Abstract
Changing antigenic structure such as with capsule polysaccharide is a common strategy for bacterial pathogens to evade a host immune system. The recent emergence of an invasive W:2a:P1.7-2,4 sequence type 11 (ST-11) strain of Neisseria meningitidis in New Zealand, an uncommon serogroup/serotype in New Zealand disease cases, was investigated for its genetic origins. Molecular typing of 107 meningococcal isolates with similar serotyping characteristics was undertaken to determine genetic relationships. Results indicated that the W:2a:P1.7-2,4 strain had emerged via capsule switching from a group C strain (C:2a:P1.7-2,4). Neither the upstream nor downstream sites of recombination could be elucidated, but sequence analysis demonstrated that at least 45 kb of DNA was involved in the recombination, including the entire capsule gene cluster. The oatWY gene carried by the W:2a:P1.7-2,4 strain contained the insertion sequence element IS1301, one of five variants of oatWY found in group W135 strains belonging to the carriage-associated ST-22 clonal complex. This suggested that the origin of the capsule genes carried by the invasive W:2a:P1.7-2,4 strain is carriage associated. These results provide novel evidence for the long-standing dogma that disease-associated strains acquire antigenic structure from carriage-associated strains. Moreover, the capsule switch described here has arisen from the exchange of the entire capsule locus.
Neisseria meningitidis group W135 is uncommon among meningococcal disease cases in New Zealand. Approximately 80 to 85% of disease cases are due to group B, 80% of which have been due to the epidemic strain B:4:P1.7-2,4 over the last 15 years (10). A further 10 to 15% of cases are due to group C, and the remainder, approximately 1 to 5%, are due to strains belonging to groups W135 and Y. The one or two group W135 isolates found causing disease per year were not from disease-associated clonal complexes. In the last 7 years worldwide, a marked increase in the number of group W135 isolates has occurred, particularly those with the strain type W:2a:P1.5,2 from the sequence type 11 (ST-11) clonal complex. Approximately 10% of cases in Europe and North America are now due to meningococci with the W135 capsular type (23, 25, 26), and there have been two large outbreaks of disease. The first, occurring at the Hajj, Saudi Arabia, in 2000, resulted in the considerable global spread of the ST-11 strain W:2a:P1.5,2 as pilgrims returned to their home countries (28). The second, in Burkina Faso in 2002, also involved ST-11 strain type W:2a:P1.5,2, although using variable-number tandem repeat analysis, this strain was found to differ from that causing disease in Saudi Arabia (29). Burkina Faso was the first country in the world to have a group W135 strain as the predominant disease-causing strain (9). In sharp contrast, only one case of disease caused by the W:2a:P1.5,2 strain type was recorded in New Zealand prior to 2003 (our unpublished data). In 2003, a new W135 strain type (W:2a:P1.7-2,4) was identified in New Zealand and subsequently isolated from 12 cases. Of particular interest was the fact that this strain carried the same PorA type as the New Zealand epidemic B strain B:4:P1.7-2,4.
N. meningitidis is known for its high rates of DNA transformation and subsequent recombination, leading to the generation of considerable antigenic diversity observed in carriage strains (5). While such levels of genetic variation are not commonly observed among strains associated with disease, “switching” of antigenic structures is not uncommon. These switches may involve capsule polysaccharide or a number of antigenic outer membrane proteins. However, these changes are generally reported only for the capsule, PorA, and PorB proteins. Capsule switching has been shown to result from recombination of the polysialyltransferase gene (siaD) or the capsule biosynthesis operon (22). Transformation and recombination leading to an altered PorA or PorB may occur either by substitution of the entire allele or by exchange of single or multiple variable regions (3, 11, 21). The W:2a:P1.7-2,4 strain identified in New Zealand was originally thought to have arisen through recombination, resulting in a porA switch from the New Zealand epidemic strain. We have investigated the genetic origins of the W:2a:P1.7-2,4 strain in an effort to determine how the epidemic PorA strain was associated with a group W135 strain.
MATERIALS AND METHODS
Meningococcal isolates and growth conditions.
The meningococcal isolates used in this study are shown in Table 1. Meningococcal isolates assigned an NZ number originate from clinical cases of disease that have been referred to New Zealand's Meningococcal Reference Laboratory, ESR, under New Zealand's national surveillance program. All strains were routinely serogrouped, serotyped, and serosubtyped (1), using monoclonal antibodies that recognize serotypes 1, 2a, 2b, 4, 14, and 15 (Netherlands Vaccine Institute, Bilthoven, The Netherlands) and serosubtypes P1.1, P1.2, P1.4, P1.5, P1.6, P1.7, P1.9, P1.10, P1.12, P1.13, P1.14 (NVI), P1.15, and P1.16 (National Institute for Biological Standards and Control, Potters Bar, United Kingdom). Isolates were also subject to genotyping to determine PorA variable region (VR) types (described below). All isolates were stored at −70°C in glycerol broth suspensions (Trypticase soy broth, 15% glycerol).
TABLE 1.
Genotyping results of major antigenic structures on the cell surface
| Reference strain | Genotypesa | No. of isolates tested |
|---|---|---|
| NZAK04051 | B:15:P1.7,16-26; F3-3; ST-32 (cc32) | 2 |
| NZAK04078 | B:4:P1.7-2,13-1; F4-1; ST-5709 (cc35) | 1 |
| NZD2-052-1 | B:4:P1.7-2,4; F1-18; ST-41 (cc41/44) | 1 |
| NZ98/254 | B:4:P1.7-2,4; F1-5; ST-42 (cc41/44) | 2 |
| NZ96/142 | B:4:P1.7-2,4; F1-5; ST-154 (cc41/44) | 1 |
| NZ02/16 | B:4:P1.7-2,4; F5-5; ST-154 (cc41/44) | 1 |
| NZ05/65 | B:4:P1.7-2,30; F1-5; ST-42 (cc41/44) | 1 |
| NZAK04038 | B:14:P1.22-12,14; F1-5; ST-437 (cc41/44) | 1 |
| NZA2-008-1 | B:nt:P1.5,2; F5-1; ST-32 (cc32) | 1 |
| NZ00/68 | C:2b:P1.5,2; F5-8; ST-8 (cc8) | 1 |
| NZ97/296 | C:2b:P1.5,2; F3-9; ST-66 (cc8) | 1 |
| NZ96/211 | C:2b:P1.7-2,4; F3-9; ST-66 (cc8) | 1 |
| NZ94/106 | C:2a:P1.5,2; F1-1; ST-11 (cc11) ★ | 2 |
| NZ99/142 | C:2a:P1.5,2; F1-30; ST-11 (cc11) | 1 |
| NZ01/303 | C:2a:P1.5,2; F3-6; ST-11 (cc11) | 1 |
| NZ93/149 | C:2a:P1.5,2; F4-1; ST-11 (cc11) | 2 |
| NZ03/288 | C:2a:P1.5,2; F5-1; ST-11 (cc11) | 1 |
| NZ05/118 | C:2a:P1.5,2; F3-6; ST-4251 (cc11) ★ | 1 |
| NZ04/155 | C:2a:P1.5-1,10-1; F3-6; ST-11 (cc11) ★ | 3 |
| NZ00/198 | C:2a:P1.5-1,10-4; F3-6; ST-11 (cc11) ★ | 12 |
| NZ03/101 | C:2a:P1.5-1,10-8; F3-6; ST-11 (cc11) ★ | 6 |
| NZAK04020 | C:2a:P1.7-2,4; F3-6; ST-11 (cc11) ★ | 1 |
| NZ03/236 | C:2a:P1.7-2,4; F5-2; ST-11 (cc11) ★ | 5 |
| NZ96/59 | C:2a:P1.7-2,4; F5-2; ST-2344 (cc-) | 1 |
| NZ03/232 | W:2a:P1.5,2; F1-1; ST-11 (cc11) | 19 |
| NZ95/204 | W:2b:P1.5,2; F1-1; ST-11 (cc11) | 1 |
| NZ05/50b | W:2a:P1.7-2,4; F-; ST-11 (cc11) ★ | 1 |
| NZ03/243 | W:2a:P1.7-2,4; F5-2; ST-11 (cc11) ★ | 14 |
| NZAK04008 | W:1:P1.18-1,3; F3-9; ST-22 (cc22) | 1 |
| NZAK04031c | W:1:P1.18-1,3; F-; ST-184 (cc22) | 1 |
| NZ01/257 | W:nt:P1.18-1,3; F4-1; ST-22 (cc22) | 9 |
| NZ00/276c | W:nt:P1.-,-; F4-1; ST-22 (cc22) | 1 |
| NZD4-208-1 | W:nt:P1.18-1,3; F5-5; ST-22 (cc22) | 1 |
| NZAK04043 | W:nt:P1.18-1,3; F1-5; ST-184 (cc22) | 1 |
| NZD1-032-1 | W:4:P1.18-1,16; F4-1; ST-184 (cc22) | 1 |
| NZ03/38 | W:14:P1.19,15; F5-8; ST-4183 (cc23) | 1 |
| NZAK04005 | W:15:P1.18,25; F5-5; ST-198 (cc198) | 1 |
| NZ97/169 | W:4:P1.19-1,26-4; F1-7; ST-1846 (cc41/44) | 1 |
| NZAK04113 | Y:14:P1.5-2,10-1; F1-6; ST-23 (cc23) | 1 |
| NZA1-348-1c | Y:a48:P1.5-1,10-1; F4-1; ST-167 (cc167) | 1 |
| NZA1-354-1 | Y:4:P1.5-1,10-4; F5-5; ST-167 (cc167) | 1 |
| NZ95/195d | Y:a48:P1.5-1,10-4; F1-5; ST-5159 (cc167) | 1 |
nt, nontypeable; ★, belongs to the ET-15 subgroup of ST-11; cc, clonal complex. Boldface indicates deletion.
A 20-amino-acid deletion in the VR resulted in no assignment of an allele name. Most likely a deletion of F5-2.
No PCR product obtainable; may denote a deletion.
Not able to be named in the usual format, instead matches porB allele 48 from http://neisseria.org/nm/typing/porb.
All strains of N. meningitidis were grown on Columbia base agar (Fort Richard Laboratories, Auckland, New Zealand) supplemented with 5% sheep's blood overnight at 36°C in an atmosphere of 5% CO2.
DNA extractions.
Genomic DNA from meningococcal isolates was harvested using the QIAamp DNA minikit (Qiagen). For microarray experiments, DNA was extracted using a phenol-chloroform-based method. Briefly, an overnight plate culture of N. meningitidis was harvested and suspended in 1 ml phosphate-buffered saline. The cells were washed; pelleted; resuspended in 500 μl of 10 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM EDTA; and heat killed for 2 to 3 h at 56°C. Sodium dodecyl sulfate and proteinase K were then added, giving final concentrations of 1% and 10 μg/ml, respectively. The solution was incubated overnight at 37°C. The DNA was purified by phenol-chloroform extraction, precipitated using 3.2 M sodium acetate and 100% ethanol, and then washed in 70% ethanol and resuspended in sterile distilled water.
PCR-based analysis and sequencing.
PCR was carried out using PCR master mix (Qiagen) with the appropriate oligonucleotide primers and under the appropriate conditions. Oligonucleotides were synthesized by Invitrogen, and sequencing was performed using an ABI 3100 sequencer (Applied Biosystems, Foster City, CA). The determination of the porA VRs was done as described previously (10), using primers porA1 and porA2 for PCR and primers porAVRseqF (CAGCCTGTACGGCGAAATC), porAVRseqR (CGCATATTTAAAGGCATAGTTCC), and porAVR3seq (GGCGAGATTCAAGCCGCC) (6) to generate the sequence. VRs were identified by submitting the sequence to the porA typing website (http://neisseria.org/perl.agdbnet/agdbnet.pl?file=poravr.xml&page=oneseq) or matching VR3 with published sequences (Scottish Meningococcus and Pneumococcus Reference Laboratory). Typing of fetA was carried out as previously described (24) using oligonucleotide primers fetAP1 (CGGCGCAAGCGTATTCGG) and fetAP2 (CGCGCCCAATTCGTAACCGTG) for PCR amplification and fetaseqF (TTCAACTTCGACAGCCGCCTT) and fetaseqR (TTGCAGCGCGTCRTACAGGCG) for sequencing. The fetA allele was determined by submitting the DNA sequence to the fetA website (http://neisseria.org/perl/agdbnet/agdbnet.pl?file=fetavr.xml). Multilocus sequence typing (MLST) was performed as described previously (15), with the addition of primers fumC-A1 and fumC-A2 to amplify fumC (10). To determine the allele types and subsequent sequence type and clonal complex, the sequence was submitted to the MLST website (http://pubmlst.org/neisseria). The fumC sequence was also analyzed for the presence of a point mutation at position 640, indicating the fumC2 allele found in ET-15 isolates of the ST-11 complex (27). New MLST and fetA alleles or porA VRs discovered were submitted to the www.neisseria.org website for allocation of allele names. Multiple sequence alignments were carried out using ClustalW (http://www.ch.embnet.org/software/ClustalW.html).
Whole-genome RFLP analysis and pulsed-field gel electrophoresis (PFGE).
Isolates used for restriction fragment length polymorphism (RFLP) analysis were resuspended in cold 1× Tris-EDTA to an optical density at 550 nm of 0.35. Salmonella serotype Branderup H9812 was resuspended to an optical density at 550 nm of 0.5. Plugs were made using 1.6% InCert agarose (Cambrex) and lysed overnight using ESP solution (0.5 M EDTA, 1% Sarkosyl, 1 mg/ml proteinase K). The plugs were washed in 1× Tris-EDTA, including an overnight washing, and then stored at 4°C until use. For RFLP analysis, a slice of each plug was digested with at least 20 U of the appropriate enzyme. Plugs were loaded into the wells of a 1% GTG agarose gel, and the gel was run overnight on a CHEF-DR II (Bio-Rad) system. The electrophoresis switching conditions used were as follows: initial time, 5 s; final time, 30 s; start ratio, 1; and run time, 22 h. The gel was stained in an ethidium bromide bath (10 mg/ml) and destained for 10 min in water. The image was analyzed using BioNumerics software (Applied Maths BVBA, Sint-Martens-Latem, Belgium), and both the position tolerance and optimization were set to 1%.
DNA-DNA microarray hybridizations.
DNA-DNA microarrays were performed, using the N. meningitidis FAM-18 serogroup C reference strain as a control against the isolates tested. Experiments were done in duplicate, and the Cy3 and Cy5 fluors were swapped in the duplicate to avoid artifacts of labeling. Approximately 1 μg of DNA from the test isolate and reference isolate was used in each experiment. Dye labeling was achieved using a random primer (Invitrogen) and a Klenow fragment to incorporate the Cy3- or Cy5-labeled dCTP into the probes. To set up the DNA-DNA hybridization, labeled test and reference DNA was mixed, denatured, and applied to prehybridized microarray slides (St. George's Hospital, London, United Kingdom). The pan-Neisseria array and procedures for hybridization and washing were followed, as previously described by the St. George's Bacterial Microarray Group (19a).
The hybridized slides were then scanned using a GenePix 4000B scanner (Axon). The scanned images were analyzed with GenePix Pro 6.0 (Axon), and statistical analysis was carried out using GeneSpring GX 7.3 (Agilent). Background-subtracted spot intensities resulting from hybridization of test DNA were divided by the intensity of the signal derived from reference DNA. Log-transformed ratios were normalized by applying the intensity-dependent data analysis technique LOWESS, using 20% of the data for smoothing. Only genes not marked absent by GenePix Pro analysis were included. If the value for the reference channel was less than 10, then a value of 10 was used instead. Genes that differed between strains were defined as having a change in fluorescence intensity greater than 1.5-fold and a P value of less than 0.05. This was determined using parametric one-way analysis of variance of the normalized intensity ratios, including those from the dye swaps. Multiple testing correction was performed using the Benjamini and Hochberg false discovery rate, set to 5%.
Microarray data accession number.
The complete set of microarray data was submitted to the Array Express database at the European Molecular Biology Laboratory—European Bioinformatics Institute (www.ebi.ac.uk/arrayexpress) (accession no. E-MEXP-1910).
Nucleotide sequence accession numbers.
The sequences of the cps cluster from group W135 strains NZ05/50 (W:2a:P1.7-2,4) and NZD1-006-1 (W:nt:P1.18-1,3) were submitted to GenBank (accession numbers EU164779 and EU164780, respectively).
RESULTS
Molecular typing of the W:2a:P1.7-2,4 isolate indicated capsule gene switching.
Molecular typing methods used included genotyping of the capsule, porB, porA, and fetA alleles, and MLST (Table 1). All but 1 of the 15 W:2a:P1.7-2,4 ST-11 isolates were indistinguishable from one another using these methods. The exception had a deletion in the fetA allele. All 19 W:2a:P1.5,2 isolates tested were indistinguishable. Other than the porA allele, the W:2a:P1.7-2,4 isolates differed from the W:2a:P1.5,2 isolates only at the fetA locus. The W:2a:P1.5,2 isolates all carried the F1-1 fetA allele despite being isolated from different countries and in different years, while the W:2a:P1.7-2,4 isolates carried the F5-2 allele. Similarly, while the W:2a:P1.7-2,4 and W:2a:P1.5,2 isolates all belonged to the ST-11 clonal complex, only the W:2a:P1.7-2,4 isolates belonged to the ET-15 subgroup. This indicated that the W:2a:P1.5,2 strain was not directly related to the W:2a:P1.7-2,4 strain and that the emergence of this new strain did not involve a porA switch in a W:2a:P1.5,2 isolate.
Analysis of the genotyping data revealed that the W:2a:P1.7-2,4 isolates exhibited the highest level of similarity to five C:2a:P1.7-2,4 isolates, consistent with a capsule switch from group C to group W135. Another two isolates with the C:2a:P1.7-2,4 strain type appear to have arisen through separate DNA exchanges. The first of these, isolate NZ96/59, appears to have resulted from an entirely separate DNA transfer, as the ST is unique and of unknown clonal complex. None of the seven MLST alleles were the same as those from the ST-11 allelic profile. The second isolate, NZAK04020, shared all the features of the other four C:2a:P1.7-2,4 isolates, except for the fetA allele identified as F3-6. Allele F3-6 was also found to be carried by the group C isolates carrying porA subtype P1,5-1,10-4, P1.5-1,10-8, or P1.5-1,10-1, suggesting a possible porA switch to result in a C:2a:P1.7-2,4 strain type.
Macrorestriction analysis further supported capsule gene switching.
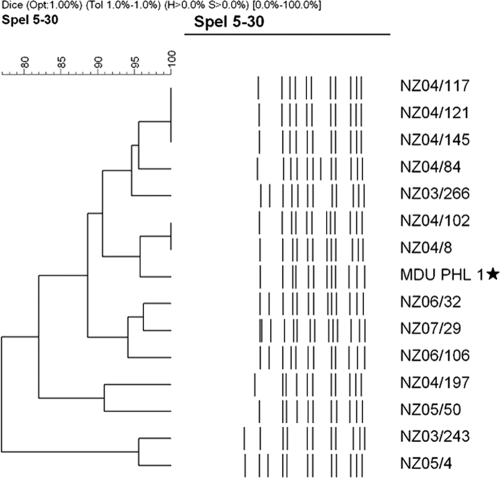
The 107 isolates were subjected to DNA macrorestriction with the enzyme SpeI, and banding patterns were generated by PFGE. The 15 W:2a:P1.7-2,4 isolates showed 12 banding patterns (Fig. 1), indicating genetic differences not detected using allele-based typing methodologies. Only two patterns were produced by multiple isolates, the first of which involved three isolates received within a month of each other from the same geographical location. The second pattern was produced by two isolates separated by 6 months and 350 kilometers. All remaining patterns corresponded to single isolates. The W:2a:P1.7-2,4 banding patterns, as a whole, showed distinct differences to those of the W:2a:P1.5,2 isolates (data not shown) and instead showed more similarity with group C isolates. In particular, the banding patterns of the newly emerged W135 isolates appeared most like those of the C:2a:P1.7-2,4 isolates, providing further evidence of a capsule switch, as deduced from the genotyping results. The remaining group W135 isolates tested, mostly strain type W:nt:P1.18-1,3 ST-22, produced distinctly different banding patterns compared with those of the ST-11 group W135 isolates (data not shown).
FIG. 1.
RFLP banding patterns of the W:2a:P1.7-2,4 isolates. The 15 isolates produced multiple banding patterns, with only two groups of identical patterns. Isolate MDU PHL 1 (★) was isolated in Australia in 2003 before the first isolation of the strain in New Zealand.
Analysis of capsule biosynthesis and downstream genes in the group W135 and C isolates.
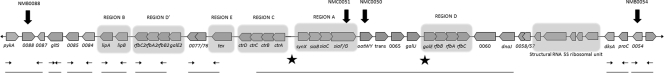
The genetic arrangement of the capsule-associated genes in N. meningitidis W135 has been previously reported and is shown, along with further sequence data from this study, in Fig. 2 (7, 22). The sequence contains several regions, including those involved in capsule biosynthesis, transport, modification, and inner core lipooligosaccharide (LOS) biosynthesis. While these regions have been fully sequenced in the group C reference strain (FAM-18), apart from the capsular biosynthesis locus, little or no sequence data exist in the public domain for group W135 isolates. To determine the possible sites of recombination (SOR), it was essential to have sequence data spanning the area of interest. A study by Swartley et al. on capsule switching in N. meningitidis concluded that the upstream SOR was within the intergenic region between the ctrA and synX genes, while the downstream SOR was probably located either upstream from or within the galE gene of the LOS biosynthesis operon (22). Therefore, these sites were used as starting points in an effort to determine the SOR in the W:2a:P1.7-2,4 strain. Where sequencing was unavailable for group W135, oligonucleotide primers were designed based on regions of conserved DNA in the group B and C reference strains. Sequencing produced 20 kb of contiguous sequence from group C and W135 variants of the 2a:P1.7-2,4 strain, but consistent differences between the two strain types could be identified throughout. Further primer pairs were designed to amplify sections of DNA further upstream and downstream, but differences were still found over a span of 35 kb. DNA microarrays were then performed on several of the isolates (for the C:2a:P1.7-2,4 strain, isolates 991.3179, NZ04/154, NZ00/291, and NZ03/236; for the W:2a:P1.7-2,4 strain, isolates NZ03/243, NZ04/145, NZ04/121, and NZ05/50). Using the group C FAM-18 reference strain as a control, seven genes were identified that were consistently different in these group C and W135 strain variants (Table 2). Two of these genes are specific to the production and O acetylation of the group C capsule (NMC0051 or siaE and NMC0050 or oatC) and were expected to differ. Another three genes identified were located at three different locations on the genome, the significance of which remains unknown but may represent further DNA recombination events. The two remaining open reading frames (ORFs) (NMC0038 and NMC0073) are homologues that encode putative outer membrane proteins and are located outside of the region of DNA previously sequenced (Fig. 2). Further sequence analysis of these homologues and surrounding ORFs indicated that there still were consistent differences between the group C and W135 strains (results not shown). The putative SOR remain undetermined. We report that at least 45 kb of DNA appears to have been recombined to result in the switching of a group C capsule to a W135 capsule.
FIG. 2.
Gene arrangement of the group W135 cps cluster. Regions show the various functional groups within the cluster, as follows: region B (lipid modification genes), region D′ (duplicate LOS biosynthesis operon), region E (putative regulator), region C (capsule transport genes), region A (capsule biosynthesis operon), and region D (LOS biosynthesis operon). Underlined sections represent sequence data collected during the course of this study. Localization of the genes identified with comparative genome hybridizations between the group W135 and group C variants are also identified. NMC0051 and NMC0050 are involved with biosynthesis and O acetylation of the capsule, respectively, and were expected to differ. NMC0038 (NMB0054) and NMC0073 (NMB0088) are located outside the region sequenced (underlined), and further analysis found there to be significant sequence differences in the genes of the two variant strains. The locations marked with a filled star signify the SOR found in a previous study (21).
TABLE 2.
List of ORFs that differ between W:2a:P1.7-2,4 and C:2a:P1.7-2,4 strains, using microarray-based comparative genome hybridizations
| ORF | Gene name | Ratio for strain with indicated genotypea
|
Gene product | |
|---|---|---|---|---|
| C:2a:P1.7-2,4 | W:2a:P1.7-2,4 | |||
| NMC0038 | 0.7242 | 2.1994 | Putative lipoprotein | |
| NMC0050 | oatC | 1.3995 | 0.0536 | O-Acetylase |
| NMC0051 | siaE | 1.2739 | 0.0454 | Polysialyltransferase |
| NMC0073 | 0.8073 | 0.6337 | Outer membrane protein P1, putative | |
| NMC0177 | 1.4797 | 2.4588 | Putative undecaprenyl diphosphate synthase | |
| NMC0259 | ruvA | 0.9472 | 1.1708 | Holliday junction DNA helicase |
| NMC1905 | atpC | 0.8005 | 1.6427 | ATP synthase F1, epsilon subunit |
Average ratio of the strain ORF to control DNA (FAM-18; C:2a:P1.5,2 ST-11).
Determining the donor of the capsule genes.
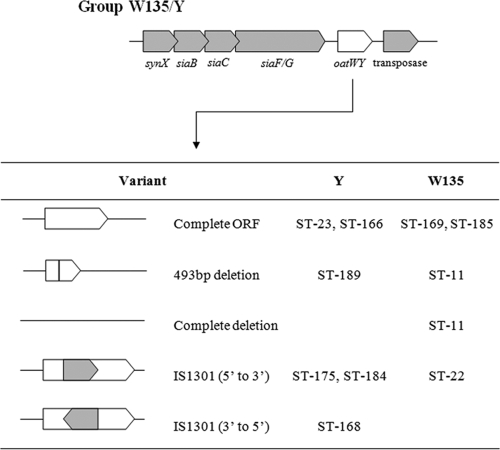
Five variants of the oatWY gene, located directly downstream of the polysialyltransferase gene, have been shown to exist in groups W135 and Y, and each variant is associated with a certain sequence type (7). Sequencing of this gene in the W:2a:P1,7-2,4 isolates revealed the presence of an insertion sequence, IS1301, with the same orientation as the gene. No deletions were present in oatWY. This particular variant is associated with group W135 strains from the ST-22 clonal complex, a nonhyperinvasive lineage and the predominant ST of group W135 carriage isolates in New Zealand. Sequence analysis of oatWY in the W:2a:P1.5,2 ST-11 isolates found all to carry a partial deletion, consistent with previous findings (7). These results suggest that the capsule genes of the new W:2a:P1.7-2,4 strain most likely came from a strain from the carriage-associated ST-22 complex and not from a strain from the disease-associated ST-11 complex.
DISCUSSION
Capsule switching in N. meningitidis is frequently reported in the literature. We report a capsule switch from group C to group W135. Molecular typing methods showed that a W:2a:P1.7-2,4 ST-11 strain was descended from a C:2a:P1.7-2,4 ST-11 strain, a result endorsed by the use of whole-genome RFLP. Sequence analysis indicated that a large tract of DNA was recombined to bring about this capsular change. Our finding endorses that of Swartley et al. (22), who reported that a capsule switch in a group B organism involved the capsule polysaccharide biosynthesis operon and not just the polysialyltransferase gene (siaD). In the group C-to-group W135 capsule switch, we describe that it appears that the entire cps cluster and its flanking region have been exchanged. This was supported by the microarray analysis when two carriage-associated group W135 isolates (both strain type W:nt:P1.18-1,3) were included in the results. Analysis of the collective DNA hybridization profiles using unsupervised gene clustering methods revealed that the capsule genes (the entire cps cluster) of the W:2a:P1.7-2,4 strain clustered with those of the W:nt:P1.18-1,3 strain, while analysis using all genes present on the array shows the W:2a:P1.7-2,4 strain clustering with those of the group C strain.
The recombination of such a large fragment of DNA is not without precedent in N. meningitidis, as the generation of a capsule null mutant was shown to have involved the replacement of the entire cps cluster, approximately 30 kb of DNA, with an allele most likely from Neisseria lactamica (8). While this represents the removal of a large amount of DNA, the capsule switch we describe may represent the largest reported amount of DNA to be recombined into an N. meningitidis genome. Linz et al. (14) studied the recombination of the tbpB gene, which encodes a surface-exposed transferrin binding protein and, like the capsule, is also under strong selection pressures. Numerous recombinants were indentified, and the range in fragment size varied from 1.9 kb to 9.9 kb, with the average size being 5.1 kb (14). While the capsule switch described in our study involves a considerably greater amount of DNA than that observed by Linz et al., two other recombination events were identified in the C:2a:P1.7-2,4 and W:2a:P1.7-2,4 strains with microarray analysis (at least 6.8 kb involving hisS and flanking genes) and sequencing (2 to 3 kb within the LOS biosynthesis operon of the cps cluster) (results not shown). These results concur with the findings of Linz et al. (14).
The capsule switching event that has given rise to an invasive group W135 strain, as demonstrated in this study, is unlikely to be the first instance of such an occurrence. It has been suggested that the hyperinvasive ST-11 W:2a:P1.5,2 phenotype may have occurred through a capsule switch from a C:2a:P1.5,2 strain (17). Nineteen W:2a:P1.5,2 strains from nine countries were examined during the course of our study, and all were indistinguishable based on the typing methods employed. In contrast, the 10 C:2a:P1.5,2 isolates studied carried one of six fetA alleles, including the allele carried by the W:2a:P1.5,2 isolate, suggesting that the W:2a:P1.5,2 strain type is a subgroup of the C:2a:P1.5,2 strain. The banding patterns produced by PFGE also show similarity between the C:2a:P1.5,2 and W:2a:P1.5,2 isolates (data not shown), while no similarities appear to exist between the W:2a:P1.5,2 isolate and other group W135 isolates.
The results presented here indicate that recombination can give rise to different strains that share the same strain type. This was evident with the two C:2a:P1.7-2,4 isolates, NZ96/59 and NZAK04020. Isolate NZ96/59 has the same capsule type, PorA and PorB types, and FetA type as the other C:2a:P1.7-2,4 isolates yet belongs to a completely new sequence type, indicating that it is genetically unrelated to other C:2a:P1.7-2,4 isolates that belong to ST-11. This was confirmed by RFLP analysis in which the NZ96/59 isolates produced a different banding pattern. In comparison, NZAK04020 and the remaining C:2a:P1.7-2,4 isolates belong to the same ET-15 subgroup of ST-11. However, molecular typing demonstrated that the fetA allele of NZAK04020 was the same as that of the C:2a:P1.5-1,10-4 and C:2a:P1.5-1,10-8 isolates, suggesting the possibility of an independent porA switch. RFLP analysis confirmed this, as the banding pattern of NZAK04020 was identical to that of a C:2a:P1.5-1,10-8 isolate. This finding suggests that recombination results in both new and previously defined meningococcal strain types and highlights the importance of using multiple typing methods to define isolates. Moreover, these recombination events support models of meningococcal strain structure which propose that transmission, and thereby virulence, are determined by the combinations of cell surface antigens (4). Isolate NZ96/59 has undergone at least three recombination events with cell surface antigens found at various loci on the genome, giving rise to a phenotype indistinguishable from that of an unrelated disease-causing strain. While random chance cannot be ruled out as the cause of these particular recombinations, a “fitness” bestowed by a combination of surface antigens appears more likely, especially given the apparent lower level of variability among disease-causing strains of meningococci (5). Current genomic studies of “disease” versus “carriage” strains have found little to indicate what may differentiate between the two states other than capsule (19). The authors suggested that the recombination of genes encoding surface-exposed proteins may lead to deletions or translocations that alter expression, thus contributing to these differences. Another possibility may be that the combinations of surface-exposed proteins may predispose a strain to transmission (virulence) or colonization (carriage) and that recombination makes these states interchangeable.
Most capsule switches investigated have involved a group B-to-group C switch or vice versa (2, 12, 13, 18, 20, 22). Differences between the group B and C capsule genes are in the siaD and siaE alleles, respectively, and are shown by the presence of the single variant of oatC in group C, indicating no association between clonal origin and capsule gene sequences. The presence of five variants (Fig. 3) of the oatWY gene in groups W135 and Y suggests that a possible origin of switched capsule genes may be deduced (7). Our studies showed that the W:2a:P1.7-2,4 isolates carry the oatWY variant found in isolates of the ST-22 complex. Such isolates are commonly identified in carriage studies in New Zealand. While not unexpected, our findings provide strong circumstantial proof to established theory that disease-causing strains acquire DNA, and thereby antigenic structure, from carriage isolates (16).
FIG. 3.
Variants of oatWY found in groups W135 and Y (7).
Acknowledgments
We thank Dominique Caugant (Norwegian Public Health Laboratory, Norway) for providing the W:2a:P1.5,2 ST-11 strains and the Microbiological Diagnostic Unit Public Health Laboratory, Department of Microbiology and Immunology, University of Melbourne, Victoria, Australia, for the provision of strains MDU PHL 1 (W:2a:P1.7-2,4) and 991.3179 (C:2a:P1.7-2,4).
This publication made use of the Neisseria MLST website (http://pubmlst.org/neisseria/), developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford (11a). The development of this site has been funded by the Wellcome Trust and the European Union.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Abdillahi, H., and J. Poolman. 1987. Whole cell ELISA for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol. Lett. 48367-371. [PubMed] [Google Scholar]
- 2.Alcala, B., L. Arreaza, C. Salcedo, M. J. Uria, L. De La Fuente, and J. A. Vazquez. 2002. Capsule switching among C:2b:P1.2,5 meningococcal epidemic strains after mass immunization campaign, Spain. Emerg. Infect. Dis. 81512-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bash, M. C., K. B. Lesiak, S. D. Banks, and C. E. Frasch. 1995. Analysis of Neisseria meningitidis class 3 outer membrane protein gene variable regions and type indentification using genetic techniques. Infect. Immun. 631484-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caugant, D. A. 2008. Genetics and evolution of Neisseria meningitidis: importance for the epidemiology of meningococcal disease. Infect. Genet. Evol. 8558-565. [DOI] [PubMed] [Google Scholar]
- 5.Caugant, D. A., K. Bovre, P. Gaustad, K. Bryn, E. Holten, E. A. Hoiby, and L. O. Froholm. 1986. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J. Gen. Microbiol. 132641-652. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, S. C., M. A. Diggle, P. Molling, M. Unemo, and P. Olcen. 2003. Analysis of PorA variable region 3 in meningococci: implications for vaccine policy? Vaccine 212468-2473. [DOI] [PubMed] [Google Scholar]
- 7.Claus, H., R. Borrow, M. Achtman, G. Morelli, C. Kantelberg, E. Longworth, M. Frosch, and U. Vogel. 2004. Genetics of capsule O-acetylation in serogroup C, W-135 and Y meningococci. Mol. Microbiol. 51227-239. [DOI] [PubMed] [Google Scholar]
- 8.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 1481813-1819. [DOI] [PubMed] [Google Scholar]
- 9.Decosas, J., and J. B. Koama. 2002. Chronicle of an outbreak foretold: meningococcal meningitis W135 in Burkina Faso. Lancet Infect. Dis. 2763-765. [DOI] [PubMed] [Google Scholar]
- 10.Devoy, A. F., K. H. Dyet, and D. R. Martin. 2005. Stability of PorA during a meningococcal disease epidemic. J. Clin. Microbiol. 43832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyet, K. H., and D. R. Martin. 2005. Sequence variation in the porB gene from B:P1.4 meningococci causing New Zealand's epidemic. J. Clin. Microbiol. 43838-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kertesz, D. A., M. B. Coulthart, J. A. Ryan, W. M. Johnson, and F. E. Ashton. 1998. Serogroup B, electrophoretic type 15 Neisseria meningitidis in Canada. J. Infect. Dis. 1771754-1757. [DOI] [PubMed] [Google Scholar]
- 13.Kriz, P., D. Giorgini, M. Musilek, M. Larribe, and M. K. Taha. 1999. Microevolution through DNA exchange among strains of Neisseria meningitidis isolated during an outbreak in the Czech Republic. Res. Microbiol. 150273-280. [DOI] [PubMed] [Google Scholar]
- 14.Linz, B., M. Schenker, P. Zhu, and M. Achtman. 2000. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol. Microbiol. 361049-1058. [DOI] [PubMed] [Google Scholar]
- 15.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden, M. C. J., B. Malorny, and M. Achtman. 1996. A global gene pool in the neisseriae. Mol. Microbiol. 211297-1298. [DOI] [PubMed] [Google Scholar]
- 17.Mayer, L. W., M. W. Reeves, N. Al-Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schmink, C. A. Noble, M. L. Tondella, A. M. Whitney, Y. Al-Mazrou, M. Al-Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. E. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J. Infect Dis. 1851596-1605. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Trallero, E., D. Vicente, M. Montes, and R. Cisterna. 2002. Positive effect of meningococcal C vaccination on serogroup replacement in Neisseria meningitidis. Lancet 360953. [DOI] [PubMed] [Google Scholar]
- 19.Schoen, C., J. Blom, H. Claus, A. Schramm-Gluck, P. Brandt, T. Muller, A. Goesmann, B. Joseph, S. Konietzny, O. Kurzai, C. Schmitt, T. Friedrich, B. Linke, U. Vogel, and M. Frosch. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 1053473-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Stabler, R. A., G. L. Marsden, A. A. Witney, Y. Li, S. D. Bentley, C. M. Tang, and J. Hinds. 2005. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology 1512907-2922. [DOI] [PubMed] [Google Scholar]
- 20.Stefanelli, P., C. Fazio, A. Neri, T. Sofia, and P. Mastrantonio. 2003. First report of capsule replacement among electrophoretic type 37 Neisseria meningitidis strains in Italy. J. Clin. Microbiol. 415783-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suker, J., I. M. Feavers, M. Achtman, G. Morelli, J. F. Wang, and M. C. Maiden. 1994. The porA gene in serogroup A meningococci: evolutionary stability and mechanism of genetic variation. Mol. Microbiol. 12253-265. [DOI] [PubMed] [Google Scholar]
- 22.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taha, M. K., D. Giorgini, M. Ducos-Galand, and J. M. Alonso. 2004. Continuing diversification of Neisseria meningitidis W135 as a primary cause of meningococcal disease after emergence of the serogroup in 2000. J. Clin. Microbiol. 424158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 1491849-1858. [DOI] [PubMed] [Google Scholar]
- 25.Tsang, R. S., S. G. Squires, and T. W. Tam. 2003. Characterization of Neisseria meningitidis strains isolated from invasive meningococcal disease cases in Canada in 2001. Can. J. Microbiol. 49633-638. [DOI] [PubMed] [Google Scholar]
- 26.Tsolia, M. N., M. Theodoridou, G. Tzanakaki, V. Vlachou, G. Mostrou, F. Stripeli, P. Kalabalikis, A. Pangalis, D. Kafetzis, J. Kremastinou, and A. Konstantopoulos. 2006. Invasive meningococcal disease in children in Greece: comparison of serogroup A disease with disease caused by other serogroups. Eur. J. Clin. Microbiol. Infect. Dis. 25449-456. [DOI] [PubMed] [Google Scholar]
- 27.Vogel, U., H. Claus, M. Frosch, and D. A. Caugant. 2000. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J. Clin. Microbiol. 38941-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilder-Smith, A., T. M. Barkham, A. Earnest, and N. I. Paton. 2002. Acquisition of W135 meningococcal carriage in Hajj pilgrims and transmission to household contacts: prospective study. BMJ 325365-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yazdankhah, S. P., B. A. Lindstedt, and D. A. Caugant. 2005. Use of variable-number tandem repeats to examine genetic diversity of Neisseria meningitidis. J. Clin. Microbiol. 431699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]