Abstract
Monoclonal antibody (MAb) 11E10 recognizes the Shiga toxin type 2 (Stx2) A1 subunit. The binding of 11E10 to Stx2 neutralizes both the cytotoxic and lethal activities of Stx2, but the MAb does not bind to or neutralize Stx1 despite the 61% identity and 75% similarity in the amino acids of the A1 fragments. In this study, we sought to identify the segment or segments on Stx2 that constitute the 11E10 epitope and to determine how recognition of that region by 11E10 leads to inactivation of the toxin. Toward those objectives, we generated a set of chimeric Stx1/Stx2 molecules and then evaluated the capacity of 11E10 to recognize those hybrid toxins by Western blot analyses and to neutralize them in Vero cell cytotoxicity assays. We also compared the amino acid sequences and crystal structures of Stx1 and Stx2 for stretches of dissimilarity that might predict a binding epitope on Stx2 for 11E10. Through these assessments, we concluded that the 11E10 epitope is comprised of three noncontiguous regions surrounding the Stx2 active site. To determine how 11E10 neutralizes Stx2, we examined the capacity of 11E10/Stx2 complexes to target ribosomes. We found that the binding of 11E10 to Stx2 prevented the toxin from inhibiting protein synthesis in an in vitro assay but also altered the overall cellular distribution of Stx2 in Vero cells. We propose that the binding of MAb 11E10 to Stx2 neutralizes the effects of the toxin by preventing the toxin from reaching and/or inactivating the ribosomes.
Escherichia coli O157:H7 and other Shiga toxin (Stx)-producing E. coli (STEC) strains cause approximately 110,000 cases of infection and over 90 deaths each year in the United States according to the Centers for Disease Control and Prevention (16). Infections with STEC can lead to diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (HUS). HUS occurs in about 6 to 15% of individuals after infection with E. coli O157:H7 (15)—but less frequently with other STEC strains (5)—and is characterized by hemolytic anemia, thrombotic thrombocytopenia, and renal failure. The development of this sequela is linked to the expression of Stxs by the bacteria (18).
The Stx family comprises two serogroups, Stx/Stx1 and Stx2, and polyclonal antisera raised against either Stx1 or Stx2 do not cross-neutralize the other toxin (29, 30). Stx is produced by Shigella dysenteriae type 1 and differs by only 1 amino acid from the Stx1 made by the prototypic STEC O157:H7 strain, EDL933. A single isolate of STEC can express Stx1 (or one of its variants), Stx2 (or one of its variants), or both toxins. Variants of each toxin type are defined by either a biological or immunological difference from the prototypical toxin (31). Stx1 variants include Stx1c and Stx1d, while the variants of Stx2 are Stx2c, Stx2d, Stx2d-activatable (Stx2dact), Stx2e, and Stx2f (reviewed in reference 18).
Stxs are complex holotoxins with a stoichiometry of five identical binding (B) subunits and a single active (A) domain. These AB5 molecules are potent cytotoxins with an N-glycosidase activity that stops protein synthesis by inactivation of the 60S ribosome (6); this activity eventually leads to eukaryotic cell death. The ∼32-kDa A subunit contains the enzymatic activity of the toxin with the active site glutamic acid residue at position 167. The A subunit is asymmetrically cleaved by trypsin or furin into an enzymatically active ∼28-kDa A1 fragment and an ∼4-kDa A2 peptide. The A2 peptide remains linked to the large enzymatic domain through a disulfide bond and is encircled by the five identical B subunits of ∼7.7 kDa. The B subunits of the Stxs typically bind to the eukaryotic glycolipid receptor globotriaosylceramide (Gb3), also known as CD77. The mature A and B subunits of Stx1 and Stx2 are approximately 68 and 73% similar at the amino acid level. The crystal structures of Stx and Stx2 have been resolved, and the two structures are remarkably similar (7, 8). Nevertheless, there are some features of these three-dimensional models that differ (summarized in reference 8).
Currently, there are no Food and Drug Administration-approved therapies in the United States to treat STEC infections. However, our research group is one of several that investigate passive immunization strategies to neutralize the Stxs associated with STEC infections (3, 4, 10, 13, 19, 20). Our passive immunization strategy is based on murine monoclonal antibodies (MAbs) developed in this laboratory that specifically bind to and neutralize Stx/Stx1 or Stx2 (21, 28). The MAb 11E10 was generated by immunization of BALB/c mice with Stx2 turned into a toxoid (“toxoided”) by treatment with formaldehyde (21). MAb 11E10 specifically recognizes the A1 fragment of Stx2 and neutralizes Stx2 for Vero cells and mice but does not bind to or neutralize Stx/Stx1 (21). The murine MAb 11E10 was modified to contain a human constant region to reduce the potential for an antibody recipient to generate an antimouse antibody response (4). This human-mouse chimeric antibody, called cαStx2, successfully underwent phase I clinical testing (3). In this report, we define the epitope on the A subunit of Stx2 recognized by the murine MAb 11E10 (and, therefore, also by cαStx2) and present evidence that the MAb blocks the enzymatic action of the toxin in vitro and also alters toxin trafficking in Vero cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, purified Stx1 and Stx2, and MAbs 11E10 and 13C4.
Bacteria were grown in Luria-Bertani (LB) broth or on LB agar (Becton Dickinson and Company, Sparks, MD) supplemented with 100 μg/ml of ampicillin as needed for selection of recombinant plasmids. Bacterial strains used in this study are listed in Table 1. Stx1 and Stx2 were purified by affinity chromatography as described previously (17), and the MAbs 11E10, 11F11 (specific for Stx2) (21), and 13C4 (specific for Stx1) (28) were produced in this laboratory and deposited with BEI Resources (Manassas, VA).
TABLE 1.
Bacterial strains, cloning vectors, and plasmids used in this study
| Strain, vector, or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Gibco BRL |
| XL10 Gold | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr]a | Stratagene |
| Bl21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| EH250 | E. coli Ount:H12 isolate; Stx2d producer | 22 |
| Cloning vectors | ||
| pBluescript II KS(−) | E. coli cloning vector (Ampr) | Stratagene |
| pTrcHis2 C | E. coli expression vector (Ampr) | Invitrogen |
| Recombinant plasmids | ||
| pCKS120 | pBR328 toxin clone of stx2c | 12 |
| pJES101 | pBluescript II KS(−) toxin clone of stx2e | 24 |
| pSQ543 | pBluescript II SK(−) toxin clone of stx2dact | 12 |
| pMJS1 | pBluescript II KS(−) toxin clone of stx1 | 27 |
| pMJS2 | pBluescript II KS(−) toxin clone of stx2 | 27 |
| pMJS3 | pBluescript II KS(−) toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 29-297) | This study |
| pMJS9 | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene with an optimized Shine-Dalgarno sequence (StxA2 amino acids 1-158) | This study |
| pMJS13 | pBluescript II KS (−) toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 29-128) | This study |
| pMJS15 | pBluescript II KS (−) toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 29-76) | This study |
| pMJS16 | pBluescript II KS (−) toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-76) | This study |
| pMJS28 | pBluescript II KS (−) toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-49) | This study |
| pMJS49 | pTrcHis2 C toxin clone of stx1 | This study |
| pMJS49A | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-49) | This study |
| pMJS49AB | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-49 and 96-100) | This study |
| pMJS49AC | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-49 and 244-259) | This study |
| pMJS49BC | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 96-100 and 244-259) | This study |
| pMJS49ABC | pTrcHis2 C toxin clone, chimeric stxA1-stxA2 gene (StxA2 amino acids 42-49, 96-100, and 244-259) | This study |
| pMJS50 | pTrcHis2 C toxin clone of stx2 | 23 |
| pMJS52 | pTrcHis2 C toxin clone of stx2c | This study |
| pMJS59 | pTrcHis2 C toxin clone of stx2d | This study |
| pMJS49ABC* | pMJS49ABC with Y77S mutation | This study |
Hte, high transformation efficiency; Amy, amylase.
Construction of chimeric toxin plasmids.
Six chimeric toxin genes that contained portions of both stxA1 and stxA2 were generated by PCR with the splicing by overlap extension protocol (9), and the PCR products were ligated into pBluescript II KS(−) (Stratagene, La Jolla, CA). A list of all plasmids used in this study is given in Table 1. The chimeric toxin genes contained the native promoters and Shine-Dalgarno sequences, and the levels of toxin expression from five of the clones were sufficient under those conditions. To increase the level of expression of the A subunit in one clone, an optimized Shine-Dalgarno sequence was added upstream of stxA2, and the toxin gene was inserted into the pTrcHis2 C expression vector (Invitrogen, Carlsbad, CA). All primers used in this study are listed in Table 2. The DNA sequence of each construct created for this study was confirmed prior to use.
TABLE 2.
Synthetic oligonucleotide primers used in this study
| Primer | Sequence (5′→3′)a | Purpose/region of homology |
|---|---|---|
| MJS1 | GATCGGATCCCCCTGTAACGAAGTTTGCGTAACAGC | stx1 upstream primer, used to generate pMJS9, pMJS13, pMJS15, pMJS16, and pMJS28 |
| MJS2 | GATCGAATTCTCGCTTACGATCATCAAAGAGATCATACC | stx1 downstream primer, used to generate pMJS10, pMJS11, pMJS13, pMJS15, pMJS16, and pMJS28 |
| MJS5 | GATCGGATCCAGCAAGGGCCACCATATCACATACCGCC | stx2 upstream primer, used to generate pMJS10 |
| MJS6 | CAGGGGAATTCACCATGCGAAATTTTTTTAACAAATGC | stx2 downstream primer, used to generate pMJS9 |
| 2A29F | GAACATATATCTCAGGGGACCAC | Used with 1A28R to generate pMJS9, pMJS13, and pMJS15 |
| 1A28R | GTGGTCCCCTGAGATATATGTTCTAATGGAGTACCTATTGCAGAGCG | Used with 2A29F to generate pMJS9, pMJS13, and pMJS15 |
| 1A159F | TTACGGTTTGTTACTGTGACAGCTGAAGC | Used with 2A158R to generate pMJS10 and pMJS11 |
| 2A158R | GCTTCAGCTGTCACAGTAACAAACCGTAAAACTGCTCTGGATGCATCTCTGGT | Used with 1A159F to generate pMJS10 and pMJS11 |
| 1A129F | CAGATAAATCGCCATTCGTTGA | Used with 2A128R to generate pMJS13 |
| 2A128R | TCAACGAATGGCGATTTATCTGCATTCCGGAACGTTCCAGCGC | Used with 1A129F to generate pMJS13 |
| 2A42F | GGTACGTCTTTACTGATGATTAACCACACCCCACCGGGCAGTTATTTTGC | Used with 1A41R to generate pMJS16 |
| 1A41R | GCAAAATAACTGCCCGGTGGGGTGTGGTTAATCATCAGTAAAGACGTACC | Used with 2A42F to generate pMJS16 |
| 1A77F | TATGTGACAGGATTTGTTAACAGGAC | Used with 2A76R to generate pMJS15 |
| 2A76R | GTCCTGTTAACAAATCCTGTCACATATAAATTATTTTGCTCAATAATCAGACGAAGATGG | Used with 1A77F to generate pMJS15 |
| 1A51 | AGGAGGACAGCTATGAAAATAATTATTTTTAGAGTGCTA | stxA1 upstream primer 1 with optimized Shine-Dalgarno sequence, used to generate pMJS49 |
| 1A52 | GATCGGATCCTAAGGAGGACAGCTATGAAAATAATT | stxA1 upstream primer 2 with optimized Shine-Dalgarno sequence, used to generate pMJS49 |
| 1BC1 | GGTGGTGGTGACGAAAAATAACTTCGCTGAATCC | stxB1 His-tagged downstream primer 1, used to generate pMJS49 |
| 1BC2 | CAGTGGTGGTGGTGGTGGTGACGAAAAATAAC | stxB1 His-tagged downstream primer 2, used to generate pMJS49 |
| BC3 | GATCGAATTCTCAGTGGTGGTGGTGGTGGTG | stxB1 His-tagged downstream primer 3, used to generate pMJS49 and pMJS52 |
| MSAF | AACCACACCCCACCGGGCAGTTATTTTGCAGTTGATGTCAGAGGG | Used with MSAR to generate pMJS28, pMJS49A, pMJS49AB, pMJS49AC, and pMJS49ABC |
| MSAR | ATAACTGCCCGGTGGGGTGTGGTTAATCATCAGTAAAGACGTACC | Used with MSAF to generate pMJS28, pMJS49A, PMJS49AB, pMJS49AC, and pMJS49ABC |
| 96100F | ACACATATATCAGTGCCAGGTACAACAGCGGTTACATTGTCTGG | Used with 96100R to generate pMJS49AB, pMJS49BC, and pMJS49ABC |
| 96100R | ACCTGGCACTGATATATGTGTAAAATCAGCAAAGCGATAAAAAACA | Used with 96100F to generate pMJS49AB, pMJS49BC, and pMJS49ABC |
| JCT1F | GTGAATGAAGAGAGTCAACCAGAATGTCCGGCAGATGGAAGAGTCCG | C region primer 1, used with JCT1R to generate pMJS49AC, pMJS49BC, and pMJS49ABC |
| JCT1R | TTCTGGTTGACTCTCTTCATTCAC | C region primer 1, used with JCT1F to generate pMJS49AC, pMJS49BC, and pMJS49ABC |
| JCT2F | GGCATTAATACTGAATTGTCATCATCAGGGGGCGCGTTCTGTTCGC | C region primer 2, used with JCT2R to generate pMJS49AC, pMJS49BC, and pMJS49ABC |
| JCT2R | ATGATGACAATTCAGTATTAATGCC | C region primer 2, used with JCT2F to generate pMJS49AC, pMJS49BC, and pMJS49ABC |
| 2A51 | AGGAGGACAGCTATGAAGTGTATATTATTTAAATGGGT | stxA2 upstream primer 1 with optimized Shine-Dalgarno sequence, used to generate pMJS11 and pMJS52 |
| 2A52 | GATCGGATCCTAAGGAGGACAGCTATGAAGTGTA | stxA2 upstream primer 2 with optimized Shine-Dalgarno sequence, used to generate pMJS11 and pMJS52 |
| C12B | GGTGGTGGTGGTCATTATTAAACTGCACTTC | stxB2 His-tagged downstream primer 1, used to generate pMJS52 |
| C22B | CAGTGGTGGTGGTGGTGGTGGTCATTATTAAA | stxB2 His-tagged downstream primer 2, used to generate pMJS52 |
| 2dF | GATCGGATCCCTGGTATCGTATTACTTCAGCC | Used with 2dR to generate pMJS59 |
| 2dR | GATCGAATTCCTGCACACTACGAAACCAGC | Used with 2dF to generate pMJS59 |
| 1Y77SF | TCAGTGACAGGATTTGTTAACAGGAC | Used with 1Y77SR to generate pMJS49ABC* |
| 1Y77SR | GTCCTGTTAACAAATCCTGTCACTGATAAATTATTTCGTTCAACAATAAGCCG | Used with 1Y77SF to generate pMJS49ABC* |
Restriction enzyme sites are underlined. Mutagenic codon sites are in boldface.
Five additional His-tagged chimeric toxins were generated from an stx1 clone that contained six histidine codons immediately downstream of the B gene. The toxins produced by these chimeras contain one, two, or three regions from the Stx2 A subunit (hereafter referred to as regions A, B, and C) that comprise the putative MAb 11E10 epitope in place of the comparable sequence in Stx1 (see Fig. 2A). Regions A, B, and C refer to amino acids 42 to 49, 96 to 100, and 244 to 259, respectively, of the Stx2 A subunit. The five chimeric toxins made were named Stx1 +A, Stx1 +AB, Stx1 +AC, Stx1 +BC, and Stx1 +ABC.
FIG. 2.
Amino acid alignment of StxA1 and StxA2 in three regions of dissimilarity, and ribbon and space-filling representations of the Stx2 structure with the three regions of the putative 11E10 epitope highlighted. (A) Amino acid sequence alignment of Stx1 and Stx2 in the three regions of dissimilarity. The green and red amino acids depict conserved and nonconserved amino acids, respectively; the dots represent identical residues. The three regions of the putative MAb 11E10 epitope are as follows: region A (StxA2 residues 42 to 49), region B (StxA2 residues 96 to 100), and region C (StxA2 residues 244 to 259). The numbering of the amino acids shown in the alignments is in respect to the StxA1 mature protein. StxA1 has an additional amino acid at position 185 compared to Stx2; this insertion of an extra amino acid in StxA1 causes region C of the epitope in Stx2A to be 1 number different from the corresponding region of Stx1. (B) Ribbon diagram of the Stx2 crystal structure that shows the Stx2 A1 and B subunits in light gray, except for three proposed regions of the MAb 11E10 epitope. Regions A, B, and C are colored green, dark blue, and light blue, respectively. Parts of region C cannot be seen in the ribbon diagram because several of the residues within region C were not resolved in the crystal structure. The A2 peptide is depicted in black, and the active site glutamic acid residue at position 167 is in red. The two cysteine residues are illustrated in pink. (C) A space-filling representation of the Stx2 crystal structure, with the same colors as in panel B, except that the cysteine residues are not colored.
Generation and purification of partially toxoided Stx1 +ABC.
The cytotoxicity of the Stx1 +ABC toxin for Vero cells was reduced 10,000-fold by changing the tyrosine residue at position 77 of the A subunit to a serine residue by the splicing by overlap extension protocol. This 4-log reduction in cytotoxicity after the Y77S mutation was introduced is similar to that previously reported for the Y77S mutation in Stx1 (2).
The Stx1 +ABC toxoid was purified with a nickel affinity column as previously described (26). The concentration of the toxoid was determined by bicinchoninic acid assay (Pierce, Rockford, IL). A silver stain of a sodium dodecyl sulfate (SDS)-polyacrylamide gel revealed that the A and B subunits of the chimeric toxoid were the two major bands present, although other minor bands were observed (data not shown).
Construction of Stx2c and Stx2d variant clones.
A clone that expressed His-tagged Stx2c was created by PCR as previously described for Stx2 (23). The stx2d clone was generated by PCR from E. coli EH250 with primers 2DF and 2DR (22). The PCR product was ligated into the expression vector pTrcHis2 C. That stx2c and stx2d were amplified correctly was confirmed by sequence analyses.
Western blot analyses.
Purified Stx1, Stx2, or sonic lysates of bacteria that expressed chimeric Stx1/Stx2 toxins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then examined by Western blotting as previously described (26). The concentrations of the A subunits in sonic lysates that contained Stx1, Stx2, or the chimeric toxins were estimated as follows. First, the specific dilutions of rabbit anti-Stx1 and anti-Stx2 rabbit polyclonal antibodies (PAbs) that detected the purified A subunits from Stx1 or Stx2, respectively, to relatively equivalent levels were determined through the use of NIH Image J software (http://rsb.info.nih.gov/nih-image). Second, the chimera-containing sonic lysates were separated by SDS-PAGE; the resulting gels were then transferred to nitrocellulose, and those blots were then probed with a mixture of rabbit anti-Stx1 and anti-Stx2 rabbit PAbs diluted as determined above. Third, the bands that corresponded to the chimeric A subunits in each lane were quantified with the NIH Image J program to determine the toxin concentration in each lysate sample. Fourth, two additional polyacrylamide gels were loaded with purified Stx1, Stx2, or samples of the chimera-containing lysates normalized to contain equivalent concentrations (as determined from step 3). The toxin preparations were then subjected to SDS-PAGE followed by Western blot analysis with a mixture of rabbit anti-Stx1 and/or rabbit anti-Stx2 PAbs or MAb 11E10. The secondary antibodies used in these Western blots were goat anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (Bio-Rad, Hercules, CA) at a dilution of 1:15,000 or goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad, Hercules, CA) at a dilution of 1:3,000, as appropriate. The bound secondary antibodies were detected by chemiluminescence with the ECL-Plus Western blotting detection kit (Amersham Bioscience, Little Chalfont, Buckinghamshire, England).
Western blots were also performed on sonic lysates of clones that expressed Stx2, Stx2c, Stx2d, Stx2dact, or Stx2e. First, the concentration of the A subunits from these toxin samples was determined as described above, except that only rabbit anti-Stx2 PAb was used as a probe. Then two additional polyacrylamide gels were loaded with equivalent quantities of the normalized samples, and Western blots were conducted with either rabbit anti-Stx2 PAb or MAb 11E10 as the primary antibody. The secondary antibodies and method of detection were the same as described above.
In vitro neutralization assays on Vero cells.
In vitro neutralization assays of sonic lysates from bacteria that contained Stx1, Stx2, the chimeric Stx1/Stx2 toxins, Stx2c, Stx2d, Stx2dact, or Stx2e were carried out with 11E10 on Vero cells (ATCC, Manassas, VA) as described previously (14, 26). MAb 11E10 neutralized Stx2 such that only 35% of the wild-type toxic activity remained. To make it easier to compare the percent neutralization levels of the toxin derivatives by MAb 11E10, the data were then normalized such that the amount of neutralization of Stx2 by 11E10 was set to 100% and the neutralization levels of the other toxins were calculated relative to that of Stx2.
Immunization and challenge of mice.
Preimmune sera were collected from CD-1 male mice that weighed 14 to 16 g (Charles River Laboratories, Boston, MA). These serum samples were used in enzyme-linked immunosorbent assays to determine if the mice had preexisting titers of antibodies to Stx1 or Stx2. None of the mice showed an immune response to either toxin at the start of the study. The mice were then divided into two groups: a sham-inoculated group (hereafter called the negative control group) and a group that was immunized with the chimeric toxoid. Mice in the negative control group were immunized intraperitoneally (i.p.) with a mixture of phosphate-buffered saline (PBS) and TiterMax, a water-in-oil adjuvant (TiterMax, USA, Inc., Norcross, GA). The second group of mice was immunized i.p. with 1 μg of toxoid mixed with TiterMax. The mice were boosted at 3-week intervals for a total of four boosts. Two weeks after the last boost, five negative control mice and five toxoid-immunized mice were challenged i.p. with 10 times the 50% lethal dose (LD50) of Stx1 (1,250 ng), and 29 negative control mice and 34 toxoid-immunized mice were challenged with 5 LD50s of Stx2 (5 ng).
In vitro protein synthesis inhibition assays.
Rabbit reticulocyte lysate, firefly luciferase mRNA, and luciferin substrate were purchased from Promega Corporation (Madison, WI). Stx2 (4 ng/μl) was combined with an equal volume of antibody (at 4 or 40 ng/μl), and 1 μl of the toxin-antibody mixture was combined with 9 μl reticulocyte lysate. The mixture was incubated at 30°C to allow toxin to inactivate ribosomes in the lysate. After 1 h, an aliquot of luciferase mRNA and amino acids that had been heated to 70°C for 2 min was added, and the solution was incubated for an additional 90 min to allow the in vitro protein synthesis to proceed. All assays were done in triplicate. Luciferase activity was measured by adding 1 μl of the lysate mixture to 20 μl of luciferin substrate in clear 96-well plates (Fisher Scientific, Pittsburgh, PA). Bioluminescent light emission was detected with a Kodak Image Station 440CF in a 10-min exposure. Luminescent signal was analyzed by summation of total signal intensity within a circular area that corresponded to a single well.
Localization of 11E10 in intoxicated cells.
Vero cells were seeded in eight-well tissue culture slides (Thermo Fisher Scientific, Rochester, NY) at a concentration of 1 × 105 cells/ml and allowed to adhere for 24 h at 37°C in an atmosphere of 5% CO2. Stx2 (0.2 ml of 10 ng/ml) was mixed with 10 ng of purified MAb 11E10 or with PBS as a negative control. The antibody-toxin or PBS-toxin solutions were incubated with Vero cells for 6 h, and then the cells were fixed with buffered formalin (Formalde-Fresh; Fisher Scientific, Pittsburgh, PA) and permeabilized with 0.001% Triton X-100 (Pierce, Rockford, IL) in PBS. All immunostaining procedures were done in PBS with 3% bovine serum albumin (Sigma, St. Louis MO). The presence of MAb 11E10 within the cells was detected with Alexa Fluor 488-labeled donkey anti-mouse IgG (Invitrogen, Carlsbad, CA). Total Stx2 within the intoxicated cells was labeled with rabbit anti-Stx2 PAb, and Alexa Fluor 488-conjugated donkey anti-rabbit IgG was used as the secondary antibody (Invitrogen, Carlsbad, CA). Stx2 and endosome double labeling was accomplished with anti-Stx2 MAb 11F11 (BEI Resources, Manassas, VA) and anti-EEA1 (C-15) goat PAb (Santa Cruz Biotechnology, Santa Cruz, CA), respectively, and Alexa Fluor-labeled secondary antibodies. After incubation with the appropriate primary and secondary antibodies, the cells were fixed with formalin for 20 min at 37°C, and the slides were mounted with SlowFade medium (Invitrogen, Carlsbad, CA). Images at ×40 magnification of the bound fluorophore-labeled secondary antibodies were obtained via an Olympus microscope with a reflected-light fluorescence attachment and a Spot CCD digital camera (Diagnostic Instrument Products, Sterling Heights, MI). Fluorescence images were processed and overlaid with Adobe Photoshop (Adobe Systems, San Jose, CA).
RESULTS
Interaction of initial chimeric toxins with MAb 11E10.
To determine the portion of Stx2 that interacts with MAb 11E10, we constructed an initial set of six chimeric toxin operons that contained different regions of the stxA2 gene inserted in place of the corresponding region of stxA1 (Fig. 1A). Western blots of purified Stx1, Stx2, or lysates from E. coli DH5α that express one of the six different chimeric Stx1/Stx2 toxins were probed with MAb 11E10. The antibody reacted strongly with Stx2 and the chimeric toxins that contained the amino acids from the following regions of the Stx2 A subunit: 29 to 297, 1 to 158, and 29 to 128 (Fig. 1B). The chimeric toxin with the minimal portion of Stx2 that was still recognized by 11E10, albeit weakly, contained just 8 amino acids from StxA2, region 42 to 49.
FIG. 1.
Illustration of Stx1 and Stx2 and the initial chimeric toxins that contain hybrid Stx1/Stx2 A subunits and recognition or neutralization of those toxins by MAb 11E10. (A) Stx1 is presented in black, and Stx2 is depicted in white. The names of the chimeric toxins are shown to the left of the respective chimeric proteins. (B) Western blot analyses of Stx1, Stx2 and the initial chimeric toxins probed with rabbit anti-Stx1 and anti-Stx2 PAb (top panel) or MAb 11E10 (bottom panel). Lanes 1 and 2 contain 25 ng of purified Stx1 or Stx2, respectively. Lanes 3 to 8 contain the following chimeric toxins: lane 3, Stx1(2A29-297); lane 4, Stx1(2A1-158); lane 5, Stx1(2A29-128); lane 6, Stx1(2A29-76); lane 7, Stx1(2A42-76); lane 8, Stx1(2A42-49). (The three apparent bands for the A subunit most likely represent, from the top band down, the full-length A subunit with the signal sequence intact [we purify toxin from whole-cell lysates], the mature A subunit without the signal sequence, and the A1 subunit.) 1°Ab, primary antibody. (C) Neutralization of the initial chimeric toxins with MAb 11E10. The neutralization data were normalized such that the level of Stx2 was set to 100% (actual neutralization, 65%) and the neutralization levels for the rest of the toxins are given as a percentage of the normalized Stx2 neutralization. The error bars represent the standard error of the normalized values.
Next, the capacity of MAb 11E10 to neutralize the toxicity of bacterial lysates that contained Stx1, Stx2, or one of the six initial chimeric toxins for Vero cells was examined. As expected, MAb 11E10 neutralized Stx2 but did not neutralize Stx1 (Fig. 1C). However, the hybrid toxins with region 29 to 297 or 1 to 158 from StxA2 were about 85% neutralized by 11E10 compared to Stx2, a result that led to the deduction that components of the 11E10 epitope lie between residues 29 and 158 of Stx2. In contrast, the chimeric toxin with amino acids 29 to 128 from Stx2 was recognized strongly in the immunoblot (Fig. 1B, lane 5) but was only neutralized to about 32% of the level of Stx2 (Fig. 1C, bar 5). Together these findings suggest that the 11E10 neutralizing epitope encompasses a larger number of amino acids than are present in the Stx1(2A29-128) chimera. The other three chimeric toxins that were weakly detected by MAb 11E10 in the Western blot analysis were not appreciably neutralized by 11E10 (less than 15%) compared to the normalized level of Stx2 neutralization.
Analyses of differences between the Stx1 and Stx2 A subunit amino acid sequences and crystal structures.
The Western blot and neutralization analyses of the first set of chimeric toxins indicated that the 11E10 epitope required at least amino acids 42 to 49 of the Stx2 A subunit for toxin detection but also revealed that additional amino acids were needed for full recognition and toxin neutralization. Therefore, the amino acid sequences of the mature A subunits from Stx1 and Stx2 were aligned to identify additional unique stretches of amino acids that might be involved in recognition and neutralization of Stx2 by 11E10. Next, the crystal structures of Stx (7) and Stx2 (8) (Protein Data Bank accession no. 1RQ4, and 1R4P, respectively) were compared using the Deep View/Swiss-PDB viewer to assess the location of regions of differences between the toxins in the three-dimensional structures and the proximity of such regions to each other. A comparison of the two crystal structures showed that the region we had identified as minimally necessary for 11E10 binding, the 8 amino acids that span residues 42 to 49 in the Stx2 A subunit, was within one of three regions of amino acid dissimilarity we noted between Stx1 and Stx2. We named this segment of the StxA2 molecule region A (Fig. 2A). When the 8 amino acids from region A were viewed in the context of the Stx2 crystal structure, they appeared to form a major bend in the toxin structure (shown in green in Fig. 2B and C) and, in addition, were found on the outside face of Stx2, near the active site cleft around amino acid 167.
A second dissimilar area between the A subunits of Stx1 and Stx2 was identified when the amino acid sequences and the crystal structures of these two toxins were compared, a segment we called region B (Fig. 2A). Region B spans five residues in the A subunit of Stx2 (96THISV100), and 4 out of the 5 amino acids in this region differ between Stx1 and Stx2 (Fig. 2A). Although region B is approximately 50 amino acids away from region A, this portion of amino acids extends toward region A in the Stx2 crystal structure (region B is shown in dark blue in Fig. 2B). The close proximity of region A to region B in a three-dimensional structure is even more apparent in a space-filling model (Fig. 2C).
The third dissimilar area between the A subunits of Stx1 and Stx2, which we named region C, overlaps the furin cleavage site around residue 246 of Stx2 (Fig. 2A). Region C was identified not only because of amino acid sequence differences between Stx1 and Stx2 in that location, but also because comparison of the crystal structures of Stx and Stx2 indicated that region C was in spatial proximity to regions A and B. From our analyses of the Stx and Stx2 crystal structures, we concluded that regions A, B, and C cluster on the same face of Stx2, relatively near the catalytic active site (best seen in Fig. 2C). Indeed, we found that the side chains of amino acids 46, 97, and 259 lie less than 15 Å apart from each other, well within a reasonable distance for antibody-antigen interaction (1).
Interaction of the second-generation chimeric toxins with MAb 11E10.
To determine whether regions A, B, and C were part of the 11E10 epitope, we produced a second set of chimeric toxins that contained various combinations of regions A, B, or C from Stx2 in place of the corresponding regions on Stx1 (Fig. 3A). Next, Western blots of Stx2 or the chimeric toxins were probed with 11E10 (Fig. 3B). MAb 11E10 detected all of the toxins that contained region A (Stx2, Stx1 +A, Stx1 +AB, Stx1 +AC, and Stx1 +ABC) (Fig. 3B). The toxins missing region A were not detected by MAb 11E10 (Stx1 and Stx1 +BC), a finding that confirms that region A is an essential component of the 11E10 epitope. However, the two chimeric toxins that incorporated regions A and B (Stx1 +AB or Stx1 +ABC) appeared to be more strongly detected by MAb 11E10 than chimeric toxins that included region A alone or A combined with region C (Fig. 3B). Collectively, these results indicate that both regions A and B are important for full 11E10 recognition of the toxin.
FIG. 3.
Illustration of Stx1 and Stx2 and second-generation chimeric toxins that contain chimeric A subunits and recognition or neutralization of those toxins by MAb 11E10. (A) Stx1 is presented in black, while Stx2 is depicted in white. The names of the chimeric toxins are shown to the left of the respective chimeric proteins, and the regions of Stx2 are listed beneath the chimeric A subunits. Regions A, B, and C refer to amino acids 42 to 49, 96 to 100, and 244 to 259, respectively, of StxA2. (B) Western blot analyses of Stx2 and the five second-generation chimeric toxins probed with rabbit anti-Stx1 (top panel) or MAb 11E10 (bottom panel). Lane 1 contains 25 ng of purified Stx2. Lanes 2 to 6 contain the following chimeric toxins: lane 2, Stx1 +A; lane 3, Stx1 +AB; lane 4, Stx1 +AC; lane 5, Stx1 +BC; and lane 6, Stx1 +ABC. 1°Ab, primary antibody. (C) Neutralization of the second-generation hybrid toxins with MAb 11E10. The level of Stx2 neutralization was normalized to 100% as in Fig. 2. The error bars represent the standard error of the normalized values.
We next assayed sonic lysates of each of the five second-generation chimeric toxins (Fig. 3A) for in vitro neutralization by MAb 11E10. The antibody neutralized the chimeric toxin that contained regions A, B, and C (Stx1 +ABC) to almost 70% of the level of Stx2 neutralization. In contrast, the chimeric toxins that contained only regions A and B (Stx1 +AB) or A and C (Stx1 +AC) were neutralized to about half the neutralization level of the Stx1 +ABC chimera (Fig. 3C). No appreciable neutralization by 11E10 was observed against the Stx1 +A or Stx1 +BC chimeric toxins (approximately 6.9 and 4.3% respectively). Since more extensive (>50%) neutralization of the chimeric toxins required regions A, B, and C from Stx2, we concluded that all three regions (A, B, and C) are necessary for neutralization by 11E10.
Western blot and in vitro neutralization assay results with Stx2 and Stx2 variants and MAb 11E10.
To determine which of the Stx2 variants could be recognized and/or neutralized by 11E10, Stx2 or the Stx2 variants (Stx2c, Stx2d, Stx2dact, and Stx2e) were analyzed by Western blotting and by neutralization of activity on Vero cells (Fig. 4). Stx2 and all of the Stx2 variants were recognized by 11E10, although Stx2e was detected to a much lesser extent (Fig. 4A). This weak detection of Stx2e by 11E10 in the Western blot format is consistent with our previous report that 11E10 was unable to detect Stx2e-producing strains by colony blotting (21). Stx2e has two conservative amino acid differences in region B as compared to Stx2 (AHISL rather than THISV). There are also several amino acid sequence differences immediately adjacent to region A (not shown). We speculate that these differences may be responsible for the reduced recognition of Stx2e by 11E10 on the Western blot.
FIG. 4.
Western blot analyses and neutralization assays with Stx2 and Stx2 variants with MAb 11E10. (A) Lane 1 contains 25 ng of purified Stx2. Lanes 2 to 5 contain the following toxins: lane 2, Stx2c; lane 3, Stx2d; lane 4, Stx2dact; lane 5, Stx2e. The Western blots were probed with either rabbit anti-Stx2 PAb (top panel) or MAb 11E10 (bottom panel). 1°Ab, primary antibody. (B) Neutralization by 11E10 of the Stx2 variants. The level of Stx2 neutralization was normalized to 100% as in Fig. 2. The error bars represent the standard error of the normalized values.
When the neutralization capacity of MAb 11E10 for the Stx2 variant toxins was evaluated, we found that 11E10 neutralized all of the Stx2 variant toxins to ≥60% of the level of neutralization of Stx2 (Fig. 4B). We were surprised at the level of neutralization observed by 11E10 of Stx2e because of the limited recognition of Stx2e by 11E10 in the Western blot format (Fig. 4B). However, the neutralization of Stx2e by 11E10 in this study agrees with our previous result that showed that 11E10 partially neutralizes Stx2e (21).
Immune and protective response of the Stx1 +ABC toxoid in mice.
We next sought to ascertain whether a toxoided derivative of the Stx1 +ABC hybrid molecule could elicit a serum-neutralizing or protective response to Stx2 in mice. Groups of mice were immunized with the chimeric toxoid or PBS as a control. Sera from five toxoid-immunized mice and five PBS-immunized mice were then evaluated for an anti-Stx1 neutralizing response. None of the sera from the PBS-immunized mice contained Stx1-neutralizing activity. As expected from previous studies, all five toxoid-immunized mice had neutralizing antibodies directed against Stx1 (27). The mean anti-Stx1 neutralization titer for the serum from these five mice was 4.0 ± 0.9 logs above background. Eleven of the sera from the remaining 34 toxoid-immunized mice had some neutralizing response to Stx2, while none of the sera from the 29 PBS-immunized mice exhibited any anti-Stx2 response (data not shown).
Two weeks after the final boost, five negative-control mice and five toxoid-immunized mice were challenged i.p. with 10 LD50s of Stx1. All of the negative-control mice died, while the toxoid-immunized mice survived the lethal challenge (Table 3), as predicted from the results of a previous study (27). In addition, the survival of the toxoid-immunized mice that were challenged with Stx1 directly correlated with the in vitro neutralizing titers from those mice.
TABLE 3.
Protection of immunized mice against a lethal challenge with Stx1 or Stx2
| Mouse group | Immunization | Stx challenge (10 LD50s)a | No. of surviving mice/total (% survival) |
|---|---|---|---|
| A | PBS | Stx1 | 0/5 |
| B | Stx1 +ABC toxoid | Stx1 | 5/5 |
| C | PBS | Stx2 | 6/29 (20.7) |
| D | Stx1 +ABC toxoid | Stx2 | 12/34 (35.3)b |
The LD50s were previously determined to be 125 and 1 ng per mouse for Stx1 and Stx2, respectively. The average weight of the mice when they were challenged was 47.1 g.
Fisher's exact test was used to compare the proportions that survived in groups C and D, and the P value was 0.2667.
Because low Stx2 neutralizing antibody titers were observed in the toxoid-immunized group, we chose to challenge the rest of the mice with only 5 LD50s of Stx2. Six out of 29 negative-control mice (20.7%) survived the challenge with Stx2, while 12 out of 34 toxoid-immunized mice (35.3%) survived (Table 3), a finding that, while not statistically significant, suggests that the chimeric toxoid may have provided some protection from Stx2.
In vitro protein synthesis inhibition assay.
Our finding that the 11E10 epitope appeared to consist of surface loops around the Stx2 active site cleft led us to hypothesize that 11E10 might neutralize Stx2 by blocking the capacity of the toxin to inhibit protein synthesis. Therefore, we assessed whether MAb 11E10 could neutralize the ribosome-inactivating effects of Stx2 in a rabbit reticulocyte protein synthesis assay to which luciferase mRNA was added. A concentration of toxin was chosen that decreased the signal from the luciferase reporter protein by approximately 60% as compared to the signal measured when no toxin was added (Fig. 5). Addition of 11E10 to the assay allowed protein synthesis to occur in the rabbit reticulocyte lysate even when Stx2 was present, whereas the isotype-matched irrelevant antibody did not (Fig. 5).
FIG. 5.
Protein synthesis inhibition measured by translation of luciferase mRNA in rabbit reticulocyte lysate. A 0.2-ng aliquot of purified Stx2 was mixed with 0, 0.2, or 2 ng 11E10 and added to reticulocyte lysates. Protein synthesis inhibition was indicated by a reduction of translation of luciferase mRNA and was measured by bioluminescence after addition of the toxin-treated lysate to luciferin substrate. A 2-ng sample of the isotype-matched irrelevant antibody 13C4 was mixed with 2 ng Stx2 as a negative control. Error bars represent the 95% confidence interval calculated from the standard error of the mean ratio. Probability values derived from a two-tailed Student's t test indicate a significant difference in bioluminescence signal between samples with and without antibody (P < 0.005).
MAb 11E10 alters the overall distribution of Stx2 in Vero cells.
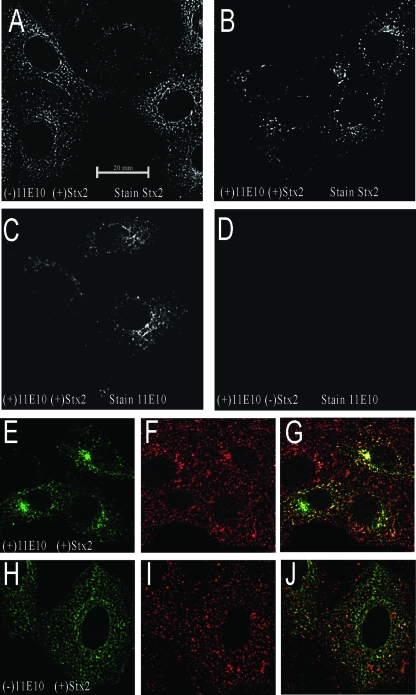
Although we found that MAb 11E10 prevented the inhibition of protein synthesis by Stx2 in the in vitro protein synthesis assay, we further hypothesized that 11E10 may prevent Stx2 from reaching ribosomes in the cytoplasm of intoxicated Vero cells. We therefore sought to determine if MAb 11E10 alters Stx2 localization in target cells. (We previously found that 11E10-bound Stx2 could bind to Vero cells and that 11E10 could attach to Stx2 bound to Vero cells [data not shown].) Stx2 was mixed with 11E10 or PBS, and the antibody-toxin or PBS-toxin mixture was incubated with Vero cells. The distribution of Stx2 in the target cells was then visualized with rabbit anti-Stx2 PAb and a fluorophore-labeled anti-rabbit IgG secondary antibody (Fig. 6). Stx2 appeared to be distributed throughout the cytoplasm in the absence of 11E10 (Fig. 6A) but seemed to remain concentrated in largely perinuclear bodies in the presence of 11E10 (Fig. 6B). When the cells incubated with the toxin-11E10 mixture were stained with anti-mouse IgG, 11E10 was observed in the same perinuclear punctate structures as Stx2 (Fig. 6C). MAb 11E10 was unable to enter cells in the absence of toxin (Fig. 6D). The localization of 11E10-bound Stx2 within punctate bodies around the nucleus suggested that the antibody/toxin complex entered the cell but did not traffic into the cytoplasm. We therefore asked if Stx2 or 11E10-bound Stx2 was localized in early endosomes by immunostaining the intoxicated cells with the early endosome marker MAb EEA-1. We found that much of the Stx2 in cells intoxicated with 11E10-bound Stx2 colocalized with the early endosome marker (Fig. 6E to G), as shown by a yellow-orange color when the staining patterns were overlapped. In contrast, when Vero cells were incubated with Stx2 alone, the toxin was found throughout the cytoplasm and only a small amount colocalized with the early endosome marker (Fig. 6H to J).
FIG. 6.
MAb 11E10 alters the overall cellular distribution of Stx2 in Vero cells. Stx2 was mixed with PBS (A and H to J) or 11E10 (B, C, and E to G) and then added to Vero cells for 6 h. As a control, 11E10 was added to Vero cells in the absence of Stx2 (D). The toxin was detected with PAb against Stx2 followed by secondary antibody conjugated with Alexa Fluor 488 (A and B), while 11E10 was detected with anti-mouse IgG conjugated with Alexa Fluor 488 (C and D). Stx2 colocalization with the early endosome marker EEA1 was assessed by double labeling of intoxicated cells. Stx2 distribution in the presence (E) and absence (H) of antibody 11E10 was visualized with anti-Stx2 MAb 11F11 and green fluorescent secondary antibody. The distribution of endosome marker EEA1 was visualized with goat anti-EEA1 and red fluorescent secondary antibodies (F and I). These staining patterns were superimposed (G and J), and colocalization of toxin with endosomes was indicated by a yellow-orange coloration.
DISCUSSION
Our results demonstrate that the MAb 11E10 epitope is conformational and comprises three nonlinear regions in the Stx2 A subunit that appear close to the active site of the toxin in the crystal structure (Fig. 2C). Our strategy to identify the 11E10 epitope involved the generation of chimeric Stx1/Stx2 toxins and was based on the assumption that placing Stx2 sequences onto the Stx1 backbone would maintain the three-dimensional tertiary structure of the antibody epitope and allow recognition by MAb 11E10. We found that the minimal region of StxA2 that allowed recognition by 11E10 in Western analysis consisted of only 8 Stx2 amino acids (42NHTPPGSY49). However, the chimeric Stx1/Stx2 with just those 8 amino acids from Stx2 (region A) was not neutralized by MAb 11E10. Because 11E10 neutralizes Stx2, we considered that the 8 amino acids we had identified consisted of a critical region of the 11E10 epitope but did not comprise the complete neutralizing epitope. We further analyzed differences in both the amino acid sequences and crystal structures of Stx1 and Stx2 to try to identify additional regions on Stx2 that might be involved in 11E10 recognition and neutralization. Through these comparisons, we identified two more segments of Stx2 that could potentially contribute to the 11E10 epitope. Indeed, we found that all three regions were required for the most complete recognition and neutralization by 11E10 when those regions were used to replace the corresponding segments on Stx1.
Our conclusion that the complete 11E10 neutralizing epitope comprises three noncontinuous regions on Stx2 is perhaps surprising because the MAb recognizes Stx2 under the putatively denaturing conditions of a Western blot. Several explanations for this latter observation are conceivable. These possibilities include that the Western reactivity is primarily due to the interaction of 11E10 with region A (42NHTPPGSY49) or that partial refolding of the A subunit occurs during the Western blot process, as we observed for MAb 13C4 in another study (26).
The sequences of the three surface loops that form the MAb 11E10 neutralizing epitope on Stx2 are conserved among the Stx2 variants. There are a few amino acids that differ within those regions in Stx2d and Stx2e, two toxins that are rarely found in human isolates (18). However, MAb 11E10 did detect and partially neutralize the cytotoxic activity of Stx2 and all of the Stx2 variants analyzed in this report (Stx2c, Stx2d, Stx2dact, and Stx2e). The finding that Stx2e is neutralized by 11E10 on Vero cells but recognized poorly in the Western format as compared to Stx2 may indicate that the sequences in region B that are different between Stx2 and Stx2e are more important for recognition on Western blotting than for neutralization. However, the fact that 11E10 has the capacity to neutralize all of these variant toxins suggests that it may be a good candidate for treating disease mediated by Stx2 and Stx2-related toxins in humans. Indeed, we have found that 11E10 is protective in a toxemia (Stx2) model of disease (25) and an orally fed mouse model of disease with a strain that produces Stx2dact (4). We are currently involved in an ongoing laboratory evaluation of the humanized version of 11E10, cαStx2, on which phase I safety testing has been completed (3).
We attempted to protect mice from Stx2 challenge by immunization with the toxoided chimeric Stx1 molecule that contained just the 29 amino acids from Stx2 that comprise the 11E10 epitope. We found that although the immunized mice raised a protective response to Stx1, only a few of the mice generated Stx2-neutralizing antibodies, and these were of low titer. The response to Stx2 may have been improved with additional boosts of the chimeric toxoid. However, it is more likely that protecting the mice from Stx2 with the 11E10 epitope in the context of the Stx1 toxoid was an unrealistic expectation given the small percentage of that hybrid molecule that actually represented Stx2.
We found that 11E10 blocked the enzymatic activity of Stx2 in vitro, a fact that we predicted based on the close proximity of the 11E10 epitope to the toxin active site. We further observed that 11E10 altered the overall distribution of the toxin inside the cell, a finding that is similar to the data on Stx2 neutralization by a different StxA2 MAb, 5C12, as reported by Krautz-Peterson et al. (11). These investigators concluded that when MAb 5C12 binds StxA2 it alters the intracellular trafficking pattern of the toxin (11). Our data indicate that once the 11E10/Stx2 complex binds to and enters the host cell, the antibody may prevent toxin trafficking to the target ribosomes in the cytosol. However, since we demonstrated that 11E10 prevented the enzymatic function of the toxin in vitro, we predict that should the A subunit of Stx2 complexed with 11E10 reach its enzymatic target in the cytosol, the toxin would be unable to kill the cell.
Acknowledgments
We acknowledge Eddy Twiddy for the purification of the Stx1 and Stx2 used in these studies and Stephen Darnell for reference database management.
This research was supported by National Institutes of Health/National Institute for Allergy and Infectious Diseases grant AI20148-26 and Uniformed Services University of the Health Sciences grants R073KD and RO73NQ.
The views and assertions made concerning the results of this article are the authors' and should not be construed as the views of the Department of Defense.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Davies, D. R., E. A. Padlan, and S. Sheriff. 1990. Antibody-antigen complexes. Annu. Rev. Biochem. 59439-473. [DOI] [PubMed] [Google Scholar]
- 2.Deresiewicz, R. L., S. B. Calderwood, J. D. Robertus, and R. J. Collier. 1992. Mutations affecting the activity of the Shiga-like toxin I A-chain. Biochemistry 313272-3280. [DOI] [PubMed] [Google Scholar]
- 3.Dowling, T. C., P. A. Chavaillaz, D. G. Young, A. Melton-Celsa, A. O'Brien, C. Thuning-Roberson, R. Edelman, and C. O. Tacket. 2005. Phase 1 safety and pharmacokinetic study of chimeric murine-human monoclonal antibody cαStx2 administered intravenously to healthy adult volunteers. Antimicrob. Agents Chemother. 491808-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards, A. C., A. R. Melton-Celsa, K. Arbuthnott, J. R. Stinson, C. K. Schmitt, H. C. Wong, and A. D. O'Brien. 1998. Vero cell neutralization and mouse protective efficacy of humanized monoclonal antibodies against Escherichia coli toxins Stx1 and Stx2, p. 388-392. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, DC.
- 5.Elliott, E. J., and R. M. Robins-Browne. 2005. Hemolytic uremic syndrome. Curr. Probl. Pediatr. Adolesc. Health Care 35310-330. [DOI] [PubMed] [Google Scholar]
- 6.Endo, Y., K. Tsurugi, T. Yutsudo, Y. Takeda, T. Ogasawara, and K. Igarashi. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 17145-50. [DOI] [PubMed] [Google Scholar]
- 7.Fraser, M. E., M. M. Chernaia, Y. V. Kozlov, and M. N. James. 1994. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 Å resolution. Nat. Struct. Biol. 159-64. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, M. E., M. Fujinaga, M. M. Cherney, A. R. Melton-Celsa, E. M. Twiddy, A. D. O'Brien, and M. N. James. 2004. Structure of Shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J. Biol. Chem. 27927511-27517. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi, R. 1989. Using PCR to engineer DNA, p. 61-70. In H. A. Erlich (ed.), PCR technology. Stockton Press, New York, NY.
- 10.Kimura, T., M. S. Co, M. Vasquez, S. Wei, H. Xu, S. Tani, Y. Sakai, T. Kawamura, Y. Matsumoto, H. Nakao, and T. Takeda. 2002. Development of humanized monoclonal antibody TMA-15 which neutralizes Shiga toxin 2. Hybrid. Hybrid. 21161-168. [DOI] [PubMed] [Google Scholar]
- 11.Krautz-Peterson, G., S. Chapman-Bonofiglio, K. Boisvert, H. Feng, I. M. Herman, S. Tzipori, and A. S. Sheoran. 2008. Intracellular neutralization of Shiga toxin 2 by an A subunit-specific human monoclonal antibody. Infect. Immun. 761931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindgren, S. W., J. E. Samuel, C. K. Schmitt, and A. D. O'Brien. 1994. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect. Immun. 62623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, Y., X. Mao, J. Li, H. Li, Y. Feng, H. Chen, P. Luo, J. Gu, S. Yu, H. Zeng, G. Guo, K. Yang, and Q. Zou. 2008. Engineering an anti-Stx2 antibody to control severe infections of EHEC O157:H7. Immunol. Lett. 121110-115. [DOI] [PubMed] [Google Scholar]
- 14.Marques, L. R., M. A. Moore, J. G. Wells, I. K. Wachsmuth, and A. D. O'Brien. 1986. Production of Shiga-like toxin by Escherichia coli. J. Infect. Dis. 154338-341. [DOI] [PubMed] [Google Scholar]
- 15.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 3521207-1212. [DOI] [PubMed] [Google Scholar]
- 16.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melton-Celsa, A. R., and A. D. O'Brien. 2000. Shiga toxins of Shigella dysenteriae and Escherichia coli, p. 385-406. In K. Aktories and I. Just (ed.), Handbook of experimental pharmacology, vol. 145. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 18.Melton-Celsa, A. R., M. J. Smith, and A. D. O'Brien. March 2005, posting date. Chapter 8.7.8, Shiga toxins: potent poisons, pathogenicity determinants, and pharmacological agents. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 19.Mukherjee, J., K. Chios, D. Fishwild, D. Hudson, S. O'Donnell, S. M. Rich, A. Donohue-Rolfe, and S. Tzipori. 2002. Human Stx2-specific monoclonal antibodies prevent systemic complications of Escherichia coli O157:H7 infection. Infect. Immun. 70612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee, J., K. Chios, D. Fishwild, D. Hudson, S. O'Donnell, S. M. Rich, A. Donohue-Rolfe, and S. Tzipori. 2002. Production and characterization of protective human antibodies against Shiga toxin 1. Infect. Immun. 705896-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perera, L. P., L. R. M. Marques, and A. D. O'Brien. 1988. Isolation and characterization of monoclonal antibodies to Shiga-like toxin II of enterohemorrhagic Escherichia coli and use of the monoclonal antibodies in a colony enzyme-linked immunosorbent assay. J. Clin. Microbiol. 262127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 363317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, C. M., J. F. Sinclair, M. J. Smith, and A. D. O'Brien. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. USA 1039667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuel, J. E., L. P. Perera, S. Ward, A. D. O'Brien, V. Ginsburg, and H. C. Krivan. 1990. Comparison of the glycolipid receptor specificities of Shiga-like toxin type II and Shiga-like toxin type II variants. Infect. Immun. 58611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauter, K. A., A. R. Melton-Celsa, K. Larkin, M. L. Troxell, A. D. O'Brien, and B. E. Magun. 2008. Mouse model of hemolytic-uremic syndrome caused by endotoxin-free Shiga toxin 2 (Stx2) and protection from lethal outcome by anti-Stx2 antibody. Infect. Immun. 764469-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, M. J., H. M. Carvalho, A. R. Melton-Celsa, and A. D. O'Brien. 2006. The 13C4 monoclonal antibody that neutralizes Shiga toxin (Stx) type 1 recognizes three regions on the Stx1 B subunit and prevents Stx1 from binding to its eukaryotic receptor globotriaosylceramide. Infect. Immun. 746992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, M. J., L. D. Teel, H. M. Carvalho, A. R. Melton-Celsa, and A. D. O'Brien. 2006. Development of a hybrid Shiga holotoxoid vaccine to elicit heterologous protection against Shiga toxins types 1 and 2. Vaccine 244122-4129. [DOI] [PubMed] [Google Scholar]
- 28.Strockbine, N. A., L. R. Marques, R. K. Holmes, and A. D. O'Brien. 1985. Characterization of monoclonal antibodies against Shiga-like toxin from Escherichia coli. Infect. Immun. 50695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strockbine, N. A., L. R. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen, S. X., L. D. Teel, N. A. Judge, and A. D. O'Brien. 2006. Genetic toxoids of Shiga toxin types 1 and 2 protect mice against homologous but not heterologous toxin challenge. Vaccine 241142-1148. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. 1991. Subcommittee meeting held at the conference Shiga-like toxin producing Escherichia coli with a special emphasis on zoonotic aspects, Giessen, Germany. World Health Organization, Geneva, Switzerland.