Abstract
Fimbriae, lipopolysaccharide (LPS), and extracellular polymeric substance (EPS) all contribute to biofilm formation by the periodontopathogen Aggregatibacter actinomycetemcomitans. To understand how individual biofilm determinants respond to changing environmental conditions, the transcription of genes responsible for fimbria, LPS, and EPS production, as well as the translation of these components, was determined in rough (Rv) and isogenic smooth (Sv) variants of A. actinomycetemcomitans cultured in half-strength and full-strength culture medium under anaerobic or aerobic conditions, and in iron-supplemented and iron-chelated medium. The transcription of tadV (fimbrial assembly), pgaC (extracellular polysaccharide synthesis), and orf8 or rmlB (lipopolysaccharide synthesis) was measured by real-time PCR. The amounts of fimbriae, LPS, and EPS were also estimated from stained sodium dodecyl sulfate-polyacrylamide gels and verified by Western blotting and enzyme-linked immunoadsorbent assay using specific antibodies. Each gene was significantly upregulated in the Rv compared to in the Sv. The transcription of fimbrial, LPS, and EPS genes in the Rv was increased approximately twofold in cells cultured in full-strength medium under anaerobic conditions compared to that in cells cultured under aerobic conditions. Under anaerobic conditions, the transcription of fimbrial and EPS enzymes was elevated in both Rv and Sv cells cultured in half-strength medium, compared to that in full-strength medium. Iron chelation also increased the transcription and translation of all biofilm determinants compared to their expression with iron supplementation, yet the quantity of biofilm was not significantly changed by any environmental perturbation except iron limitation. Thus, anaerobic conditions, nutrient stress, and iron limitation each upregulate known biofilm determinants of A. actinomycetemcomitans to contribute to biofilm formation.
Aggregatibacter (Actinobacillus) actinomycetemcomitans is a gram-negative, capnophilic bacterium associated with localized aggressive periodontitis and refractory forms of chronic periodontitis (16-18, 51, 58, 76). A. actinomycetemcomitans is also associated with several systemic infections, including endocarditis, cardiovascular disease, and abscesses (8, 39, 47, 51, 62, 68). The adhesion of A. actinomycetemcomitans to oral surfaces is essential for its colonization and persistence within the oral cavity. Adhesion and subsequent biofilm formation are strategies that A. actinomycetemcomitans has evolved to overcome the constant pressure provided by clearance mechanisms in the oral cavity.
Biofilms are adherent bacterial communities that display a distinctive physiological state that protects bacteria from hostile environmental conditions, such as attack by the immune system and antimicrobials (6, 10, 11). Biofilm development is a complex process initiated by cell adhesion to a surface, followed by subsequent cell division and growth and formation of surface microcolonies. Initial adhesion of A. actinomycetemcomitans to an abiotic surface is mediated by type IVb-like, bundle-forming fimbriae encoded by flp-1, part of the flp operon (also known as the tight adherence [tad] locus) (13, 22-24, 28, 29, 36, 43, 49, 53, 56). In addition to fimbriae, nonproteinaceous, nonfimbrial adhesins also contribute to biofilm formation by A. actinomycetemcomitans (28). The cells within the biofilm are encased in a protective extracellular polymeric substance (EPS) matrix composed of dissolved nutrients, secreted enzymes, carbohydrate polymers, and extracellular DNA (28, 32, 38). The primary component of EPS is a high-molecular-weight hexosamine (N-acetyl-d-glucosamine) that is structurally and functionally related to Escherichia coli (poly-N-acetylglucosamine [PGA]) and Staphylococcus epidermidis (polysaccharide intercellular adhesin [PIA]) (38). It has been shown that A. actinomycetemcomitans PGA-like polysaccharide is not essential for the adhesion of cells to a surface but functions primarily in intercellular aggregation (38). A. actinomycetemcomitans strains are classified into six serotypes (a to f) based on the structurally and immunologically distinct O-polysaccharide component of lipopolysaccharide (LPS) (1, 37, 46, 75). A recent study has shown that the serotype a antigen promotes the adhesion of A. actinomycetemcomitans to abiotic surfaces (19).
Primary clinical isolates of A. actinomycetemcomitans have a rough colony phenotype (53, 56). In broth culture, fimbriated, rough variants (Rv) produce adherent microcolonies with a clear supernatant that spontaneously converts to a less adherent, smooth variant (Sv) with a cloudy supernatant during the course of in vitro culture (13, 14, 30). On abiotic surfaces in vitro, Rv produce a biofilm of scattered, dense microcolonies, while Sv produce a thinner, more homogenous biofilm without microcolonies (21, 35). The switch from the rough to smooth colony phenotype is due, in part, to random point mutations in the promoter region located upstream of flp-1, the first gene of the flp operon (66). Restoration of the Rv phenotype is achieved through introduction of the wild-type flp promoter from the Rv into the Sv of A. actinomycetemcomitans (67). In an in vivo study of colonization, the Rv of A. actinomycetemcomitans was able to persist in the oral cavity of rats much longer (15) and caused more bone loss than did Sv (57). However, though the Sv is able to invade oral epithelial cells in vitro better than the Rv can (45), the phenotype of cells invading buccal epithelial cells in situ is presently unknown (5, 54). Thus, the virulence potential of these colony variants in vivo has not yet been fully determined, warranting the study of both variants. Furthermore, the Sv of several strains of A. actinomycetemcomitans are routinely used in vitro to study immunomodulary effects on host cells, toxin expression, LPS, and the expression of other adhesins in part because of the ease of enumeration of the Sv planktonic cells. The Sv also provides an opportunity to study the role of other adhesins and cell components without the presence of fimbriae. Thus, by comparing the expression of fimbriae, EPS, and LPS in the Rv with that in the Sv, potential coordinate expression of these biofilm determinants may be better observed.
Although A. actinomycetemcomitans is usually isolated from the subgingival crevice, it can also be isolated from other sites in the oral cavity. There are a number of microenvironments within the oral cavity that vary in nutrient availability, iron level, and oxygen concentration. These environmental conditions may influence the biofilm forming capability of A. actinomycetemcomitans. Mechanisms to explain how nutrient availability, iron, and oxygen tension influence the expression of biofilm determinants have yet to be elucidated. We postulate that A. actinomycetemcomitans biofilm determinants are differentially expressed in response to changes in nutrient availability, iron, and oxygen tension. Thus, the goal of this study was to compare the transcription and translation of fimbrial, LPS, and EPS genes in the Rv and Sv of A. actinomycetemcomitans strains grown under different growth conditions and to determine these effects on biofilm formation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The A. actinomycetemcomitans strains D7 (obtained from Casey Chen, University of Southern California) and HK1651 (ATCC 700685) were isolated as Rv from the subgingival plaque of patients with aggressive periodontitis. The spontaneous isogenic Sv from each strain was obtained by repeated broth passage from the Rv until the cells produced a turbid suspension showing the absence of cells tightly adherent to the culture vessel wall. Strains were cultured at 37°C in an anaerobic chamber (<0.2% O2, 5% CO2, 10% H2, 85% N2) or in aerobic (microaerophilic, <21% O2) conditions in a candle jar in Bacto tryptic soy broth supplemented with 0.6% (wt/vol) yeast extract (Becton, Dickinson and Co.) and 0.04% NaHCO3 (TSBY; full-strength [1×]), with 1.5% Bacto agar added as needed. TSBY was diluted 1:2 with distilled water to prepare half-strength (0.5×) medium. Iron chelation was achieved through the addition of 2,2′-dipyridyl (Sigma) to TSBY to a final concentration of 300 μM, while iron supplementation was achieved through the addition of FeCl3 to 300 μM in TSBY just prior to use. Bacteria were also cultured in brain heart infusion (BHI) broth (Becton, Dickinson and Co.) with iron chelator or iron supplement added as described above for biofilm experiments. To ensure maintenance of phenotypes, each strain of A. actinomycetemcomitans was cultured weekly from frozen stocks onto TSBY agar prior to use.
