Abstract
Shiga toxin 1 (Stx1) is located in the periplasmic fraction, while Stx2 is found in the extracellular fraction, suggesting that enterohemorrhagic Escherichia coli (EHEC) contains a specific Stx2 release system. Both stx1 and stx2 are found within the late operons of Stx-encoding phages. Stx2 production is greatly induced by mitomycin C, suggesting that stx2 can transcribe from the late phage promoter of the Stx2-encoding phage. However, the Stx1 promoter adjacent to stx1 is a dominant regulatory element in Stx1 production. With the deletion of phage lysis genes of the Stx2-encoding phage, Stx2 remains in the bacterial cells. Further, we demonstrate that the Stx2-encoding phage, but not the Stx1-encoding phage, is spontaneously induced at extremely low rates. These results indicate that spontaneously specific Stx2-encoding phage induction is involved in specific Stx2 release from bacterial cells. Furthermore, to examine whether another system for specific Stx2 release is present in EHEC, we analyze the stx-replaced mutants. As expected, Stx2 derived from the Stx1 promoter is located in both the extracellular and cell-associated fractions, while mutant Stx2 (B subunit, S31N) derived from the Stx1 promoter is found only in the cell-associated fraction. These results indicate that EHEC has another Stx2 release system that strictly recognizes the serine 31 residue of the B subunit. Overall, we present evidence that specific Stx2 release from bacterial cells is involved in both the Stx2-encoding phage induction system and another Stx2 release system.
Enterohemorrhagic Escherichia coli (EHEC) consists of multiple serotypes, among which O157:H7 is the one most commonly linked to epidemic and sporadic disease in humans throughout Japan, North America, and parts of Europe. EHEC O157:H7 infections are a primary cause of hemorrhagic colitis and the hemolytic-uremic syndrome (15, 21, 22). The pathogenesis of EHEC infections is associated with the production of Shiga toxins that are similar to the Shiga toxin that is produced by Shigella dysenteriae type 1 (41). EHEC produces two Shiga toxins, Stx1 and Stx2, and ∼56% of their nucleotide sequences are identical (18). These holotoxins are bipartite molecules composed of an enzymatically active 32-kDa A subunit noncovalently associated with a pentamer (37.5 kDa) composed of 7.5-kDa B subunits. The A subunit is an N-glycosidase that enters the cytosol and cleaves off a single adenine residue from the 28S rRNA of the 60S ribosomal subunit, ultimately inhibiting protein synthesis (9). The B pentamer mediates holotoxin binding to the well-characterized glycolipid receptor Gb3 that is present on certain eukaryotic cells (1, 29).
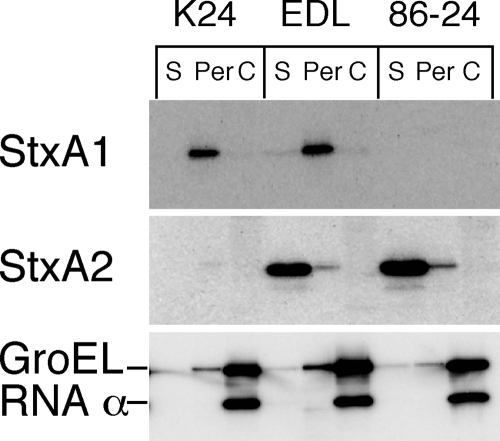
Although the molecular structures of Stx1 and Stx2 in EHEC are similar (10, 38), their cellular distributions are different. Stx1 is almost completely cell associated, whereas Stx2 is found in the extracellular fraction (35, 39). This difference in distribution is due to the different translocations across the outer membrane in EHEC, because Stx1 is located in the periplasmic fraction of the bacterial cells (51) (Fig. 1). In particular, the observation of different localizations in both Stx1- and Stx2-producing EHEC has suggested that EHEC strains contain a specific Stx2 system for release from bacterial cells. A previous study found that the B subunit dictated the cytotoxic localization of Shiga toxin within bacterial cells (48). Recently, we reported that the alteration of the serine 31 residue to an asparagine residue in the B subunit of Stx2 led to the inhibition of the extracellular localization of mutant Stx2 (37). These results suggest that the serine 31 residue of the B subunit of Stx2 contains secretion-related information that is recognized by a hypothetical Stx2 secretion system in EHEC.
FIG. 1.
Distribution of Stx1 and Stx2 in EHEC. Stx1-producing EHEC K24, Stx1- and Stx2-producing EHEC EDL933 (EDL), and Stx2-producing EHEC 86-24 were cultured in LB broth for 12 h at 37°C and fractionated into supernatant (S), periplasmic (Per), and cytoplasmic (C) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using anti-Stx1 antiserum or anti-Stx2A MAb VT2-32. The anti-GroEL and anti-RNA polymerase α subunit antibodies were used as experimental controls.
On the other hand, stx1 and stx2 in EHEC are located within Stx-encoding phages that are related to the λ phage, which is well characterized in both its genome arrangement and transcription patterns (Fig. 2). In the lysogenic state of the λ phage, the repressor silences the transcription of most phage genes (33). The removal of repression, which can occur when DNA damage activates the bacterial SOS response (24), leads to a cascade of regulatory events, beginning with the expression of the N transcription antitermination protein (33). Terminator readthrough mediated by the N protein results in the expression of delayed early genes that encode products involved in replication and prophage excision, as well as the Q antitermination protein (11). Q acts at a site, qut, within the phage late promoter, modifying RNA polymerase to a highly processive form that reads through downstream terminators (50). Thus, late gene expression by lambdoid prophages is a consequence of prophage induction. stx1 and stx2 are found within the late operons of the Stx-encoding phages (6, 16, 19, 20, 30, 34, 35, 42), suggesting that they transcribe from the late phage promoter along with lysis and morphogenesis genes following prophage induction (27, 32). Furthermore, it was reported that Stx1 release, not Stx2 release, from E. coli was related to the mechanism of phage induction by mitomycin C treatment (44). These results suggest that this mechanism also contributes to the specific release of Stx2 to the extracellular fraction in EHEC under non-phage-inducing conditions.
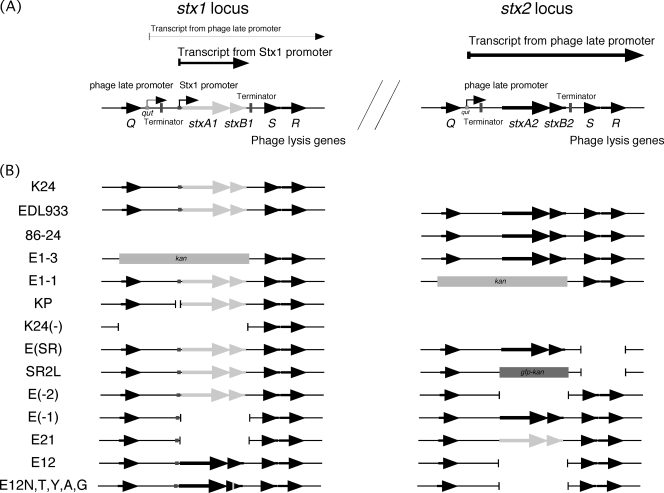
FIG. 2.
Genome arrangement and transcription patterns surrounding the stx genes of the Stx-encoding phages (A) and gene structures of mutant strains (B). This diagram is based on the characterized Stx-encoding phages (44, 45). Shown are relevant genes, promoters, terminators, and transcripts for the expression and distribution of Shiga toxins. Following prophage induction, repressor cleavage allows for increased early transcription and then the production of antitermination Q protein. It facilitates transcription initiating at the late promoter by readthrough at the terminator located downstream of the Q gene. On the other hand, transcription initiating at the Stx1 promoter is inducible in low-iron conditions.
Although the gene arrangements around stx1 and stx2 of Stx-encoding phages in EHEC are similar, their high-expression conditions reportedly are somewhat different (4, 5, 7, 14, 17). Stx2 production is activated by phage-inducing agents, such as mitomycin C, while Stx1 production is increased under low-iron conditions. Moreover, the promoters of Stx1 and Stx2 expression in Stx-encoding phages were different. Previous studies identified a functional Stx1 promoter, regulated by the environmental iron concentration, adjacent to the stx1 gene in Stx1-encoding phages (4, 5), while prophage induction and the resulting transcription from the phage late promoter of the Stx2-encoding phage were important in Stx2 expression (45). Thus, these differences in the mechanisms of gene expression between stx1 and stx2 in EHEC also may contribute to the difference in localization between Stx1 and Stx2, because phage lysis genes of Stx-encoding phages as well as stx1 and stx2 are transcribed by the phage late promoter during phage induction.
Therefore, to examine the mechanism of Stx2 release from EHEC in this study, we determined that the specific induction of the Stx2-encoding phage and another specific Stx2 release system are involved in Stx2 release from EHEC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The bacterial strains and plasmids used in this study are listed in Table 1. The oligonucleotides used are listed in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| K24 | EHEC O157; stx1 | 37 |
| EDL933 | EHEC O157; stx1stx2 | 31 |
| 86-24 | EHEC O157; stx2 | 13 |
| E1-3 | EDL933; ΔQ-stx1::kan | This study |
| E1-1 | EDL933; ΔQ-stx2::kan | This study |
| KP | K24; Δstx1 promoter | This study |
| K24(−) | K24; ΔQ-stx1 | This study |
| E(SR) | EDL933; ΔSR | This study |
| SR2L | E(SR); Δstx2::gfp-kan | This study |
| E(−2) | EDL933; Δstx2 | This study |
| E(−1) | EDL933; Δstx1 | This study |
| E21 | E(−1); Δstx2::stx1 | This study |
| E12 | E(−2); Δstx1::stx2 | This study |
| E12N | E(−2); Δstx1::stx2 (B subunit, S31N) | This study |
| E12T | E(−2); Δstx1::stx2 (B subunit, S31T) | This study |
| E12Y | E(−2); Δstx1::stx2 (B subunit, S31Y) | This study |
| E12A | E(−2); Δstx1::stx2 (B subunit, S31A) | This study |
| E12G | E(−2); Δstx1::stx2 (B subunit, S31G) | This study |
| E(etpD−) | EDL933; ΔetpD | This study |
| BL21 | F−ompT hsdSB(rB− mB−) dcm gal | Promega |
| DH5α(λpir) | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR (lacZYA-argF)U169 λpir+ | 8 |
| pRed/ET(amp) | Red/ET expression plasmid; Ampr | Gene Bridges GmbH |
| pFRT-Kan | Suicide plasmid, FRT-flanked PGK-gb2-neo cassette | Gene Bridges GmbH |
| pFT-A | FLP expression plasmid; Ampr | National BioResource Project (NIG, Japan), 26 |
| pFKS1-31 | stx1 in pFRT-Kan | This study |
| pFKS2-31 | stx2 in pFRT-Kan | This study |
| pFKSN22 | stx2 mutant (B subunit, S31N) in pFRT-Kan | This study |
| pFKST6 | stx2 mutant (B subunit, S31T) in pFRT-Kan | This study |
| pFKSY1 | stx2 mutant (B subunit, S31Y) in pFRT-Kan | This study |
| pFKSA17 | stx2 mutant (B subunit, S31A) in pFRT-Kan | This study |
| pFKSG2 | stx2 mutant (B subunit, S31G) in pFRT-Kan | This study |
| pFLG2 | gfp in pFRT-Kan | This study |
| pAcGFP1 | GFP expression plasmid | Clontech |
| pUC118 | Cloning vector | Takara |
| pUF5 | Promoterless plasmid from pUC118 | This study |
| p1Plac3 | stx1-gfp transcriptional fusion in pUF5 | This study |
| pTrcHis2A | Ptrc, lacIq; codons for six histidine residues; Ampr | Invitrogen |
| pAH-5 | (RBR-Stx1B)-stx2B fusion with codons for six histidine residues in pTrcHis2A | 37 |
| pS31N-6 | (RBR-Stx1B)-stx2B (S31N) fusion codons for six histidine residues in pTrcHis2A | 37 |
| pS31T-1 | (RBR-Stx1B)-stx2B (S31T) fusion codons for six histidine residues in pTrcHis2A | This study |
| pS31Y-4 | (RBR-Stx1B)-stx2B (S31Y) fusion codons for six histidine residues in pTrcHis2A | This study |
| pS31A-3 | (RBR-Stx1B)-stx2B (S31A) fusion codons for six histidine residues in pTrcHis2A | This study |
| pS31G-5 | (RBR-Stx1B)-stx2B (S31G) fusion codons for six histidine residues in pTrcHis2A | This study |
| pTRS1 | stcEH in pTrcHis2A | This study |
(RBR-Stx1B), ribosomal binding region of Stx1B; FRT, FLP recombination target.
Cell fractionation.
EHEC was cultured in Luria-Bertani (LB) broth left unsupplemented or supplemented with 2,2′-dipyridyl, an iron chelator, at 37°C for 12 h. For mitomycin C treatment, overnight cultures grown in LB broth were diluted 1:200 in 40 ml of LB broth and grown for 3 h at 37°C, and then an appropriate concentration of mitomycin C was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. Cells were pelleted by centrifugation at 10,000 × g for 5 min, and the supernatant obtained was used as the supernatant fraction. The pellet was suspended in an equal volume of ice-cold phosphate-buffered saline (PBS), pH 7.2, and sonicated for 30 s on ice. After sonication, the cell homogenate was centrifuged at 10,000 × g for 5 min, and the supernatant obtained was used as the cell-associated fraction. When the periplasmic fraction was prepared, the pellet was suspended in an equal volume of 0.5 M sucrose, 5 mM EDTA, and 50 mM Tris-HCl (pH 8.0) containing 0.6 mg/ml of lysozyme, and then it was incubated at 30°C for 30 min. After incubation, the suspension was centrifuged at 10,000 × g for 5 min, and the supernatant obtained was used as the periplasmic fraction. The pellet was resuspended in an equal volume of ice-cold PBS and was sonicated for 30 s on ice. After sonication, the cell homogenate was centrifuged at 10,000 × g for 5 min, and the supernatant obtained was used as the cytoplasmic fraction. For conditions of low iron concentrations, 2,2′-dipyridyl, an iron chelator, was used at 0.2 mM. For the low-level overproduction of the B subunit in transformed EHEC 86-24, IPTG (isopropyl-β-d-thiogalactopyranoside) was used at 1 μM.
SDS-PAGE and immunoblotting.
Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to polyvinyl difluoride membranes. The membranes were incubated in antiserum or monoclonal antibody (MAb), followed by incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G and/or HRP-conjugated anti-mouse immunoglobulin G. Antibody-antigen complexes were detected using the ECL detection kit (Amersham Biosciences K.K., Tokyo, Japan) by an LAS-1000 luminescent image analyzer (Fujifilm, Tokyo, Japan).
Antibodies.
Polyclonal antisera for Stx1 and Stx2 were prepared as described previously (28, 52). The anti-Stx1 and anti-Stx2 antisera primarily reacted with the A subunits of Stx1 and Stx2, respectively, and hardly reacted with the B subunits of the respective proteins. A MAb (VT2-32) for the A subunit of Stx2 was prepared as described previously (36). Polyclonal antiserum against StcE was prepared via the immunization of a rabbit. The Japanese white rabbit initially was subcutaneously immunized with 100 μg of histidine-tagged StcE (designated StcEH) in 200 μl of PBS emulsified with an equal volume of Freund's complete adjuvant. Two weeks after the initial immunization, a booster injection of 100 μg of StcEH with Freund's incomplete adjuvant was administered. Identical booster doses were given at 2-week intervals. Bleedings for antiserum were performed 7 days after the third booster injection. Anti-His Tag (27E8; a mouse MAb), anti-GroEL (SPS-870; a mouse MAb), and anti-α subunit of RNA polymerase (W0003; a mouse MAb) were obtained from Cell Signaling Technology (Beverly, MA), Stressgen Bioreagents (Ann Arbor, MI), and NeoClone Biotechnology International (Madison, WI), respectively.
Strain construction.
Mutant strains are derivatives of EHEC EDL933 and K24 and were produced using a Red/ET recombination system (Gene Bridges GmbH, Heidelberg, Germany). Briefly, PCR primers containing 50 bp immediately upstream and downstream of the target locus were used to amplify the DNA fragment containing the FLP recombination target-flanked PGK-gb2-neo cassette found in appropriate plasmids (see Table S2 in the supplemental material). The resulting linear PCR product was electroporated with the appropriate parent strain that previously had been transformed with the plasmid pRed/ET(amp). Recombinants containing Kanr elements in place of the target locus were selected on Kan plates and confirmed by PCR. The deletion of Kanr elements was accomplished by transformation with plasmid pFT-A (26), resulting in kanamycin-sensitive mutants that were verified by PCR. Plasmids pRed/ET(amp) and pFT-A are temperature sensitive and were cured by overnight growth at 37°C.
To construct a template plasmid using a Red/ET recombination system, PCR products were cloned into the NotI site of plasmid pFRT-Kan. The stx1 and stx2 genes were amplified by PCR from the genomic DNA of EHEC EDL933 using the primer sets P616 and P617 as well as P618 and P619, and the gfp gene was amplified by PCR from the plasmid DNA of pAcGFP1 using the primer sets P652 and P588. These fragments were cloned by the ligation of a NotI fragment into pFRT-Kan to generate plasmids pFKS1-31, pFKS2-31, and pFLG2, respectively.
To construct the stx2 gene encoding the mutant B subunit (S31N, T, Y, A, and G), a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used (Table 1). Sequences derived by PCR were verified by the ABI 310 sequencer. To construct plasmids expressing histidine-tagged StxB2 (StxB2-H) and mutant StxB2-H, in which six histidine residues were added at the carboxyl termini, a two-step PCR-based strategy was adopted (37; also see Table S3 in the supplemental material). Sequences derived by PCR were verified by the ABI 310 sequencer.
Plasmid construction of stx1-gfp transcriptional fusion plasmid.
To construct a promoterless plasmid, the PvuII-EcoO109I fragment was obtained from pUC118 to remove the lac promoter region. A multiple cloning site fragment was amplified by PCR using M4 and RV primers from plasmid DNA of pUC118. The PvuII-EcoO109I fragment was blunted and ligated to the multiple cloning site fragment to yield the pUF5 plasmid. To construct the stx1-gfp transcriptional fusion plasmid, a two-step PCR-based strategy was adopted. A 346-bp Stx1 promoter fragment, which did not include the phage late promoter region of the Stx1-encoding phage, and a gfp fragment were obtained by PCR from genomic DNA of EHEC EDL933 and plasmid DNA of pAcGFP1 using P570 and P647 as well as P646 and P643, respectively. The resulting fragment mixture was used as a template for reamplification using P605 and P588. The DNA fragment was blunted and then inserted into the SmaI site in pUF5 to yield p1lac3. Sequences derived by PCR were verified by the ABI 310 sequencer.
Plasmid construction and purification of StcEH.
To construct the plasmid expressing StcEH, the stcE gene was amplified by PCR from the genomic DNA of EHEC EDL933 using the primers P564 and P565. A DNA fragment was cleaved with NcoI and SalI and then was inserted into these sites in pTrcHis2A to yield pTRS1 expressing StcEH. Sequences derived by PCR were verified by the ABI 310 sequencer.
This plasmid was transformed into E. coli BL21. To purify protein from this strain, a 1-liter culture was grown at 30°C to an approximate optical density (at 600 nm) of 0.5. One milliliter of a 1 M IPTG solution was added, and the culture was incubated for 3 h at 30°C. Cells were pelleted by centrifugation, resuspended in 12 ml of PBS, and sonicated on ice. Cellular debris was removed by centrifugation at 12,000 × g for 30 min. The supernatant was separated by affinity chromatography using an Ni2+-loaded HiTrap chelating HP column (1 ml; Amersham Biosciences K.K., Tokyo, Japan) equilibrated with PBS, and StcEH was eluted by a 0 to 0.5 M gradient of imidazole. The peak fractions of StcEH were dialyzed against PBS, concentrated, and finally stored at −80°C. The protein concentration was determined by the Bradford method using the protein assay reagent (Bio-Rad, Richmond, CA).
Fluorescence microscopy.
E. coli growing at 37°C was fixed directly in growth medium by the addition of paraformaldehyde and glutaraldehyde in PBS to final concentrations of 2.6% (vol/vol) and 0.04% (vol/vol), respectively. The mixture was incubated for 10 min at room temperature and for a further 50 min on ice. Fixed cells were washed three times in PBS at room temperature by centrifugation and resuspended in PBS containing 0.2 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) and then were mounted on glass slides.
RESULTS
Stx-encoding phage induction in EHEC.
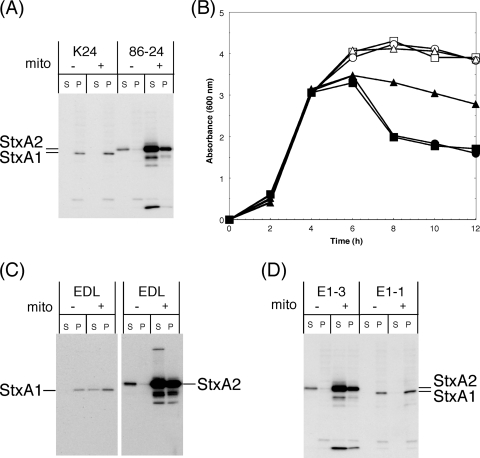
Following mitomycin C treatment, Stx2 production was greatly increased in EHEC 86-24, but Stx1 production was not increased in EHEC K24 (Fig. 3A). This result indicates that mitomycin C effectively induces the Stx2-encoding phage but is not effective at inducing the Stx1-encoding phage in EHEC. To confirm that this result was consistent with that using both Stx1- and Stx2-producing EHEC, we constructed derivatives of EHEC EDL933, mutant E1-3 and E1-1 strains, in which regions including Q-stx1 and Q-stx2, respectively, within the Stx-encoding phages each were replaced by a kanamycin cassette to inhibit the transcription of late phage genes. After phage induction by mitomycin C, the absorbance of cultures was greatly decreased in EDL933 and mutant E1-3 but was not greatly decreased in mutant E1-1 (Fig. 3B). Western blotting revealed that, after mitomycin C treatment, Stx2 production was greatly increased in EDL933 and mutant E1-3, but Stx1 production was not sufficiently increased in EDL933 and mutant E1-1 (Fig. 3C, D). This finding indicates that, after mitomycin C treatment, the Stx2-encoding phage induction leads to an increase in Stx2 production in both Stx1- and Stx2-producing EHEC.
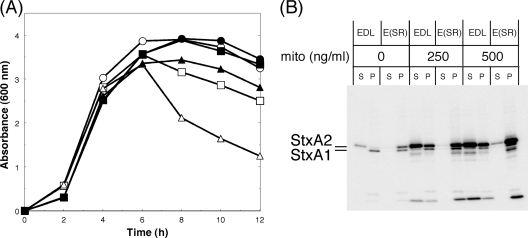
FIG. 3.
Induction of Stx1 and Stx2 production (A, C, and D) and the growth curve (B) for mitomycin C treatment in EHEC and mutants. (A, C, and D) Overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then mitomycin C (final concentration, 500 ng/ml) was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. EHEC strains were fractionated into supernatant (S) and cell-associated (P) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using anti-Stx1 and anti-Stx2 antiserum mixtures (A and D) or anti-Stx1 antiserum or anti-Stx2A MAb VT2-32 (C). (B) Growth was monitored by determining the optical density of the culture at 600 nm. Squares, EDL933 (EDL); circles, E1-3 (ΔQ-stx1::kan); triangles, E1-1 (ΔQ-stx2::kan) (Fig. 2); closed, with mitomycin C (mito) treatment; open, without mitomycin C treatment.
Stx1 production was not effectively induced by mitomycin C treatment in two EHEC strains. Therefore, to confirm the generality of Stx1-encoding phage induction by mitomycin C in EHEC, we analyzed the increase in Stx1 production by mitomycin C treatment in 38 additional EHEC strains (21 Stx1- and Stx2-producing and 17 Stx1-producing EHEC strains K1 to K40, except for strains K15 and K24) (37) by Western blot analysis. Mitomycin C sufficiently increased Stx1 production in only 2 of 38 Stx1-producing strains: EHEC serotype O157 K23 and serotype O26 K37. Interestingly, in K23 and K37, 75 and 28% of total Stx1 production occurred in supernatant fractions, respectively (37). Therefore, our observations indicate that 5% of Stx1-encoding phages in EHEC strains are efficiently inducible by mitomycin C treatment under these conditions.
Role of Stx1 promoter in EHEC.
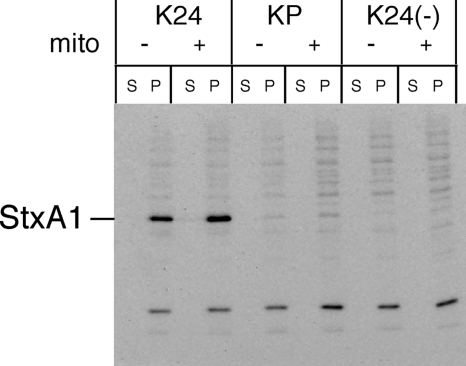
According to these results and previous studies (43, 49), Stx1 is expressed independently of the regulatory system governing phage gene expression in almost all EHEC. Thus, to examine that the Stx1 promoter (4) adjacent to stx1 was a dominant regulatory element in Stx1 production in EHEC, we constructed derivatives of Stx1-producing K24, called the KP strain, in which a region including the Stx1 promoter was deleted. This deletion left intact stx1A, stx1B, Q, and the phage late promoter of the Stx1-encoding phage. However, the Fur binding site overlaps with the −10 sequence of the Stx1 promoter region (4, 5). Therefore, the Fur binding site adjacent to stx1 also was deleted from KP. Western blot analysis revealed that K24 produced Stx1, while KP failed to produce detectable levels of Stx1 (Fig. 4). Following induction by mitomycin C, K24 produced Stx1 at the same level, while the KP and K24(−) strains again failed to produce observable levels of Stx1 (Fig. 4). Furthermore, we obtained the same results using derivatives of both Stx1- and Stx2-producing EDL933 (data not shown) and under iron-depleted conditions (data not shown). These results indicate that the phage induction system of the Stx1-encoding phage in EHEC hardly contributed to Stx1 production with or without mitomycin C treatment, and the Fur protein did not repress the transcription of the phage late operon of the Stx1-encoding phage.
FIG. 4.
Role of Stx1 promoter on Stx1 production in EHEC. Overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then mitomycin C (mito; final concentration, 500 ng/ml) was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. EHEC strains were fractionated into supernatant (S) and cell-associated (P) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using anti-Stx1 antiserum. Strain characteristic: KP, Δstx1 promoter; K24(−), ΔQ-stx1 (Fig. 2).
Role of lysis genes in Stx2-encoding phage for Stx2 release in EHEC.
Since stx2 was located directly upstream of the phage lysis genes in Stx2-encoding phages, the downstream lysis genes might serve as the primary mechanism for coordinating the release of Stx2 during a spontaneous phage induction in EHEC (45, 46). Therefore, to confirm whether or not the phage lysis S and R genes in the Stx2-encoding phage were involved in the specific release of Stx2 in EHEC under non-phage-inducing conditions, we constructed the SR-deficient mutant E(SR) strain. We compared the absorbance of the culture of the mutant E(SR) to that of EDL933 after phage induction with mitomycin C, and we found that mutant cultures continued to grow after the absorbance of wild-type cultures began to decrease (Fig. 5A). However, the viable cell number in the culture of the mutant E(SR) with mitomycin C treatment was significantly lower than that of EDL933 without mitomycin C treatment (data not shown). This result shows that although the bacterial lysis of mutant E(SR) by mitomycin C is inhibited by the absence of the lysis genes of the Stx2-encoding phage, the induced cells may not be viable. Western blot analysis revealed that Stx2 was almost completely cell associated in mutant E(SR) (Fig. 5B). Following induction with mitomycin C, Stx2 was increased and found to be cell associated in mutant E(SR) at 500 ng/ml mitomycin C (Fig. 5B). This indicates that a spontaneous Stx2-encoding phage induction under non-phage-inducing conditions led to specific Stx2 release via phage-mediated lysis in EHEC.
FIG. 5.
Role of phage lysis genes on Stx2 localization in EHEC. Growth curve (A) and Stx1 and Stx2 production levels (B) of EHEC and the mutant. Overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then mitomycin C (mito; final concentration, 500 ng/ml) was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. (A) The growth was monitored by determining the optical density of the culture at 600 nm. (B) EHEC EDL933 (EDL) and E(SR) were fractionated into supernatant (S) and cell-associated (P) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using an anti-Stx1 and anti-Stx2 antiserum mixture. (A) Open symbols, EDL933; closed symbols, E(SR) (ΔS, ΔR) (Fig. 2); circles, without mitomycin C treatment; squares, with 250 ng/ml of mitomycin C; triangles, with 500 ng/ml of mitomycin C.
Rate of Stx2-encoding phage induction in EHEC.
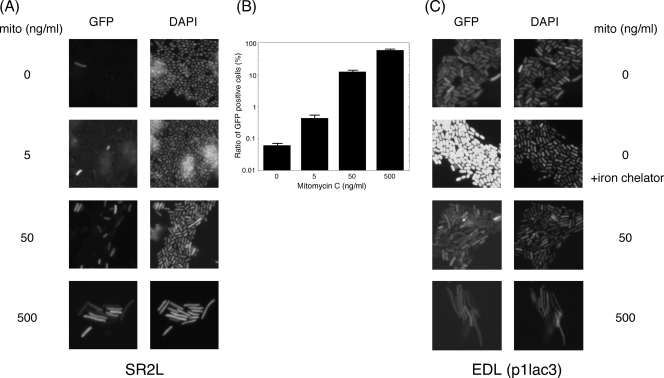
Although specific Stx2 release from bacterial cells was involved in Stx2-encoding phage-mediated lysis during a spontaneous phage induction in EHEC under non-phage-inducing conditions, Stx1 remained within the bacterial cells in both Stx1- and Stx2-producing EHEC. These findings suggested that the Stx2-encoding phage-induced subpopulation of the total EHEC population was responsible for a significant level of Stx2 production and that the Stx2 production rate of the induced subpopulation in the total EHEC population was extremely low in EHEC. Therefore, to examine the rate of the Stx2-encoding phage in EHEC induced under non-phage-inducing conditions, we constructed a derivative of mutant E(SR), called the SR2L strain, in which a region including the endogenous stx2 in the Stx2-encoding phage genome was replaced with gfp to express green fluorescent protein (GFP) within the bacterial cells depending on the phage late promoter of Stx2-encoding phage. Fluorescence microscopy revealed that, under non-phage-inducing conditions, mutant SR2L expressed GFP into the bacterial cells at extremely low rates (0.06%) (Fig. 6A, B). This result is in agreement with a study by Livny and Friedman (25), who used a selectable in vivo expression technology to calculate that about 0.005% of E. coli K-12 lysogens of Stx1-encoding phage H-19B from EHEC serotype O26 are spontaneously induced per generation. As for induction with mitomycin C, the expression rates of GFP-positive cells in the total EHEC population were increased (Fig. 6B). At 500 ng/ml of mitomycin C, approximately 60% of total bacterial cells were GFP positive by fluorescence microscopy. However, the brightness of fluorescence from GFP in each bacterial cell was almost the same regardless of the mitomycin C concentration (Fig. 6A). This indicates that mitomycin C increased the proportions of cells activated by the induction of Stx2-encoding phage. Consequently, mitomycin C increased Stx2 production in the total EHEC population.
FIG. 6.
Expression patterns from the phage late promoter of the Stx2-encoding phage and the Stx1 promoter in the EHEC population. (A) Overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then an appropriate concentration of mitomycin C (mito) was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. SR2L (ΔS, ΔR, Δstx2::gfp-kan) (Fig. 2) cells were fixed. (B) The rates of GFP-positive cells were calculated by fluorescence microscopy. Mean values derived from four independent cultures are shown along with standard deviations. (C) p1lac3-transformed EDL933 strains [EDL(p1Plac3)] were grown in LB broth left unsupplemented or supplemented with an iron chelator for 12 h at 37°C. After treatment with mitomycin C, overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then an appropriate concentration of mitomycin C was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. Cells were fixed and observed by fluorescence microscopy. As mitomycin C was added, bacterial cells became elongated, but when the iron chelator was used, the bacteria did not change shape.
On the other hand, to examine the transcription patterns of the Stx1 promoter in the Stx1-encoding phage, we constructed the transcriptional fusion of the Stx1 promoter to the gfp reporter gene. The Stx1 promoter-dependent GFP fluorescence plasmid p1Plac3-transformed EDL933 strain uniformly expressed bright GFP fluorescence in each cell (Fig. 6C). This agrees with the flow cytometry analysis of the promoter region for the Stx1 expression of Stx1-encoding phage in H-19B (2). Upon supplementation with an iron chelator, the uniform brightness in each cell was increased (Fig. 6C), while after induction by mitomycin C the brightness of fluorescence in each bacterial cell did not change (Fig. 6C). These results indicate that, unlike the case with the Stx2 expression pattern, Stx1 was uniformly expressed in each cell. Therefore, when only a small fraction of bacterial cells spontaneously undergo lysis by Stx2-encoding phage induction under non-phage-inducing conditions, Stx1 remains to be localized in the cell-associated fraction of the total population.
Role of Stx2-encoding phage in Shiga toxin release in EHEC.
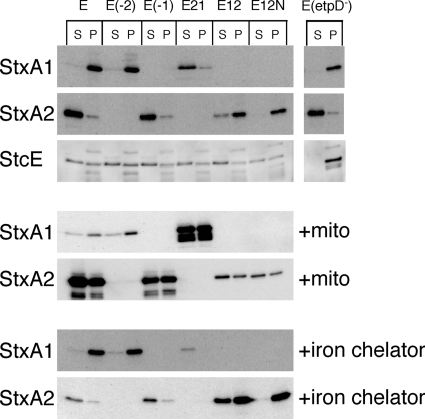
We revealed that the extracellular distribution of Stx2 in EHEC under non-phage-inducing conditions also was related to the Stx2-encoding phage-mediated lysis system. Thus, to determine whether or not the Stx2-encoding phage-mediated lysis system was specific to the release of Stx2 protein in EHEC, we constructed the stx-replaced mutant E21 strain. In this mutant, the region including the endogenous stx1 was deleted, and the endogenous stx2 in the Stx2-encoding phage genome was replaced with stx1. Stx1 derived from the phage late promoter of the Stx2-encoding phage was found in the supernatant fraction in mutant E21, while Stx1 derived from the Stx1 promoter was located in the cell-associated fraction in the mutant E(−2) strain (Fig. 7). This result indicates that the release of Shiga toxin in EHEC depends on the spontaneous Stx2-encoding phage induction regardless of the presence of stx2. Following induction by mitomycin C, Stx1 production derived from the phage late promoter of the Stx2-encoding phage was increased in mutant E21 (Fig. 7). The activity of the Stx1 promoter within the Stx1-encoding phage is regulated by the environmental iron concentration in EHEC (4, 5). As expected, a low medium iron concentration did not increase Stx1 production derived from the phage late promoter of the Stx2-encoding phage in mutant E21, although Stx1 production derived from the Stx1 promoter was increased in E(−2) (Fig. 7).
FIG. 7.
Distribution of Stx1 and Stx2 in EHEC EDL933 and isogenic derivatives containing the regions of stx deletion, etpD deletion, or stx replacement genes. EHEC strains were grown in LB broth left unsupplemented or supplemented with an iron chelator for 12 h at 37°C. After treatment with mitomycin C (mito), overnight cultures grown in LB broth were diluted 1:200 in LB broth and grown for 3 h at 37°C, and then mitomycin C (final concentration, 500 ng/ml) was added at mid-log phase. Further growth was allowed for 5 h at 37°C until the early stationary phase. EHEC strains were fractionated into supernatant (S) and cell-associated (P) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using anti-Stx1 antiserum, anti-Stx2A MAb VT2-32, or anti-StcE antiserum. EDL, EDL933. Strain characteristic(s): E(−2), Δstx2; E(−1), Δstx1; E21, Δstx1 Δstx2::stx1; E12, Δstx1::stx2 Δstx2; E12N, Δstx1::stx2 (B subunit, S31N) Δstx2; E(etpD−), ΔetpD (Fig. 2).
Stx2 secretion system in EHEC.
We previously reported that the serine 31 residue of the B subunit of Stx2 was important for Stx2 release in EHEC (37). Thus, to examine how another specific Stx2 release system was involved in Stx2 release in EHEC, we constructed the stx-replaced mutant E12 and E12N strains. In the mutant E12, the region including the endogenous stx2 was deleted, and the endogenous stx1 in the Stx1-encoding phage genome was replaced by stx2. Similarly, the endogenous stx1 was replaced by the mutant stx2 (B subunit, S31N) in mutant E12N. Stx2 and mutant Stx2 (B subunit, S31N), derived from the Stx1 promoter, were found in the cell-associated fraction in mutants E12 and E12N, while Stx2 derived from the phage late promoter of the Stx2-encoding phage was located in the supernatant fraction in the mutant E(−1) strain (Fig. 7). Following induction by mitomycin C, the production of Stx2 and that of mutant Stx2 (B subunit, S31N) derived from the Stx1 promoter were not increased in mutants E12 and E12N. A low medium iron concentration increased the production of Stx2 and mutant Stx2 (B subunit, S31N) derived from the Stx1 promoter by mutants E12 and E12N. Notably, even though the promoter for gene expression was the same, Stx2 in mutant E12 also was detected in the supernatant fraction under iron-depleted conditions, whereas mutant Stx2 (B subunit, S31N) in mutant E12N was not under iron-depleted conditions. These results indicate that Stx2 also is specifically transported to the extracellular fraction by another pathway in EHEC, and that this pathway is induced to express under a low concentration of iron. However, this pathway might not be the etp type II secretion system on the pO157 virulence plasmid in EHEC O157, because the etpD-deficient mutant E(etpD−) and EHEC O111, which did not retain the pO157 virulence plasmid, almost completely released Stx2 into the extracellular fraction (Fig. 7) (37). Furthermore, EHEC O157 produces StcE protease that specifically cleaves C1 esterase inhibitor (23), and StcE is secreted by the etp type II secretion system in EHEC EDL933 (23). Thus, to confirm the effect on the etp type II secretion pathway when Stx2 was localized into periplasmic space, EDL933 and mutant strains were cultured in LB broth at 37°C for 12 h, and StcE proteins were detected in supernatant and cell-associated fractions by anti-StcE antiserum. EDL933, E(−2), E(−1), E21, E12, and E12N released the same levels of StcE into culture medium, whereas the etpD-deficient mutant E(etpD−) did not (Fig. 7). However, under iron-depleted conditions, EDL933 and mutant strains failed to produce detectable levels of StcE (data not shown). These results suggested that the localization of Stx2 into periplasmic space did not affect the etp type II secretion pathway in EHEC.
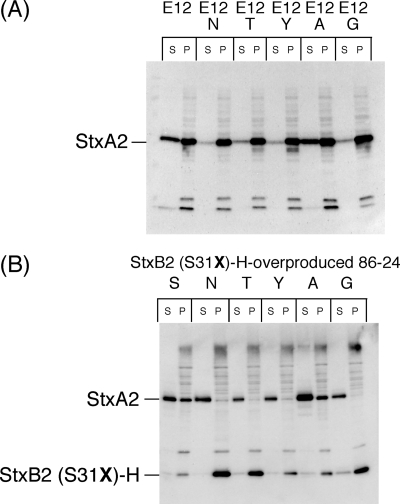
The alteration of the serine 31 residue to an asparagine residue in the B subunit of Stx2 led to the inhibition of mutant Stx2 release in mutant E12N, suggesting that the hypothetical regulatory molecule(s) of the release system for Stx2 release in EHEC specifically interacted with the serine 31 residue of the B subunit of Stx2. Thus, to further investigate the specificity of the Stx2 release system in EHEC, we constructed the mutant E12T, E12Y, E12A, and E12G strains. In these mutants, the regions including the endogenous stx2 in the Stx2-encoding phage genome were deleted, and the endogenous stx1 in the Stx1-encoding phage genome was replaced with the mutants stx2 (B subunit, S31T), stx2 (B subunit, S31Y), stx2 (B subunit, S31A), and stx2 (B subunit, S31G). The alteration to a threonine residue, a tyrosine residue, and a glycine residue in the B subunit of Stx2 also led to the inhibition of mutant Stx2 release in mutant E12T, E12Y, and E12G (Fig. 8A). However, the alteration to an alanine residue, which was the most structurally similar to the serine residue, led to the release of mutant Stx2 (B subunit, S31A) in mutant E12A (Fig. 8A). We previously reported that the overproduction of histidine-tagged StxB2 (StxB2-H) in EHEC led to the inhibition of Stx2 release (37). Therefore, to confirm that Stx2B-H (S31A)-overproduced EHEC showed inhibited Stx2 release, we generated point-mutated Stx2B-H expression plasmids and transformed them in EHEC 86-24 strains. As a result, Stx2B-H (S31A)-overproduced 86-24, as well as Stx2B-H-overproduced 86-24, showed inhibited Stx2 release, but Stx2B-H (S31T)-, Stx2B-H (S31Y)-, and Stx2B-H (S31G)-overproduced 86-24 showed a loss of Stx2 release inhibition (Fig. 8B). Therefore, another Stx2 release system strictly recognized the serine 31 residue of the B subunit of Stx2 in EHEC.
FIG. 8.
Distribution of mutant Stx2 derived from the Stx1 promoter in stx-replaced mutants (A) and effect of overproduced histidine-tagged mutant StxB2 on Stx2 secretion in EHEC 86-24 (B). (A) Cultures were grown in LB broth supplemented with an iron chelator for 12 h at 37°C. (B) Mutant StxB2-H-overproduced EHEC 86-24 strains were grown in LB broth supplemented with 1 μM IPTG for 12 h at 37°C. Cells were fractionated into supernatant (S) and cell-associated (P) fractions. The samples were analyzed by SDS-PAGE and immunoblotting using anti-Stx2 antiserum (A) or an anti-Stx2 antiserum and anti-His Tag antibody mixture (B). Strain characteristic(s): E12, Δstx1::stx2 Δstx2; E12N, Δstx1::stx2 (B subunit, S31N) Δstx2; E12T, Δstx1::stx2 (B subunit, S31T) Δstx2; E12Y, Δstx1::stx2 (B subunit, S31Y) Δstx2; E12A, Δstx1::stx2 (B subunit, S31A) Δstx2; E12G, Δstx1::stx2 (B subunit, S31G) Δstx2 (Fig. 2). S, StxB2-H-overproduced 86-24; N, StxB2 (S31N)-H-overproduced 86-24; T, StxB2 (S31T)-H-overproduced 86-24; Y, StxB2 (S31Y)-H-overproduced 86-24; A, StxB2 (S31A)-H-overproduced 86-24; G, StxB2 (S31G)-H overproduced 86-24.
DISCUSSION
In this study, our experiments demonstrate that the Stx2-encoding phage induction system and another Stx2 release system comprise two specific Stx2 mechanisms of release from bacterial cells in EHEC. Although these two systems are different mechanisms for Stx2 release in EHEC, each system has strict specificity for Stx2 release from bacterial cells in EHEC.
Specificity of induction of Stx2-encoding phage in EHEC.
We showed that Stx1 production was important for the activation of the Stx1 promoter adjacent to stx1 by a low iron concentration, and that the late promoter of the Stx1-encoding phage did not contribute to Stx1 production regardless of mitomycin C treatment in EHEC K24 and EDL933. On the other hand, Stx2-encoding phage induction was important for expressing Stx2 (45), and the Stx2 promoter adjacent to stx2 apparently was not a dominant regulatory element in Stx2 production in EHEC (40). These results suggest that Stx2 is produced by a subpopulation that occurs in the process of Stx2-encoding phage induction in EHEC. Furthermore, our observations of EHEC strains demonstrated that a marked increase in Stx1 production by mitomycin C treatment was not common; these results were consistent with previous reports (43, 49). Therefore, these findings indicate that the rate of spontaneous phage induction and the sensitivity against phage-inducing agents, such as mitomycin C, of Stx2-encoding phage generally are higher than those of Stx1-encoding phage in EHEC.
Mitomycin C treatment induced Stx1 production in EHEC K23 and K37, and we previously reported that Stx1 also was detected in supernatant fractions under non-phage-inducing conditions in these strains (37). These results suggest that Stx1-encoding phages in these strains are spontaneously induced under non-phage-inducing conditions, the same as with Stx2-encoding phages. The growth of EHEC serotype 026 H-19, which retained Stx1-encoding phage H-19B, in low iron concentrations or in prophage-inducing conditions results in increased Stx1 production, and stx1 in H-19B was transcribed from both the iron-regulated Stx1 promoter and the phage late promoter of H-19B (44). Thus, stx1 in EHEC K23 and K37 also may be transcribed from two different promoters under non-phage-inducing conditions.
Irrespective of mitomycin C treatment, the recA mutant of EHEC showed significantly reduced Stx2 production and was devoid of Stx2-encoding phage synthesis (12). Furthermore, nitric oxide, which inhibits the SOS response in host strains, reduces Stx2 production and the synthesis of Stx2-encoding phage in EHEC regardless of mitomycin C treatment (43). Thus, these facts indicate that the rate of Stx2-encoding phage induction depends on the host conditions of the SOS response. However, the reason for the different induction mechanisms between the Stx1- and Stx2-encoding phage in EHEC remains unclear.
The lysis S and R genes of both Stx-encoding phages in EDL933 were located about 3 kb downstream of the stx1 and stx2 genes, respectively (31). R encoded an endolysin that degrades the peptidoglycan of the cell wall (3). R protein was transported across the cytoplasmic membrane by the S gene product, which creates membrane lesions (3). Mutant E(SR) showed almost no decrease in the absorbance of culture with mitomycin C treatment. This suggests that the Stx2-encoding phage is a major phage that is inducible by mitomycin C treatment in EDL933, and that the advance of the lytic process for phage induction may be enough to express lysis S and R genes in the cell. To estimate the rate of the induction of the Stx2-encoding phage in EHEC, we used the mutant E(SR) as the host strain. However, when using wild-type EDL933, we found no GFP-positive cells with mitomycin C treatment (data not shown). The lysis S and R genes of the Stx2-encoding phage in EHEC could be transcribed from the late phage promoter with stx2 following Stx2-encoding phage induction (46).
Since Stx1 was not detected in the supernatant fraction in almost all EHEC cells, the transcription of the phage lysis genes of the Stx1-encoding phage was independent of the transcription of stx1 initiating at the Stx1 promoter adjacent to stx1. These results suggest that the readthrough of transcription initiating at the Stx1 promoter does not result in the sufficient expression of the downstream phage lysis genes of the Stx1-encoding phage.
The discrepancy in which Stx2 was exported in the extracellular fraction by the lysis process, whereas Stx1 was located completely in bacterial cells in both Stx1- and Stx2-producing EHEC, was resolved to examine the rate of spontaneous phage induction in the Stx2-encoding phage in EHEC. We previously reported that Stx2 production levels without mitomycin C treatment were more than 10-fold higher than those of Stx1 in various EHEC strains (37). Furthermore, in fluorescence microscopy, we did not detect the fluorescence of GFP in the gfp-replaced mutant strain, in which the stx1 of the Stx1-encoding phage was replaced (data not shown). These results suggest that the specific activity of the late promoter in the Stx2-encoding phage during phage induction is much stronger than that of the Stx1 promoter adjacent to stx1 in the Stx1-encoding phage in EHEC.
In the mutant E21, stx2 was replaced by stx1, and Stx1 derived from the phage late promoter of the Stx2-encoding phage was found in the supernatant fraction. This indicates that Shiga toxin genes located in late phage operons of spontaneous inducible phages, such as Stx2-encoding phages in EHEC, can be nonspecifically transcribed from the late phage promoter, and consequently lysis occurs in, and toxin is released from, bacterial cells.
We analyzed the distribution and induction between Stx1 and Stx2 in various EHEC strains (37). In general, we found that Stx2 in EHEC is predominantly extracellular and its levels are increased by mitomycin C treatment, whereas Stx1 is mostly cell associated and its levels are not increased by mitomycin C treatment. Therefore, these observations suggest that the Stx1-encoding phage is characterized by the phage that retained the iron-regulated Stx1 promoter adjacent to stx1, while the Stx2-encoding phage is characterized by the spontaneous inducible phage that retained stx2.
Specificity of another Stx2 release system in EHEC.
We demonstrated that there was another Stx2 release system in EHEC by the difference in the distribution of StxA2 between mutants E12 and E12N. Another Stx2 release system in EHEC might interact with the region containing the 31st amino acid residue of the B subunit of Stx2, because only Stx2 and mutant Stx2 (B subunit, S31A) were released from the periplasmic space to the extracellular fraction in mutants E12 and E12A. The B subunit structure of Stx2 is a pentamer in which each monomer comprises two three-stranded antiparallel β-sheets and an α-helix (10). The serine 31 residue of the B subunit is located on the carboxyl-terminal end of the first three-stranded β-sheets (10, 37). Each side chain of the serine 31 residue of the B subunit monomer of Stx2 face the Gb3-binding surface of B-pentamer (10, 37). Moreover, the overexpression of StxB2-H specifically affects the system of Stx2 release in EHEC, and the B subunit pentamer is necessary for the inhibition of Stx2 release in EHEC (37). Therefore, if a specific Stx2 secretion system is present in EHEC, the side chain structure of the 31st amino acid residue of the B subunit of Stx2 may be important for the recognition of a hypothetical regulatory molecule(s) in the Stx2 secretion machinery in EHEC, and the hypothetical assembled regulatory molecules of the system for Stx2 secretion might specifically interact with the Gb3 binding surface of the assembled B subunits of Stx2. Since low-iron conditions in mutant E12 led to increased Stx2 release, the expression of Stx2 release system genes might be regulated by Fur in EHEC, as is the case with stx1 in the Stx1-encoding phage (5). Iron is limiting in the human host, and bacterial pathogens respond to this environment by activating genes required for bacterial virulence. Therefore, the expression of the Stx1 protein and Stx2 release machinery may be advantageous in the case of EHEC infection. However, the Stx2 system of release from periplasmic space in EHEC remains unclear.
Stx2 is released to the extracellular fraction by two different mechanisms, while Stx1 may be released from dead or stationary-phase cells in vivo to cause the human diseases. However, prophage induction during growth in mouse intestine was demonstrated by CTXphage, which carries the cholera toxin genes in Vibrio cholerae (47). Therefore, this mechanism also might occur with Stx-encoding phages in EHEC, thereby inducing cell lysis and allowing the release of the toxin into the intestinal environment, although we did not observe the spontaneous induction of Stx1-encoding phages in EHEC in vitro under non-phage-inducing conditions. Thus, it will be important to examine the in vivo mechanism of the production and release of Shiga toxins from bacterial cells in EHEC; in other words, in the case of EHEC infection.
Supplementary Material
Acknowledgments
We are grateful to Ayako Kiuti and Reiko Komine for their technical assistance.
This research was supported by Grants-in-Aid for Scientific Research (19590440) from the Ministry of Education, Culture, Sports, Science and Technology and Japan Society for the Promotion of Science (JSPS).
Editor: B. A. McCormick
Footnotes
Published ahead of print on 20 April 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Acheson, D. W. K., A. Donohue-Rolfe, and G. T. Keusch. 1991. The family of Shiga and Shiga-like toxins, p. 415-433. In J. E. Alouf, and J. H. Freer (ed.), Sourcebook of bacterial protein toxins. Academic Press Ltd., London, United Kingdom.
- 2.Aertsen, A., R. Van Houdt, and C. W. Michiels. 2005. Construction and use of an stx1 transcriptional fusion to gfp FEMS Microbiol. Lett. 24573-77. [DOI] [PubMed] [Google Scholar]
- 3.Bläsi, U., and R. Young. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21675-682. [DOI] [PubMed] [Google Scholar]
- 4.Calderwood, S. B., F. Auclair, A. Donohue-Rolfe, G. T. Keusch, and J. J. Mekalanos. 1987. Nucleotide sequence of the Shiga-like toxin genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 844364-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 1694759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Grandis, S., J. Ginsberg, M. Toone, S. Climie, J. Friesen, and J. Brunton. 1987. Nucleotide sequence and promoter mapping of the Escherichia coli Shiga-like toxin operon of bacteriophage H-19B. J. Bacteriol. 1694313-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, S. J., and J. B. Kaper. 1997. Role of type 1 fimbriae in EPEC infections. Microb. Pathog. 23113-118. [DOI] [PubMed] [Google Scholar]
- 9.Endo, Y., K. Tsurugi, T. Yutsudo, Y. Takeda, T. Ogasawara, and K. Igarashi. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 17145-50. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, M. E., M. Fujinaga, M. M. Cherney, A. R. Melton-Celsa, E. M. Twiddy, A. D. O'Brien, and M. N. James. 2004. Structure of Shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J. Biol. Chem. 27927511-27517. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18191-200. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 2713-23. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109705-712. [DOI] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Hofmann, S. L. 1993. Southwestern Internal Medicine Conference: Shiga-like toxins in hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura. Am. J. Med. Sci. 306398-406. [DOI] [PubMed] [Google Scholar]
- 16.Huang, A., J. Friesen, and J. L. Brunton. 1987. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J. Bacteriol. 1694308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hull, A. E., D. W. Acheson, P. Echeverria, A. Donohue-Rolfe, and G. T. Keusch. 1993. Mitomycin immunoblot colony assay for detection of Shiga-like toxin-producing Escherichia coli in fecal samples: comparison with DNA probes. J. Clin. Microbiol. 311167-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, M. P., J. W. Newland, R. K. Holmes, and A. D. O'Brien. 1987. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2147-153. [DOI] [PubMed] [Google Scholar]
- 19.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 1471929-1936. [DOI] [PubMed] [Google Scholar]
- 20.Karch, H., H. Schmidt, C. Janetzki-Mittmann, J. Scheef, and M. Kroger. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262600-607. [DOI] [PubMed] [Google Scholar]
- 21.Keusch, G. T., and D. W. Acheson. 1997. Thrombotic thrombocytopenic purpura associated with Shiga toxins. Semin. Hematol. 34106-116. [PubMed] [Google Scholar]
- 22.Lansbury, L. E., and H. Ludlam. 1997. Escherichia coli O157: lessons from the past 15 years. J. Infect. 34189-193. [DOI] [PubMed] [Google Scholar]
- 23.Lathem, W. W., T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr, and R. A. Welch. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45277-288. [DOI] [PubMed] [Google Scholar]
- 24.Little, J. W. 1995. The SOS regulatory system, p. 453-479. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landis, Georgetown, TX.
- 25.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 511691-1704. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Morales, F., A. C. Borges, A. Martinez, K. T. Shanmugam, and L. O. Ingram. 1999. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction. J. Bacteriol. 1817143-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 281255-1267. [DOI] [PubMed] [Google Scholar]
- 28.Noda, M., T. Yutsudo, N. Nakabayashi, T. Hirayama, and Y. Takeda. 1987. Purification and some properties of Shiga-like toxin from Escherichia coli 0157:H7 that is immunologically identical to Shiga toxin. Microb. Pathog. 2339-349. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien, A. D., and R. K. Holmes. 1987. Shiga and Shiga-like toxins. Microbiol. Rev. 51206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226694-696. [DOI] [PubMed] [Google Scholar]
- 31.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 32.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 1811767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ptashne, M. 1992. A genetics swith, 2nd ed. Cell Press, Blackwell Scientific Publications, Cambridge, MA.
- 34.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Distinctiveness of the genomic sequence of Shiga toxin 2-converting phage isolated from Escherichia coli O157:H7 Okayama strain as compared to other Shiga toxin 2-converting phages. Gene 30935-48. [DOI] [PubMed] [Google Scholar]
- 35.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Genome analysis of a novel Shiga toxin 1 (Stx1)-converting phage which is closely related to Stx2-converting phages but not to other Stx1-converting phages. J. Bacteriol. 1853966-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., T. Hamabata, A. Yoshiki, T. Hori, S. Ito, Y. Takeda, and H. Hayashi. 2003. An association of 27- and 40-kDa molecules with glycolipids that bind A-B bacterial enterotoxins to cultured cells. Biochim. Biophys. Acta 1612186-194. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu, T., S. Kawakami, T. Sato, T. Sasaki, M. Higashide, T. Hamabata, T. Ohta, and M. Noda. 2007. The serine 31 residue of the B subunit of Shiga toxin 2 is essential for secretion in enterohemorrhagic Escherichia coli. Infect. Immun. 752189-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein, P. E., A. Boodhoo, G. J. Tyrrell, J. L. Brunton, and R. J. Read. 1992. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 355748-750. [DOI] [PubMed] [Google Scholar]
- 39.Strockbine, N. A., L. R. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung, L. M., M. P. Jackson, A. D. O'Brien, and R. K. Holmes. 1990. Transcription of the Shiga-like toxin type II and Shiga-like toxin type II variant operons of Escherichia coli. J. Bacteriol. 1726386-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 51817-1822. [DOI] [PubMed] [Google Scholar]
- 42.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 684856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vareille, M., T. de Sablet, T. Hindre, C. Martin, and A. P. Gobert. 2007. Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA 10410199-10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44957-970. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 1832081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 703985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 2721910-1914. [DOI] [PubMed] [Google Scholar]
- 48.Weinstein, D. L., M. P. Jackson, L. P. Perera, R. K. