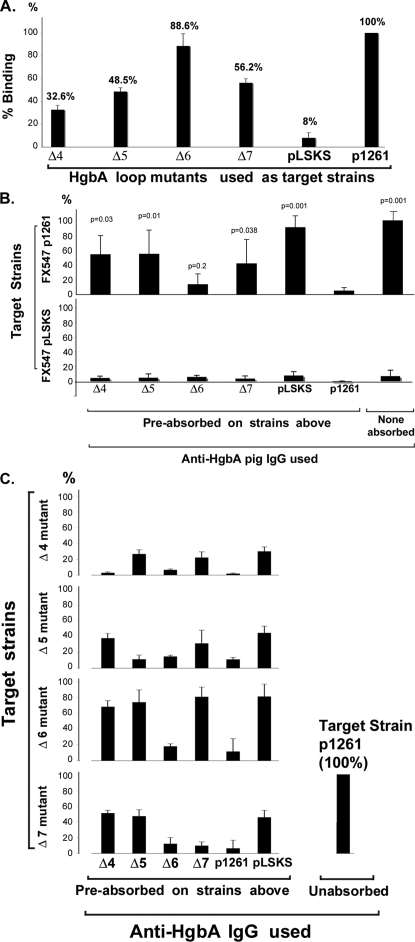
FIG. 4.
Use of loop mutants to identify immunogenic surface-exposed loops of HgbA. (A) Loop mutants bind less IgG. Unabsorbed IgG was bound to the indicated H. ducreyi strains, and antibody binding was measured as described in Materials and Methods. A decrease in antibody binding in Δ loop clones was interpreted as the possible loss of immunogenic epitopes. p1261, FX547/pUNCH1261 expressing full-length HgbA protein; pLSKS, FX547 containing empty vector pLSKS; Δ4, Δ5, Δ6, and Δ7, clones containing deletions in loops 4, 5, 6, and 7, respectively (Fig. 1). Data represent the mean values from four separate experiments. (B) Absorption of anti-HgbA IgG with certain loop mutants identifies immunogenic exposed loops. Unabsorbed or absorbed pig anti-HgbA IgG, as indicated, was tested for binding to FX547/pUNCH1261 (upper panel) or FX547/pLSKS (lower panel). Shown is the percent IgG binding of the indicated Δ loop-absorbed IgG compared to that of unabsorbed FX547/pUNCH1261, which was defined as 100%. P values indicate comparison of the IgG absorbed against the indicated mutant versus IgG absorbed against FX547/pUNCH1261. Data represent the mean values from three separate experiments. (C) Evidence that IgG was exhaustively absorbed and additional evidence that mutant proteins were surface exposed. To show that absorption was complete, we examined the binding of absorbed IgG to the absorbing mutant, selected other mutants, and controls. Note that anti-HgbA IgG absorbed against deletion clones Δ4, Δ5, and Δ7 lost binding against the homologous mutants, respectively, indicating complete absorption. In contrast, IgG absorbed against deletion clones Δ4, Δ5, and Δ7 retained binding to heterologous mutants that contained those loops, respectively, indicating that these mutant proteins were surface exposed. Data represent the mean values from three separate experiments.
In whole-cell immunoprecipitation experiments, anti- nHgbA antibodies were mixed with intact H. ducreyi, and after a brief period of binding, unbound antibodies were washed away (see Materials and Methods for complete details). Thus, this method should only detect surface-exposed epitopes on exported OMPs. Moreover, since these antibodies recognize primarily conformational epitopes, this should be a rigorous test of conformation (1). All 11 loop deletion proteins were immunoprecipitated, suggesting at least a partial amount of proper folding and export (Fig. 3B). Similar results were seen in whole-cell dot blots probed with anti-nHgbA IgG (Fig. 3E). When Sarkosyl-insoluble OMPs were prepared from H. ducreyi strains, all mutant HgbA proteins fractioned with H. ducreyi OMPs, except the HgbA protein lacking loop 5 (Fig. 3C). This finding could have been due to different detergent solubility or instability of the loop 5 protein molecules that were exported. Clearly, the loop 5 protein was surface exposed based on whole-cell immunoprecipitation and dot blot assays (Fig. 3) and in later binding studies (Fig. 4), suggesting proteolysis as a possible reason for its absence in the OMP preparation.
To test the ability of the HgbA loop deletion proteins to bind Hb-agarose, OMPs were solubilized with Zw and affinity purification was performed. Examination of the eluates from Hb-agarose revealed that 7 of 11 mutant proteins bound WT levels of Hb, 2 bound reduced levels (loop 2 and 6 mutant proteins), and 2 were absent (loop 5 and 7 mutant proteins). Since the loop 5 mutant protein was not present in the OMP starting material, it was also not found in the eluate and no conclusion can be drawn. To circumvent this problem, we performed additional affinity experiments with total cellular proteins (as opposed to OMPs, as described above) from loop 5 and 7 mutants solubilized in Zw. In these total cellular protein affinity experiments, the WT positive control protein expressed from pUNCH1261 bound the Hb-agarose but neither the loop 5 nor the loop 7 mutant protein bound Hb-agarose (data not shown). We concluded that 11 loop mutants were exported and detected on the surface by antibody, 10 of 11 fractionated in Sarkosyl detergent as expected, and that 9 of these 11 mutant proteins bound Hb-agarose to various degrees. To better localize the regions of loops 5 and 7 involved in Hb binding, we constructed new hgbA clones with smaller deletions and repeated the analysis.
Generation and characterization of second-generation Δ loop 5 and Δ loop 7 hgbA mutants.
The newly constructed loop 5 and 7 mutants, designated 5n and 7n (see Fig. 1 and 2 for details), contained deletions of 43 and 16 amino acids, respectively. In comparison to the full-length protein, the mobility of HgbA from mutants 5n and 7n (right-hand panel of Fig. 3) was not obviously different from WT mobility, presumably because fewer amino acids were missing (Fig. 3A). Localization studies (immunoprecipitation, OMP localization, and dot blot assays) revealed surface exposure of the 5n and 7n mutant proteins (Fig. 3B, C, and E). The Δ loop 5n HgbA protein bound Hb-agarose at barely detectable levels, whereas the Δ loop 7n HgbA protein failed to bind Hb-agarose (Fig. 3D). Neither the Δ loop 5n nor the Δ loop 7n mutant bound Hb in the dot blot format (Fig. 3F). We concluded that the residues absent in these two new loop mutants are required for the binding of Hb.
Identification of immunogenic surface-exposed loops of HgbA.
Previously we reported that immunization of pigs with purified nHgbA generated antibodies that recognized the surface of H. ducreyi (1). In the present study, we used this IgG to examine the surface exposure of HgbA loops from our two-dimensional model (Fig. 1). We also sought to determine which loops of HgbA contained epitopes for antibodies with biological activity. First, we screened loop mutants by examining whether there was a decrease in the binding of (unabsorbed) pig anti-HgbA IgG to each of the first-generation loop mutants compared to FX547/pUNCH1261 expressing the full-length protein. Three loop clones, loop 4, loop 5, and loop 7 clones, showed greater than 40% reduced IgG binding compared to that of FX547/pUNCH1261 when using an approximately threefold excess of IgG (Fig. 4A). Eight other loop mutants, represented in Fig. 4A by the loop 6 mutant, retained most of their binding relative to that of the control. Similar qualitative results were seen in the dot blot assays (Fig. 3E). These results could be interpreted in two ways. One possibility is that, in the context of the original nHgbA immunogen, a loop was immunogenic and loss of that loop in the whole-cell antigen resulted in a decreased signal (Fig. 3E and 4A). Another possible reason for the reduced antibody binding observed is that a particular mutant protein was not properly exported or surface exposed in H. ducreyi.
Absorption of anti-nHgbA IgG against loop mutants.
Our rationale in these absorption studies was to use our panel of first-generation loop mutants to absorb pooled pig anti-nHgbA IgG and generate loop-specific IgG. The concept is that if a loop were immunogenic in the nHgbA vaccine study and surface exposed, then after extensive absorption with a particular loop mutant, the only remaining antibodies to surface-exposed epitopes should be directed against epitopes deleted in that construct. If absorbed IgG retained binding to FX547/pUNCH1261 expressing full-length HgbA but lost binding to the particular mutant used for absorption, it indicated that we had sufficiently absorbed the antibody with the mutant. Such a result would indicate that the sequences missing in that loop elicited antibodies and confirm that that loop was surface exposed on the WT protein. Based on preliminary whole-cell binding titration experiments, we used 2.3 μg of absorbed IgG per well. By using excess IgG, this allowed us to robustly detect antibodies specific to a single loop after any antibodies to the other 10 loops were removed during absorption.
After absorption on each individual loop mutant, only IgG absorbed on loop mutants 4, 5, and 7 retained binding to the WT protein. Other H. ducreyi clones, represented by the Δ loop 6 clone, did not retain binding to WT (Fig. 4B, top panel). The majority of antibodies were specific for HgbA, since only a small percentage of antibodies bound FX547/pLSKS (Fig. 4A, B, and C). One interpretation of these results is that the Δ loop 4, 5, and 7 proteins were immunogenic in the context of the holo-nHgbA immunogen. Alternatively, if the Δ loop 4, 5 or 7 protein was not properly exported or folded or if incomplete (insufficient) absorption was accomplished, similar results might be observed. To distinguish between these possibilities, we performed additional studies.
To prove that each mutant protein was properly exported and folded and that the absorption was complete, each absorbed IgG was also tested for binding against its homologous absorbing mutant and other controls (Fig. 4C). As shown for IgGs specific to loops 4, 5, and 7, each IgG preparation lost activity against its absorbing mutant but not to other mutant strains that retained the loop in question. For example, IgG absorbed on loop mutant 4 failed to bind loop mutant 4 after absorption but bound to loop mutants 5, 6, and 7. These data confirm the surface exposure of the mutant proteins shown previously by whole-cell immunoprecipitation and dot blot experiments in Fig. 3, since they were bound by heterologous anti-loop IgG.
Having shown the specificity of the absorbed IgGs, we tested each for biological function. Previously, we reported the bactericidal activity of swine anti-nHgbA antibodies for 35000HP was modest, resulting in approximately 50% killing, and required high concentrations (10% serum or 500 μg/ml IgG) (1). After the anti-nHgbA IgG was absorbed against the loop mutants, none of the IgG retained bactericidal activity for strain 35000HP (data not shown).
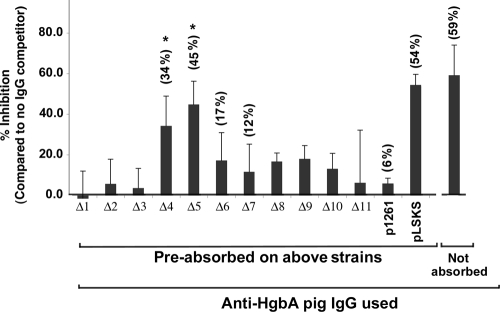
Next we tested the ability of the absorbed IgG and control IgG to inhibit the Hb binding to purified nHgbA in an ELISA (Fig. 5). In control experiments, unabsorbed IgG or IgG absorbed against FX547/pLSKS blocked binding of Hb to nHgbA whereas absorption against FX547/pUNCH1261 abrogated binding. IgG absorbed on loop 4 or 5 mutants, but not others (Fig. 5 and data not shown), partially blocked Hb binding to immobilized nHgbA.
FIG. 5.
Ability of anti-loop IgG to inhibit binding of Hb to HgbA in an ELISA format. Native HgbA purified from H. ducreyi under nondenaturing conditions was immobilized on an ELISA plate, the plate was blocked, and the indicated IgGs were added. Digoxigenin-labeled human Hb was then added, and binding was monitored with an alkaline phosphatase-conjugated anti-digoxigenin secondary antibody. Values obtained from wells that received no antibody were defined as 100% Hb binding. Data are mean values compiled from five separate experiments. Statistical comparisons were done between each IgG absorbed on loop mutants and absorption on the FX547/pUNCH1261 control expressing full-length HgbA protein. *, P = 0.008 for both Δ loop 4 and 5 IgGs (Mann-Whitney rank sum t test).
Evidence for surface exposure of loop 6.
We treated whole cells of H. ducreyi 35000HP with limiting amounts of protease (18) and identified cleaved HgbA peptides in Western blot assays (data not shown). HgbA peptides were subjected to Edman degradation. Although a number of trypsin peptides were generated and sequenced, most corresponded to the N-terminal sequence of mature HgbA (the plug domain) and were not informative. However, one of the trypsin-generated peptides matched the amino acids found at the apex of loop 6 (Fig. 1), and it was (X)NTYATQD, where X represents a mixed signal from the first Edman cycle. Taken together, the above data are consistent with surface exposure of loops 4, 5, 6, and 7.
Effect of loop mutations on the ability to utilize heme from Hb for growth.
We tested the ability of H. ducreyi containing the loop deletions to grow on agar medium containing Hb (100 μg/ml) as a sole source of heme. None of the loop deletion constructs or the negative control (FX547/pLSKS [empty vector]) was able to grow, whereas the positive control (WT construct FX547/pUNCH1261) grew well (data not shown).
DISCUSSION
We sought to identify specific surface-exposed loops of HgbA that are important for Hb binding and/or utilization. We found that deletion of putative surface-exposed loop 5 or 7 rendered the mutant HgbA proteins unable to bind Hb in two separate assays. Deletions in nine other loops had little or no effect on Hb binding. We anticipated that a number of mutants might not be properly exported to the outer membrane, and we were somewhat surprised that we could make fairly large deletions and still maintain export of mutant proteins and Hb binding. For example, elimination of the plug region (approximately 150 amino acids; data not shown) or much of the largest loop, loop 4 (124 amino acids; Fig. 1 and 3), affected neither HgbA export nor Hb binding. Since Hb binding by HgbA requires some form of proper conformation (10), these findings suggest that the mutant proteins were folded well enough to retain partial function.
We generated monospecific IgG directed at certain loops of HgbA. We found that loops 4, 5, and 7 were immunogenic in the intact native HgbA immunogen. It should be noted that there are limitations to these studies. These limitations include the possibility that the other HgbA loops were nonimmunogenic or immunogenic but not surface exposed; if so, antibodies would not be detected. Conversely, a loop could be both immunogenic and surface exposed but if we deleted an incorrect, unexposed region of the loop, absorption experiments would not detect antibodies. Thus, it is possible or even likely that additional loops are surface exposed and/or immunogenic, but with the limited set of deletions we made we were unable detect them.
Additional limitations exist for IgG that retained binding to the parent strain. First, if there had been insufficient absorption, antibodies would be present to still bind the mutant. However, antibodies specific for loops 4, 5, and 7 bound WT HgbA and also bound deletion mutants containing their respective loops but did not bind their homologous absorbing mutants. This indicated sufficient absorption and is consistent with surface exposure of these three mutant proteins. A second limitation is that if a particular clonal IgG is directed at a conformational epitope involving more than one loop, then such antibodies might not be completely absorbed and might still bind the parent and perhaps certain mutants. Nevertheless, the sera were informative for certain loops, suggesting surface exposure of some parts of loops 4, 5, and 7.
With monospecific loop antibodies, anti-loop 4 and 5 inhibited binding of human Hb to immobilized, purified nHgbA. The facts that loop 5 deletions abrogated Hb-agarose binding and that anti-loop 5 IgG inhibited Hb binding are consistent with the involvement of this loop in Hb binding and warrant additional detailed studies.
HgbA loop 6 is a relatively conserved loop among various Hb receptors from diverse genera and contained only two substitutions within the sequenced hgbA genes from 10 strains of H. ducreyi (31). The tip of loop 6 was cleaved from whole H. ducreyi by protease, suggesting that loop 6 is surface exposed. The lack of additional trypsin cleavage products could be caused by several factors. First, small trypsin digestion products could have been run off of the polyacrylamide gel prior to Western blotting and not visible in this assay. Second, the antibody used to detect fragments reacts primarily with the N-terminal plug domain and may not have reacted well enough with C-terminal fragments to detect them. Third, although the primary sequence suggests trypsin sites in loops 4, 5, and 7, the tertiary structure of loops 4, 5, and 7 may impede trypsin accessibility or lack the flexibility necessary for protease cleavage (16). Similar results were seen with the gonococcal transferrin receptor TbpA, another outer membrane TB-DR. TbpA contains 129 putative trypsin cleavage sites but only yields two distinct trypsin cleavage products (corresponding to two putative surface-exposed loops) when the protein is in its membrane-bound state (6).
We tested the ability of each mutant to grow on GCB agar containing 100 μg/ml (nominal, after filtration) human Hb. None of the FX547 clones expressing mutant proteins was able to grow under these stringent conditions. In some experiments, a haze of growth was noted after 48 h of incubation but any limited growth observed did not form colonies upon subculture. Positive controls FX547/pUNCH1261 and 35000HP, both expressing full-length hgbA, grew well, and individual colonies could be repeatedly subcultured on this medium. This suggests that the removal of heme from Hb and its subsequent transport across the outer membrane are parts of a complex event. It is known from our previous studies that growth on Hb requires an intact TonB system (13). Perhaps minor disturbances in the HgbA protein disrupt this interaction and account for the observed growth defects.
Hb receptors in other bacteria.
Numerous TB-DRs have been identified and experimentally proven to be receptors for Hb in other gram-negative bacteria. Even more putative Hb receptors have been proposed from the comparison of predicted primary protein sequences from proven Hb receptors with TB-DRs from genome sequencing projects. Some Hb receptors undergo phase and/or antigenic variation, such as those from Neisseria (7, 24) and H. influenzae (8, 23). In the case of H. influenzae, a single strain may contain three loci encoding Hb receptors, with each strain capable of expressing multiple phase-variable, antigenically distinct Hb receptors, either simultaneously or separately. H. influenzae Hb receptors are about 45% identical to HgbA of H. ducreyi. The significant differences within the various H. influenzae proteins are almost entirely confined to the tips of putative surface-exposed loops. Since a substantial amount of variation exists at these loops, apparently some parts the loops are not required for Hb binding or heme internalization from Hb. Many of the deletions we made in H. ducreyi are analogous to the variable H. influenzae regions and may explain why some H. ducreyi HgbA mutant proteins tolerated the loop deletions.
The HmbR Hb receptor of the meningococcus was previously subjected to deletion mutagenesis (21), and the following discussion compares our HgbA results with the results of HmbR expressed in meningococci. The deletion of loops 1, 2, 3, 4, 6, 8, 9, 10, and 11 of HgbA did not prevent Hb binding. Similarly, deletion of loops 6, 8, and 10 in HmbR did not affect Hb binding, and deletions in loops 2 and 5 decreased Hb binding (deletions of loops 1, 4, 9, and 11 were not reported for HmbR). Both proteins with deletions in loop 7 failed to bind Hb. The different results found for Hb binding in some cases can probably be attributed to several variable experimental conditions and perhaps structural differences between the two proteins.
The ability to utilize Hb was impaired in all of the HgbA loop mutants tested, whereas deletion of HmbR loops 5, 8, and 10 did not affect HmbR Hb utilization. Our utilization assay uses the minimum amount of Hb in GCB base medium, and it contains unchelated free Fe. This medium can support the growth of strain 35000HP and is a rigorous test. In contrast, the meningococcal experiments used GCB medium containing Hb and an iron chelator. This may partially account for the differences observed. Lastly, in all of these deletion studies, it is possible that the deletions affected the three-dimensional folding of the proteins and that the Hb-binding phenotypes are the result of misfolding. Thus, one can never be sure that binding sites in affected mutants reside in the deleted sequences.
In summary, our results suggest that the central domain of HgbA is critical to Hb binding by HgbA and has important vaccine implications. The fact that loops 4 and 5 elicited the highest titer of antibodies in protected animals that inhibited binding of Hb to HgbA suggests that perhaps a vaccine only targeting the central domain might be an effective way to generate protective antibodies. Further studies are necessary to confirm these preliminary structural findings.
Supplementary Material
Acknowledgments
We thank Marcia Hobbs and Chris Thomas for critical comments on the manuscript. We thank Annice Rountree for excellent technical support.
The work presented was supported by 5-R01-AI 05393 from NIH to C.E.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 18 May 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Afonina, G., I. Leduc, I. Nepluev, C. Jeter, P. Routh, G. Almond, P. E. Orndorff, M. Hobbs, and C. Elkins. 2006. Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA protects against infection in the swine model of chancroid. Infect. Immun. 742224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton, W. L., I. W. Macklean, P. D. Bertram, and A. R. Ronald. 1981. Haemin requirements in Haemophilus with special reference to H. ducreyi. Academic Press, Inc., New York, NY.
- 3a.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 1781684-1687. [DOI] [PubMed] [Google Scholar]
- 4.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 1811049-1054. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, M., M. E. Garrido, M. Llagostera, A. M. Perez De Rozas, I. Badiola, and J. Barbe. 2002. Characterization of the Pasteurella multocida hgbA gene encoding a hemoglobin-binding protein. Infect. Immun. 705955-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulton, I. C., M. K. Yost, J. E. Anderson, and C. N. Cornelissen. 2000. Identification of discrete domains within gonococcal transferrin-binding protein A that are necessary for ligand binding and iron uptake functions. Infect. Immun. 686988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. J., C. Elkins, and P. F. Sparling. 1998. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect. Immun. 66987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope, L. D., Z. Hrkal, and E. J. Hansen. 2000. Detection of phase variation in expression of proteins involved in hemoglobin and hemoglobin-haptoglobin binding by nontypeable Haemophilus influenzae. Infect. Immun. 684092-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutro, S. M., G. Wood, and P. Totten. 1999. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect. Immun. 673317-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 631241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C., K. B. Barkley, N. H. Carbonetti, A. J. Coimbre, and P. F. Sparling. 1994. Immunobiology of purified recombinant outer membrane porin protein I of Neisseria gonorrhoeae. Mol. Microbiol. 141059-1075. [DOI] [PubMed] [Google Scholar]
- 12.Elkins, C., C. J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus of Haemophilus ducreyi. Infect. Immun. 632194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins, C., P. A. Totten, B. Olsen, and C. E. Thomas. 1998. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect. Immun. 66151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkins, C., K. Yi, B. Olsen, C. Thomas, K. Thomas, and S. Morse. 2000. Development of a serological test for Haemophilus ducreyi for seroprevalence studies. J. Clin. Microbiol. 381520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond, G. W., C. J. Lian, J. C. Wilt, and A. R. Ronald. 1978. Comparison of specimen collection and laboratory techniques for isolation of Haemophilus ducreyi. J. Clin. Microbiol. 739-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard, S. J., F. Eisenmenger, and J. M. Thornton. 1994. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 3757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leduc, I., K. E. Banks, K. R. Fortney, K. B. Patterson, S. D. Billings, B. P. Katz, S. M. Spinola, and C. Elkins. 2008. Evaluation of the repertoire of the TonB-dependent receptors of Haemophilus ducreyi for their role in virulence in humans. J. Infect. Dis. 1971103-1109. [DOI] [PubMed] [Google Scholar]
- 18.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 723418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto, B. R., A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18217-233. [DOI] [PubMed] [Google Scholar]
- 20.Patterson, K., B. Olsen, C. Thomas, D. Norn, M. Tam, and C. Elkins. 2002. Development of a rapid immunodiagnostic test for Haemophilus ducreyi. J. Clin. Microbiol. 403694-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins-Balding, D., M. T. Baer, and I. Stojiljkovic. 2003. Identification of functionally important regions of a haemoglobin receptor from Neisseria meningitidis. Microbiology 1493423-3435. [DOI] [PubMed] [Google Scholar]
- 22.Plummer, F. A., J. N. Simonsen, D. W. Cameron, J. O. Ndinya-Achola, J. K. Kreiss, M. N. Gakinya, P. Waiyaki, M. Cheang, P. Piot, A. R. Ronald, and E. N. Ngugi. 1991. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 163233-239. [DOI] [PubMed] [Google Scholar]
- 23.Ren, Z., H. Jin, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J. Bacteriol. 1815865-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson, A. R., and I. Stojiljkovic. 1999. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 1812067-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79818-826. [PMC free article] [PubMed] [Google Scholar]
- 27.Steen, R., and G. Dallabetta. 2004. Genital ulcer disease control and HIV prevention. J. Clin. Virol. 29143-151. [DOI] [PubMed] [Google Scholar]
- 28.Stojiljkovic, I., J. Larson, V. Hwa, S. Anic, and M. So. 1996. HmbR outer membrane proteins of pathogenic neisseriae: iron-regulated, hemoglobin-binding proteins with high degrees of primary structure conservation. J. Bacteriol. 1784670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, C. E., B. Olsen, and C. Elkins. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 664254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Totten, P. A., and W. E. Stamm. 1994. Clear broth and plate media for the culture of Haemophilus ducreyi. J. Clin. Microbiol. 322019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, C. D., I. Leduc, B. Olsen, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 732387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.