RNA isolation and analysis.
Total RNA was prepared by a modification of the sodium dodecyl sulfate (SDS) lysis/CsCl cushion procedure, as previously described (22). RNA was quantified spectrophotometrically at A260 and A280 using a NanoDrop spectrophotometer and the quality evaluated on agarose formaldehyde gels.
Synthesis of cDNA and real-time PCR.
Total RNA (5 μg) was pretreated with Turbo DNase (Ambion) according to the manufacturer's protocol to remove any contaminating DNA. RNA was quantified using the Quanti-iT RNA assay kit (Molecular Probes); fluorescence was measured using an FLX-800 microplate fluorescence reader (Bio-Tek Instruments, Inc.). A 2-μg aliquot of DNase-treated RNA was used as the template to synthesize first-strand cDNA per the manufacturer's protocol (Invitrogen). A negative control lacking reverse transcriptase was included for each RNA template.
Two-step real-time quantitative PCR (RT-qPCR) was performed to quantify gene transcription. PrimerExpress software (version 2.0; Applied Biosystems) was used to design primers and 6-carboxyfluorescein dye-labeled TaqMan minor groove binder probes. The primers and probes used for RT-qPCR (Applied Biosystems) are listed in Table 1. Reactions were carried out according to the manufacturer's instructions. All reactions were run in triplicate and repeated from three independent biological replicates. Controls included (i) amplification with a primer/probe set for 16S rRNA, (ii) amplification with a reaction mixture without a template, and (iii) amplification without reverse transcriptase. ROX was used as an internal (passive) reference dye to normalize the signal of the reporter. Reaction plates were processed on an Applied Biosystems 7500 real-time PCR system, using the default thermal cycling conditions as follows: hot-start AmpliTaq Gold polymerase was activated at 95°C for 10 min and was followed by 40 cycles of denaturation for 15 s at 95°C and annealing and extension for 60 s at 60°C. Amplification data were analyzed using ABI Prism SDS 2.1 software (Applied Biosystems). Relative quantification of gene expression was performed by the threshold cycle (ΔΔCT) method, with 16S rRNA expression serving as an endogenous control to normalize target expression within each sample (70). By comparing the values at the threshold cycle, the expression of a given gene under one growth condition relative to that under another growth condition could be determined. Statistical significance was determined by Student's paired t test.
TABLE 1.
Real-time qPCR primers/probes
| Gene | TaqMan primers/probe | Sequence | Source |
|---|---|---|---|
| tadV | tadV-For | GCGGCCAAGTTTTTTCTTTTTCTT | This study |
| tadV-Rev | GCAATCCGTTTTCTTTAATTGATTTACG | ||
| MBG probe | CCGGATTGGGACTAATT | ||
| pgaC | D7R-For | TGGTTCAAGCCTTAGAGCAAGATC | This study |
| D7R-Rev | CGGTTACGTACACGCGGATTA | ||
| MBG probe-Rev | CCTGTGGTAGCAGCATAT | ||
| orf8 | D7R-For | AACGTATTAGAAGATTGGTGCCTGTT | This study |
| D7R-Rev | CCTCCCATGACCCAATCGAAA | ||
| MBG probe-Rev | ATGGATACAACCAAAACAAG | ||
| rmlB | HK1651-For | AACGTATTAGAAGATTGGTGCCTGTT | This study |
| HK1651-Rev | CCTCCCATGACCCAATCGAAA | ||
| MGB probe-For | AAACCACGCCTTATTC | ||
| 16S rRNA gene | HK1651-For | ACGCGAAGAACCTTACCTACTCT | This study |
| HK1651-Rev | CCTAAGGCACAAACCACATCTCT | ||
| MGB probe-For | CATCCGAAGAAGAACTC |
Preparation of fimbriae.
Whole-cell lysates of A. actinomycetemcomitans were used as the source of fimbriae for analysis by SDS-polyacrylamide gel electrophoresis (PAGE). These lysates were prepared by a modification of the method of Hitchcock and Brown (27). Bacterial cells grown in full- and half-strength TSBY at 37°C for 21 h, either anaerobically or in a candle jar, were harvested by centrifugation, washed once with cold phosphate-buffered saline (PBS; pH 7.2), resuspended in an equal volume of PBS, and standardized to an optical density at 600 nm (OD600) of 0.5 in 1 ml. The bacteria were solubilized by adding 0.5 ml of concentrated digestion buffer (0.1875 M Tris hydrochloride [pH 6.8], 6% SDS, 30% glycerol) followed by heating at 100°C for 5 min. Lysates were analyzed by SDS-PAGE followed by silver staining (44) and Western blotting probed with anti-D7R-flp1 peptide antibody.
Isolation of EPS.
EPS is composed primarily of extracellular polysaccharide. Isolation of extracellular polysaccharide was performed as described previously with minor modifications (38). Bacterial cells grown in full- and half-strength TSBY at 37°C for 21 h, either anaerobically or in a candle jar, were harvested by centrifugation, washed twice with PBS, and standardized to an OD600 of 0.5 in 1 ml. Cells were harvested by centrifugation, resuspended in 0.5 ml of 50 mM sodium acetate buffer (pH 5.8)-100 mM NaCl, and then transferred to a 1.5-ml microcentrifuge tube. An equal volume of 10 mM Tris-HCl (pH 8.0)-5 mM EDTA-0.5% SDS was added, and the tube was vortexed briefly and incubated at 100°C for 5 min. After centrifugation, the pellet containing the crude exopolysaccharide was resuspended in 0.5 ml of 25 mM Tris-HCl (pH 8.0)-5 mM β-mercaptoethanol-0.5% SDS and 0.5 ml of 2× SDS-PAGE sample buffer and incubated at 100°C for 10 min. EPS samples (20 μl) were separated by SDS-PAGE on a 12% gel consisting of an extra-long (10 mm) stacking gel and a 35-mm-long resolving gel. Silver staining, Western blotting, and enzyme-linked immunoadsorbent assay (ELISA) were used to determine the relative EPS expression levels. To optimize the EPS antibody dilution, a dot assay was performed by applying 15 μl of isolated EPS from D7 Rv and Sv with Staphylococcus aureus ATCC 83205 EPS and E. coli LE392 EPS as positive controls to Immobilon-P polyvinylidene difluoride membranes (Millipore) and probed with anti-CifA antibody following the Western blotting protocol.
Preparation of LPS.
LPS was extracted using the hot aqueous phenol method as previously described (37, 69). Bacterial cells grown in full- and half-strength TSBY broth at 37°C for 21 h, either anaerobically or in a candle jar, were harvested by centrifugation, washed twice with PBS, resuspended in an equal volume of PBS, and standardized to an OD600 of 0.5 in 1 ml and processed according to the protocol. LPS samples (25 μl each) were separated by SDS-PAGE on a 14% gel. LPS was detected by silver staining as described by Hitchcock and Brown (27) and Western blotting probed with serotype-specific antibody.
Polyclonal antiserum against D7R-Flp1 peptide.
A synthetic peptide (CFNDLTSTISSASVKK) derived from the last 16 amino acids of the Flp-1 fimbrial protein of strain D7 Rv was conjugated to keyhole limpet hemocyanin as previously described (40, 49) and used to immunize rabbits to obtain polyclonal antisera to the D7R-Flp1 peptide (Sigma-Genosys, TX).
Western blot analysis.
Samples separated by SDS-PAGE were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were blocked overnight with 3% bovine serum albumin in Tris-buffered saline containing 0.05% Tween 20 (TBST), washed three times with TBST, and incubated with primary antibody (strain-specific antibody to A. actinomycetemcomitans fimbriae, as follows: rabbit anti-D7R-Flp1, or rabbit anti-RcpB; goat anti-CifA raised to the deacetylated form of poly-N-acetyl-glucosamine conjugated to S. aureus clumping factor A for EPS of A. actinomycetemcomitans [provided by Gerald B. Pier, Channing Laboratory, Department of Medicine, Brigham and Women's Hospital]; rabbit anti-75 [serotype a-specific antibody] against whole cells of A. actinomycetemcomitans strain SUNYaB 75 [obtained from J. Zambon, University at Buffalo, State University of New York]; or rabbit anti-Y4 [serotype b-specific antibody] against whole cells of A. actinomycetemcomitans strain Y4 [provided by J. Zambon]) appropriately diluted in blocking buffer. After washing with TBST, membranes were incubated with secondary antibody [goat anti-rabbit immunoglobulin G (IgG) alkaline phosphatase-conjugated antibody (Bio-Rad) for fimbriae and LPS, or rabbit anti-goat IgG F(ab′)2 alkaline phosphatase-conjugated antibody (Chemicon) for EPS] diluted 1:1,000 in blocking buffer. Antibodies bound to target antigens were detected and visualized by applying the Sigma Fast BCIP/NBT alkaline phosphatase substrate (Sigma).
ELISA.
ELISA was performed as previously described by Kaplan et al. (37), with several modifications. All steps were carried out at room temperature. The wells of a flat-bottom, uncoated polystyrene Nunc-Immuno microtiter plate were coated overnight with 50 μl of purified fimbriae, EPS, or LPS (in triplicate), washed three times with distilled water, and blocked for 30 min with blocking buffer (PBS containing 0.05% Tween 20, 1 mM EDTA, and 1% bovine serum albumin). The wells were washed two times with distilled water prior to the addition of primary antibody (rabbit anti-D7R-Flp1 for fimbriae, goat anti-CifA goat antibody for EPS, rabbit anti-75 [serotype a-specific] or anti-Y4 [serotype b-specific] antibody for LPS) diluted 1:500 in blocking buffer. After a 2-h incubation, the wells were washed two times with PBS containing 0.05% Tween 20 (PBS-Tween), blocked for an additional 10 min in blocking buffer, and washed again with PBS-Tween. Then secondary antibody [goat anti-rabbit IgG alkaline phosphatase-conjugated antibody (Bio-Rad) for fimbriae and LPS, or rabbit anti-goat IgG F(ab′)2 alkaline phosphatase-conjugated antibody (Chemicon) for EPS] diluted 1:1,000 in blocking buffer was added to each well, and the plate was incubated an additional 2 h. The wells were washed two times with PBS-Tween, blocked for an additional 10 min, and washed again with PBS-Tween. To develop color, substrate solution of p-nitrophenyl phosphate (in 10% diethanolamine-0.05 mM MgCl2 [pH 9.8]) was added to each well, and the plate was incubated for 45 min. The end product was measured at 405 nm in a Beckman model AD 340 microplate reader. Statistical significance was determined by Student's paired t test.
Microtiter plate biofilm assay.
Biofilm growth and quantification was determined using a modification of a standard microtiter plate biofilm assay, as described previously (21). Briefly, frozen stock cultures were plated onto TSBY agar and incubated in a candle jar for 72 h at 37°C. For iron studies, one to five isolated colonies were transferred to BHI broth and grown overnight anaerobically. Cultures were standardized to an OD600 of 0.050 in BHI prior to medium exchange; cells were harvested by centrifugation and resuspended in BHI, BHI with FeCl3, or BHI with 2,2′-dipyridyl and applied to 96-well, flat-bottom, untreated polystyrene microtiter plates (Nalge Nunc International). The plates were then incubated anaerobically for 21 h. For medium and oxygen tension experiments, cultures were standardized to an OD600 of 0.050 in 1× TSBY, and cells were harvested by centrifugation and resuspended in either 1× or 0.5× TSBY prior to application to microtiter plates. The plates were covered with sealing tape and incubated for 21 h, either anaerobically or in a candle jar, at 37°C. Growth was monitored at OD595 using a microplate reader (Beckman Coulter AD340). Culture supernatants were decanted and unbound bacteria removed by washing with PBS [pH 7.2]. Biofilm cells were fixed with methanol and air dried prior to staining with 0.1% crystal violet (Sigma) for 5 min. Dye was decanted and wells were washed with distilled water until negative control wells were clear. After air-drying the plates, ethanol was used to solubilize the crystal violet. Plates were mixed briefly, and the absorbance of the dye was quantified at 595 nm using a microplate reader. Each sample was tested in three biological replicates with six replicates per assay. One-way analysis of variance was used to determine the statistical significance of growth and biofilm data. The Mann-Whitney test was used for analysis of biofilm/growth ratios.
RESULTS
Correlation of biofilm determinant transcription with colony phenotype.
The Rv and Sv of A. actinomycetemcomitans strains D7 and HK1651 cultured in full-strength TSBY under anaerobic conditions at 37°C were analyzed by RT-qPCR to assess the differential expression of biofilm determinant genes for fimbriae, EPS, and LPS. The 14-gene flp operon (flp-1 to tadG) encoding genes involved in the synthesis, assembly, and export of the flp fimbriae of A. actinomycetemcomitans is expressed as a polycistronic message encoding at least the first seven genes (flp-1 to tadZ) (22). The first two genes of the operon are flp-1, encoding the major fimbrial protein subunit Flp-1, and flp-2, encoding a homolog of Flp-1 (16, 35, 36). To examine fimbrial expression, the third gene in the flp operon, tadV, encoding a prepilin peptidase necessary for fimbrial subunit assembly (61, 66), contained the optimal sequence for the design of a TaqMan probe/primer set. The polycistronic transcription of this gene cluster enabled tadV transcription to be used as an unambiguous surrogate for flp-1 transcription. By RT-qPCR, the transcription of tadV from the Rv of strains D7 and HK1651 was greater by ∼1,500- and 500-fold, respectively, compared to that from their isogenic Sv (P < 0.01).
Polysaccharides such as PIA or PGA polymers containing N-acetyl-d-glucosamine (GlcNAc) residues with β-(1,6) linkage (33, 65) are a major part of the bacterial biofilm EPS matrix. PIA is produced by gram-positive bacteria, such as Staphylococcus aureus (31). Similarly, PGA is produced by various gram-negative bacteria, including E. coli, Pseudomonas fluorescens, and Yersinia pestis (31). A cluster of four genes (pgaABCD) involved in the production of exopolysaccharide in A. actinomycetemcomitans is homologous to E. coli pgaABCD (38). The pgaC gene is thought to be a glycosyltransferase gene based on its similarity to icaA of S. epidermidis (38). The expression of pgaC was chosen as a measure of EPS levels in A. actinomycetemcomitans. Transcriptional analysis using RT-qPCR showed that pgaC expression was significantly upregulated (∼13-fold) in the Rv compared to levels in the Sv for both the D7 and HK1651 strains (P < 0.01).
LPS (endotoxin) is composed of a lipid moiety (lipid A) and an O-polysaccharide chain, which serves as the basis for the A. actinomycetemcomitans serotyping system (2, 20, 37, 41, 50, 55, 77). LPS is encoded by different genes, depending on the serotype (37, 59). A 14-gene cluster consisting of the genes orf1 to orf14 encodes the LPS of the serotype a strains, such as D7 (59). Transcriptional analysis of LPS expression by D7 was analyzed by RT-qPCR of the orf8 gene of the LPS cluster, which encodes an enzyme that acetylates lipopolysaccharide (59). Similarly, LPS from the serotype b strain HK1651 is produced by the rmlBADC gene cluster that encodes four enzymes responsible for the four-step biosynthesis of dTDP-l-rhamnose from glucose-1-phosphate (37). To examine the expression of LPS by HK1651, a real-time probe was designed to rmlB that encodes glucose-1-phosphate thymidyl-transferase. Transcription of the orf8 gene in strain D7 was significantly increased ∼15-fold in the Rv compared to that in the Sv (P < 0.01). Likewise, transcription of the rmlB gene in strain HK1651 was significantly upregulated ∼13-fold in the Rv compared to that in the Sv (P < 0.01).
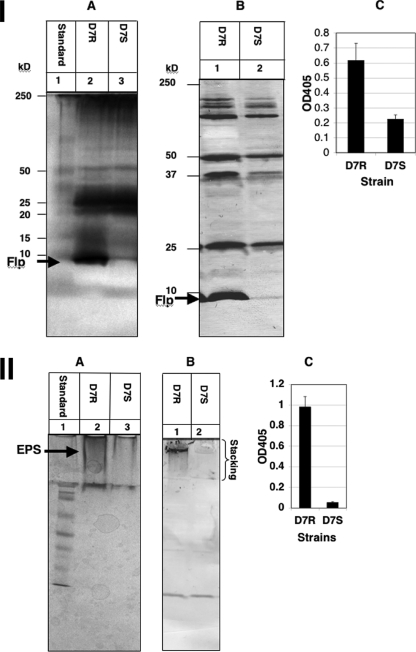
Since the transcriptional expression levels of the biofilm determinant genes were similar for strains D7 and HK1651, comparison of translational products (fimbriae, EPS and LPS) was determined from the Rv and Sv from strain D7 only. Due to the inherent imprecision of the translational assays requiring staining, only relative quantification of expressed molecules could be demonstrated. Fimbrial preparations were standardized for protein content using the bicinchoninic acid assay, while EPS and LPS preparations were prepared from cultures standardized to an OD600 of 0.5 prior to extraction. As shown in Fig. 1, silver-stained SDS-PAGE gels, Western blots, and ELISA results (P < 0.05) showed considerably more fimbriae (panel I) and EPS (panel II) produced by the Rv than by the Sv, confirming the transcriptional analysis data. The lower differential fimbrial expression by the Rv and Sv as determined by ELISA is likely due to the presence of cross-reacting antibodies in the polyclonal antiserum reacting with other proteins in the enriched, but not purified, fimbrial preparation. Based on silver staining alone, there was minimal difference in LPS expression between the Rv and Sv (data not shown). The antibodies used for LPS detection were to whole cells and were cross-reactive between serotypes. Adsorption of sera with whole cells of other serotypes to make serotype-specific sera depleted the LPS reactivity to nondetectable levels, both in Western blot analysis and in ELISA.
FIG. 1.
Phenotype comparison. (I) Fimbrial expression. Standardized fimbrial preparations from the Rv (D7R) and the Sv (D7S) of strain D7 grown anaerobically in 1× TSBY and separated by SDS-PAGE on 14% gels. A, silver stain; B, Western blot probed with anti-D7R-flp1 peptide antibody; and C, ELISA with anti-D7R-flp1 peptide antibody. (II) EPS expression. Standardized EPS preparations from the Rv and Sv of strain D7 grown anaerobically in 1× TSBY and separated by SDS-PAGE on 12.5% gels. A, silver stain; B, Western blot probed with anti-CifA antibody; and C, ELISA with anti-CifA antibody.
Effect of oxygen tension on biofilm determinant expression.
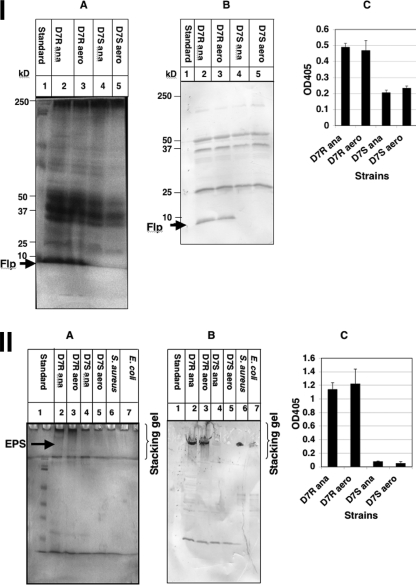
The Rv of strain D7 was grown in full-strength TSBY at 37°C under either aerobic (microaerophilic) or anaerobic conditions. Transcriptional analysis by RT-qPCR was performed using probes specific for fimbriae, EPS, and LPS, as described above. Transcription of tadV from the Rv strain was significantly increased (∼2-fold) in anaerobically grown cells compared to aerobically grown cells (P < 0.05). However, no significant difference was seen in the amount of fimbriae expressed in cell lysates from cells grown anaerobically versus aerobically examined by stained SDS-PAGE gels or in Western blot analysis or ELISA (P > 0.05) using anti-D7R-flp1 peptide antibody (Fig. 2, panel I).
FIG. 2.
Effect of atmosphere conditions on fimbrial and EPS expression. (I) Fimbrial expression. Standardized fimbrial preparations from the Rv and Sv of strain D7 grown in 1× TBSY, either anaerobically (ana) or aerobically (aero), and separated by SDS-PAGE on 14% gels. A, silver stain; B, Western blot probed with anti-D7R-flp1 peptide antibody; and C, ELISA with anti-D7R-flp1 peptide antibody. (II) EPS expression. Standardized EPS preparations from the Rv and Sv of strain D7 grown in 1× TSBY, either anaerobically or aerobically, and separated by SDS-PAGE on 12.5% gels. A, silver stain; B, Western blot probed with anti-CifA antibody; and C, ELISA with anti-CifA antibody.
Although A. actinomycetemcomitans is known to produce EPS, little is known about the environmental conditions that control the expression of these polysaccharides. Transcriptional analysis indicated that EPS enzyme expression was slightly increased (∼2-fold) in cells grown under anaerobic conditions compared to those grown aerobically. Silver-stained SDS-PAGE gels, Western blot analysis, and ELISA (P > 0.05) using goat anti-CifA antibody prepared against poly-N-acetyl-glucosamine of S. aureus showed no significant difference in production of EPS between cells grown anaerobically and those grown aerobically from strain D7 (Fig. 2, panel II).
Expression of LPS-related enzyme transcripts under anaerobic conditions also increased (∼2.8-fold) compared to aerobically grown A. actinomycetemcomitans strain D7, although this difference was not statistically significant. Also, no clear difference in LPS expression by the Rv was evident when cells were grown under anaerobic and aerobic conditions (data not shown).
Comparison of biofilm determinant expression under nutrient stress.
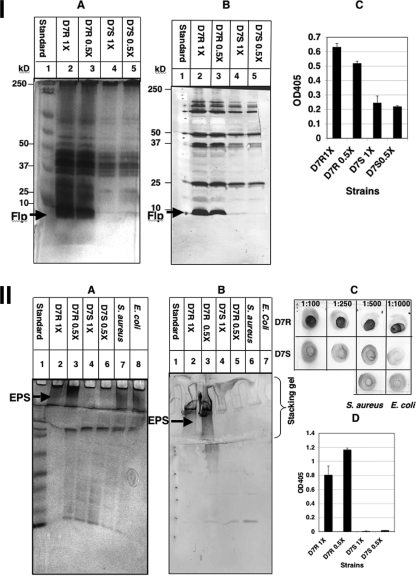
Previously, it was observed that slightly more biofilm was formed by planktonic Sv strains under nutrient stress conditions (in diluted culture medium) than by cells cultured in undiluted growth medium (21). Nutrient limitation appears to provoke biofilm formation by forcing planktonic bacteria into sessile growth (6, 43, 74). In order to analyze the effect of nutrient stress on the production of biofilm determinants by the Rv and Sv of strain D7, the expression of biofilm determinant genes by cells cultured anaerobically at 37°C in 1× and 0.5× TSBY was examined. All three biofilm determinant genes (fimbrial, EPS, and LPS) were upregulated in cells grown in 0.5× medium compared to when observed in cells grown in 1× medium. In 0.5× medium, significant increases in transcription were measured in tadV in the Sv (∼2.9-fold) and in the EPS enzyme in both the Rv (∼3.5-fold) and Sv (∼2.6-fold) strains (P < 0.05), whereas increased fimbrial transcription (1.2-fold) by the Rv and LPS transcription by both the Rv (1.6-fold) and Sv (1.2-fold) were not statistically significant.
Differential expression of fimbriae (Fig. 3, panel I) or LPS (data not shown) between cells cultured anaerobically at 37°C in 1× and 0.5× TSBY was not obvious. Since the Sv normally produces few if any fimbriae, it is not surprising that a 2.9-fold increase in transcription does not result in any noticeable increase in fimbria production. An increase in EPS production (Fig. 3, panel II) by D7 Rv cells, especially, grown in 0.5× medium was clearly evident using silver-stained SDS-PAGE gels and Western blots. The antibody to Staphylococcus PIA (anti-CifA) produced a stronger reaction with EPS isolated from A. actinomycetemcomitans than with the S. aureus strain used as a positive control (Fig. 3, panel IIC). Based on these data, an increase in transcription of at least 3.5-fold was required before an increase of biofilm determinant expression was evident at the translational level using silver-stained gels, Western blots, and ELISA.
FIG. 3.
Effect of medium on fimbriae and EPS expression. (I) Fimbrial expression. Standardized fimbrial preparations from the Rv and Sv of strain D7 grown anaerobically, either in 1× or 0.5× TSBY, and separated by SDS-PAGE on 14% gels. A, silver stain; B, Western blot probed with anti-D7R-flp1 peptide antibody; and C, ELISA with anti-D7R-flp1 peptide antibody. (II) EPS expression. Standardized EPS preparations from the Rv and Sv of strain D7 grown anaerobically, either in 1× or 0.5× TSBY, and separated by SDS-PAGE on 12.5% gels. A, silver stain; B, Western blot probed with anti-CifA antibody; C, dot assay; and D, ELISA with anti-CifA antibody.
Effect of iron on biofilm determinant expression.
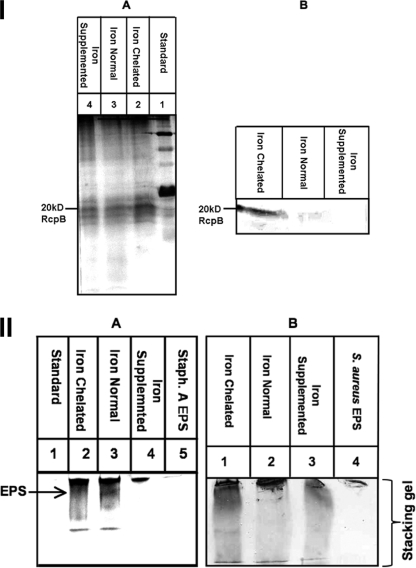
The effect of different iron concentrations on the expression of biofilm determinant genes of the A. actinomycetemcomitans strain HK1651 Rv was determined. Transcription of the fimbrial (tadV), EPS (pgaC), and LPS (rmlB) genes was significantly upregulated 4.4-, 8.9-, and 8.2-fold, respectively (P < 0.05), under iron-chelated conditions compared to under iron-supplemented conditions. These results were confirmed at the translational level, as shown in Fig. 4. The rcpB (rough colony protein B) gene encoding a secretin is part of the flp fimbrial operon polycistronic message. The expression of RcpB, as another flp-1 fimbrial surrogate, was elevated under iron-chelated conditions compared to under normal or iron-supplemented growth conditions (Fig. 4A). Flp-1 was not used as a target in these translational studies due to lack of reproducible expression in this strain. EPS expression was also higher in bacteria grown under iron-chelated condition than in bacteria grown under iron-supplemented conditions (Fig. 4B).
FIG. 4.
Effect of iron concentration on fimbrial and EPS expression. Standardized fimbrial and EPS preparations from the Rv of strain HK1651 grown anaerobically in iron-chelated, iron-normal, and iron-supplemented TSBY. EPS from S. aureus was used as a positive control. (I) RcpB (fimbria) expression. Fimbrial preparation separated by SDS-PAGE on a 14% gel. A, silver stain; B, Western blot probed with anti-RcpB antibody. (II) EPS expression. EPS preparation separated by SDS-PAGE on a 12.5% gel. A, silver stain; B, Western blot with anti-CifA antibody.
Effect of environmental perturbation on in vitro biofilm formation.
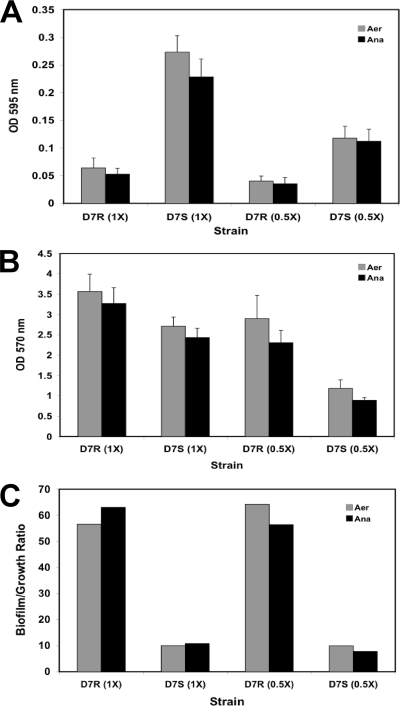
While there was no significant difference in growth of D7 Rv (1× or 0.5×) or Sv (0.5×) whether grown under aerobic (microaerophilic) or anaerobic conditions (Fig. 5A), there was significantly more biofilm (Fig. 5B) for all variants grown under aerobic (microaerophilic) conditions regardless of nutrient concentration (P < 0.05). Yet when biofilm was standardized to growth, no significant difference between microaerophilic and anaerobic conditions was observed for any variant (Fig. 5C). Similarly, when growth was compared in 1× and 0.5× media, all variants grew better (P < 0.05) and produced more biofilm (P < 0.05) in 1× medium regardless of oxygen tension (Fig. 5A and B). When the biofilm was standardized to growth (Fig. 5C), only D7 Sv under anaerobic conditions produced significantly more biofilm in 1× medium than in 0.5× medium (P < 0.001). Thus, although generally more biofilm is formed in richer medium and under aerobic (microaerophilic) conditions, once biofilm was standardized to growth, generally there was no significant difference in the quantities of biofilm formed.
FIG. 5.
Effect of oxygen tension and nutrient stress on biofilm formation. A. actinomycetemcomitans D7 Rv (D7R) and Sv (D7S) strains grown under aerobic (Aer; microaerophilic) or anaerobic (Ana) conditions in 1× and 0.5× BHI media. Growth (A), biofilm (B), and biofilm/growth ratio (C). The biofilm assay was performed in triplicate.
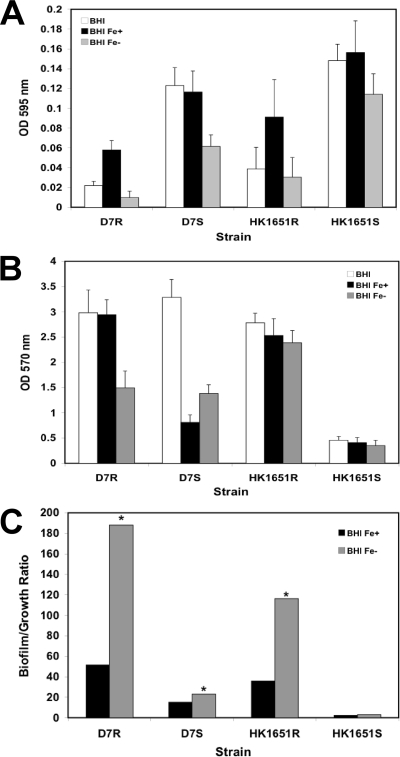
As expected, both Rv and Sv of both strains grew significantly slower (P < 0.05) under iron-chelated conditions than under iron-supplemented conditions (Fig. 6A). However, the quantity of biofilm in iron-chelated and iron-supplemented media varied by variant and strain. D7 Rv produced significantly less and D7 Sv produced significantly more biofilm (P < 0.001) under iron-chelated conditions than they did under iron-supplemented conditions. In contrast, no significant differences were observed for either the Rv or Sv of HK1651 (Fig. 6B). Comparing biofilm formations by phenotype under iron-chelated conditions, D7 Rv and Sv were not significantly different in quantity, whereas HK1651 Rv produced significantly more biofilm than did the Sv (P < 0.001). These results reflect strain differences in broth phenotype; the supernatants of the Sv's of D7 and HK1651 were uniformly turbid; however, D7 produced more slime on the surface of the culture vessel than did HK1651, resulting in a biofilm quantity comparable to that of the Rv. Interestingly, when biofilm was normalized to growth, the Rv of both the D7 and HK1651 strains under iron-chelated conditions produced significantly more biofilm (P < 0.001) than did those under iron-supplemented conditions. Also, significantly more biofilm was produced under iron-chelated conditions by the Sv of D7 (P < 0.005), but not by the Sv of HK1651 (Fig. 6C).
FIG. 6.
Effect of iron on biofilm formation. A. actinomycetemcomitans strains D7 and HK1651 grown in BHI medium, BHI with iron supplement (Fe+), and BHI with iron chelation (Fe−). Growth (A), biofilm (B), and biofilm/growth ratio (C). Biofilm assay was performed in triplicate. *, significance (P < 0.001).
DISCUSSION
In vitro, the Rv strains of A. actinomycetemcomitans grow in broth culture as biofilms with few planktonic cells. In contrast, Sv strains produce a thin, minimally adherent slime on the vessel wall, with most cells growing planktonically. This is the first study to assess the relative contributions of fimbriae, EPS, and LPS from each phenotype to the formation of biofilm under various environmental conditions at the transcriptional and translational levels. The Rv strains expressed significantly higher levels of each of the selected biofilm determinants than did the Sv strains when strains were grown in rich medium (full-strength TSBY) under an anaerobic atmosphere. The most dramatic difference observed was the greater expression of fimbriae in Rv than that in Sv strains. This is consistent with the findings of previous studies employing other methods of analysis (22, 36, 53).
EPS was also significantly increased in Rv, compared to in Sv strains, at the transcriptional and translational levels. These data support previous results showing greater extracellular polysaccharide production by Rv strains than that by Sv strains (21). This phenomenon has been observed for other bacteria, such as Vibrio cholerae O1 strain TSI-4, which produces more EPS material on the surface of the rugose variant than it does on the surface of the Sv (64). Although EPS apparently does not play a major role in surface adhesion by A. actinomycetemcomitans, it does function in intercellular aggregation or microcolony formation, a predominant feature of the Rv strains (38). The higher levels of EPS expressed by the Rv of A. actinomycetemcomitans likely contribute to greater resistance to antibiotics and macrophage killing of Rv strains, compared to Sv strains (21, 63).
LPS is the major integral component of the bacterial outer membrane and consists of three components: O-antigen polysaccharide, core oligosaccharide, and lipid A. Although the lipid A portion of LPS is known to produce many biological effects, recent studies show that the O antigen may play an important role in colonization of host tissues and drug resistance (3, 4, 25, 48, 60). However, it is not clear if LPS plays a role in biofilm formation by A. actinomycetemcomitans. Therefore, we assessed the association of LPS with biofilm formation by evaluating the expression of LPS synthetic enzyme gene transcript levels in the Rv and Sv of two different serotypes, as well as under different environmental conditions. Though the transcription of LPS was significantly increased in the Rv compared to that in the Sv, differences in expression could not be demonstrated using the methods employed. LPS has been recently suggested to be involved in biofilm formation as serotype a strain-specific polysaccharide antigen shown to promote the adhesion of A. actinomycetemcomitans cells to surfaces (19).
It is known that differential expression of components such as fimbriae and other adhesins can be influenced by environmental factors such as pH, NaCl, temperature, and oxygen or iron concentration (12, 21, 24, 26, 52, 56). Here, we characterized the effect of atmosphere and nutrient limitation on the expression of biofilm determinants of A. actinomycetemcomitans. Anaerobiosis has been shown to modulate the levels of fimbriae and other surface proteins of A. actinomycetemcomitans (56). It has been proposed that the exposure of A. actinomycetemcomitans to intermittent aerobic conditions (due to production of oxygen by neutrophils and phagocytes in the oral environment) might affect the expression of colonization determinants and, ultimately, the invasion of host cells (42, 72). Interestingly, the present study found all three selected biofilm determinant genes to be transcribed at a higher level in anaerobically grown cells than in aerobically grown cells. However, the increase in the expression of fimbriae under anaerobic conditions was not evident when we attempted to quantitate fimbrial preparations from broth-grown organisms applied to SDS-PAGE and Western blotting. This finding was contrary to previous direct observations of agar-grown organisms by electron microscopy (56). It may be that organisms grown in broth under nonagitation conditions are able to promote the anaerobiosis of the microaerophilic cultures, obviating any potential difference in fimbrial expression. Other investigators found that anaerobic conditions induced the expression of PIA in S. aureus and S. epidermidis (7). The present study also found that anaerobic conditions appeared to increase the expression of EPS at the transcriptional level, although this difference was not statistically significant. We were not able to observe a clear difference between expression of EPS under anaerobic conditions and that under aerobic conditions.
When biofilm determinant expression levels in A. actinomycetemcomitans in 0.5× and 1× media were compared, the differential expression of EPS was the most significant finding. It has been reported that growth of certain V. cholera strains in nutrient-limited medium resulted in significant EPS expression and a change in colony morphology from normal translucent variant to rugose variant (64). Wrangstadh et al. have also shown that nutrient limitation induced the expression and release of EPS in Pseudomonas sp. strain S9, resulting in distinct effects on the adhesion and aggregation of the bacterial cells (71). No similar change in colony phenotype was observed for A. actinomycetemcomitans. However, consistent with other bacteria, A. actinomycetemcomitans expressed higher levels of EPS in 0.5× medium. These results suggest that nutrient stress signals greater EPS production.
In this study, we demonstrated that iron-restricted conditions upregulated biofilm determinants in A. actinomycetemcomitans. The induction of colonization factors such as fimbriae, EPS, and LPS under iron restriction resembles the environment of an oral cavity prior to infection. Since clinical isolates usually have a rough phenotype, the finding that iron chelation appeared to affect biofilm formation by the Rv more than that by the Sv suggests that iron limitation may be an important factor regulating colonization in vivo. The induction of biofilm formation in low-iron growth conditions has been observed for other bacteria, including S. aureus (34), S. epidermidis (9), and P. aeruginosa PAO1 (73).
Taken together, these data demonstrate that environmental factors influence the expression of certain biofilm determinant genes in A. actinomycetemcomitans. Of the conditions tested here, only iron limitation significantly influenced biofilm quantity when normalized to growth. Given the important medical consequences of biofilm formation, understanding the behavior of periodontopathic bacteria under different environmental conditions may be important for the development of effective therapies that target these biofilm determinants. It is possible that mechanisms that regulate the expression of these biofilm determinant genes occur at the transcriptional or posttranscriptional levels. Studies are under way to uncover the possible regulatory mechanisms that govern biofilm formation in A. actinomycetemcomitans.
Acknowledgments
We thank Paul Bronson for his assistance in protein analysis and Robert Dunford for assistance with statistical analysis.
This research was supported in part by USPHS training grant DE007034 from the National Institute of Dental and Craniofacial Research.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Amano, K., T. Nishihara, N. Shibuya, T. Noguchi, and T. Koga. 1989. Immunochemical and structural characterization of a serotype-specific polysaccharide antigen from Actinobacillus actinomycetemcomitans Y4 (serotype b). Infect. Immun. 572942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asikainen, S., C. Chen, and J. Slots. 1995. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol. Immunol. 1065-68. [DOI] [PubMed] [Google Scholar]
- 3.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1361-92. [DOI] [PubMed] [Google Scholar]
- 4.Camprubí, S., S. Merino, J. F. Guillot, and J. M. Tomas. 1993. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Klebsiella pneumoniae. Microb. Pathog. 14433-440. [DOI] [PubMed] [Google Scholar]
- 5.Colombo, A. V., C. M. da Silva, A. Haffajee, and A. P. Colombo. 2007. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J. Periodontal Res. 42236-243. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49711-745. [DOI] [PubMed] [Google Scholar]
- 7.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 694079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, M., A. D. Badley, F. R. Cockerill, J. M. Steckelberg, and W. R. Wilson. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 4825-33. [DOI] [PubMed] [Google Scholar]
- 9.Deighton, M., and R. Borland. 1993. Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect. Immun. 614473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 582014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 441063-1076. [DOI] [PubMed] [Google Scholar]
- 14.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 1451335-1347. [DOI] [PubMed] [Google Scholar]
- 15.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 461065-1078. [DOI] [PubMed] [Google Scholar]
- 16.Fine, D. H., J. B. Kaplan, S. C. Kachlany, and H. C. Schreiner. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42114-157. [DOI] [PubMed] [Google Scholar]
- 17.Fine, D. H., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrandiz, C. Nasri, M. McKiernan, and J. Gunsolley. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 453859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fives-Taylor, P. M., D. H. Meyer, K. P. Mintz, and C. Brissette. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20136-167. [DOI] [PubMed] [Google Scholar]
- 19.Fujise, O., Y. Wang, W. Chen, and C. Chen. 2008. Adherence of Aggregatibacter actinomycetemcomitans via serotype-specific polysaccharide antigens in lipopolysaccharides. Oral Microbiol. Immunol. 23226-233. [DOI] [PubMed] [Google Scholar]
- 20.Gmür, R., H. McNabb, T. J. van Steenbergen, P. Baehni, A. Mombelli, A. J. van Winkelhoff, and B. Guggenheim. 1993. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol. Immunol. 8116-120. [DOI] [PubMed] [Google Scholar]
- 21.Haase, E. M., T. Bonstein, R. J. Palmer, Jr., and F. A. Scannapieco. 2006. Environmental influences on Actinobacillus actinomycetemcomitans biofilm formation. Arch. Oral Biol. 51299-314. [DOI] [PubMed] [Google Scholar]
- 22.Haase, E. M., J. O. Stream, and F. A. Scannapieco. 2003. Transcriptional analysis of the 5′ terminus of the flp fimbrial gene cluster from Actinobacillus actinomycetemcomitans. Microbiology 149205-215. [DOI] [PubMed] [Google Scholar]
- 23.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 672901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakalehto, E., J. Pesola, L. Heitto, A. Narvanen, and A. Heitto. 2007. Aerobic and anaerobic growth modes and expression of type 1 fimbriae in Salmonella. Pathophysiology 1461-69. [DOI] [PubMed] [Google Scholar]
- 25.Harvill, E. T., A. Preston, P. A. Cotter, A. G. Allen, D. J. Maskell, and J. F. Miller. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 686720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes, E. T., J. C. Wilks, P. Sanfilippo, E. Yohannes, D. P. Tate, B. D. Jones, M. D. Radmacher, S. S. BonDurant, and J. L. Slonczewski. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue, T., R. Shingaki, N. Sogawa, C. A. Sogawa, J. Asaumi, S. Kokeguchi, and K. Fukui. 2003. Biofilm formation by a fimbriae-deficient mutant of Actinobacillus actinomycetemcomitans. Microbiol. Immunol. 47877-881. [DOI] [PubMed] [Google Scholar]
- 29.Inoue, T., I. Tanimoto, H. Ohta, K. Kato, Y. Murayama, and K. Fukui. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42253-258. [DOI] [PubMed] [Google Scholar]
- 30.Inouye, T., H. Ohta, S. Kokeguchi, K. Fukui, and K. Kato. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 5713-17. [DOI] [PubMed] [Google Scholar]
- 31.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izano, E. A., M. A. Amarante, W. B. Kher, and J. B. Kaplan. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izano, E. A., H. Wang, C. Ragunath, N. Ramasubbu, and J. B. Kaplan. 2007. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J. Dent. Res. 86618-622. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, M., A. Cockayne, P. H. Williams, and J. A. Morrissey. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 1878211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9429-437. [DOI] [PubMed] [Google Scholar]
- 36.Kachlany, S. C., P. J. Planet, R. Desalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40542-554. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 695375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 1868213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozarov, E. V., B. R. Dorn, C. E. Shelburne, W. A. Dunn, Jr., and A. Progulske-Fox. 2005. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 25e17-e18. [DOI] [PubMed] [Google Scholar]
- 40.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 1821364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakio, L., S. Paju, G. Alfthan, T. Tiirola, S. Asikainen, and P. J. Pussinen. 2003. Actinobacillus actinomycetemcomitans serotype d-specific antigen contains the O antigen of lipopolysaccharide. Infect. Immun. 715005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Listgarten, M. A. 1972. Normal development, structure, physiology and repair of gingival epithelium. Oral Sci. Rev. 13-67. [PubMed] [Google Scholar]
- 43.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merril, C. R., D. Goldman, and M. L. Van Keruen. 1982. Simplified silver protein detection and image enhancement methods in polyacrylamide gels. Electrophoresis 317-23. [Google Scholar]
- 45.Meyer, D. H., P. K. Sreenivasan, and P. M. Fives-Taylor. 1991. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect. Immun. 592719-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano, K., H. Inaba, R. Nomura, H. Nemoto, K. Tamura, E. Miyamoto, H. Yoshioka, K. Taniguchi, A. Amano, and T. Ooshima. 2007. Detection and serotype distribution of Actinobacillus actinomycetemcomitans in cardiovascular specimens from Japanese patients. Oral Microbiol. Immunol. 22136-139. [DOI] [PubMed] [Google Scholar]
- 47.Nakano, K., H. Nemoto, R. Nomura, H. Inaba, H. Yoshioka, K. Taniguchi, A. Amano, and T. Ooshima. 2009. Detection of oral bacteria in cardiovascular specimens. Oral Microbiol. Immunol. 2464-68. [DOI] [PubMed] [Google Scholar]
- 48.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nika, J. R., J. L. Latimer, C. K. Ward, R. J. Blick, N. J. Wagner, L. D. Cope, G. G. Mahairas, R. S. Munson, Jr., and E. J. Hansen. 2002. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun. 702965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paju, S., P. Carlson, H. Jousimies-Somer, and S. Asikainen. 2000. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotype, genotype, and antimicrobial susceptibility. J. Clin. Microbiol. 3879-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piliero, P., P. E. Harnick, G. S. Rosenbaum, N. C. Klein, and B. A. Cunha. 1989. Infection due to Actinobacillus actinomycetemcomitans. Rev. Infect. Dis. 111033-1034. [DOI] [PubMed] [Google Scholar]
- 52.Roosendaal, B., P. M. van Bergen en Henegouwen, and F. K. de Graaf. 1986. Subcellular localization of K99 fimbrial subunits and effect of temperature on subunit synthesis and assembly. J. Bacteriol. 1651029-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosan, B., J. Slots, R. J. Lamont, M. A. Listgarten, and G. M. Nelson. 1988. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol. Immunol. 358-63. [DOI] [PubMed] [Google Scholar]
- 54.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 692700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saarela, M., S. Asikainen, S. Alaluusua, L. Pyhala, C. H. Lai, and H. Jousimies-Somer. 1992. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol. Immunol. 7277-279. [DOI] [PubMed] [Google Scholar]
- 56.Scannapieco, F. A., S. J. Millar, H. S. Reynolds, J. J. Zambon, and M. J. Levine. 1987. Effect of anaerobiosis on the surface ultrastructure and surface proteins of Actinobacillus actinomycetemcomitans (Haemophilus actinomycetemcomitans). Infect. Immun. 552320-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 1007295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 291013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki, N., Y. Nakano, Y. Yoshida, H. Nakao, Y. Yamashita, and T. Koga. 2000. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta 1517135-138. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, P. W. 1983. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 4746-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomich, M., D. H. Fine, and D. H. Figurski. 2006. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 1886899-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Winkelhoff, A. J., and J. Slots. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontol. 2000 20122-135. [DOI] [PubMed] [Google Scholar]
- 63.Venketaraman, V., A. K. Lin, A. Le, S. C. Kachlany, N. D. Connell, and J. B. Kaplan. 2008. Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microb. Pathog. 45173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 643648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 1862724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Y., and C. Chen. 2005. Mutation analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene 35161-71. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., A. Liu, and C. Chen. 2005. Genetic basis for conversion of rough-to-smooth colony morphology in Actinobacillus actinomycetemcomitans. Infect. Immun. 733749-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westling, K., and M. Vondracek. 2008. Actinobacillus (Aggregatibacter) actinomycetemcomitans (HACEK) identified by PCR/16S rRNA sequence analysis from the heart valve in a patient with blood culture negative endocarditis. Scand. J. Infect. Dis. 40981-983. [DOI] [PubMed] [Google Scholar]
- 69.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure, vol. 5. Academic Press, New York, NY.
- 70.Winer, J., C. K. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 27041-49. [DOI] [PubMed] [Google Scholar]
- 71.Wrangstadh, M., P. L. Conway, and S. Kjelleberg. 1986. The production and release of an extracellular polysaccharide during starvation of a marine Pseudomonas sp. and the effect thereof on adhesion. Arch. Microbiol. 145220-227. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto, M., K. Saeki, and K. Utsumi. 1991. Isolation of human salivary polymorphonuclear leukocytes and their stimulation-coupled responses. Arch. Biochem. Biophys. 28976-82. [DOI] [PubMed] [Google Scholar]
- 73.Yang, Y., P. K. Sreenivasan, R. Subramanyam, and D. Cummins. 2006. Multiparameter assessments to determine the effects of sugars and antimicrobials on a polymicrobial oral biofilm. Appl. Environ. Microbiol. 726734-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 686283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshida, Y., Y. Nakano, Y. Yamashita, and T. Koga. 1998. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect. Immun. 66107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 121-20. [DOI] [PubMed] [Google Scholar]
- 77.Zambon, J. J., J. Slots, and R. J. Genco. 1983. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect. Immun. 4119-27. [DOI] [PMC free article] [PubMed] [Google Scholar]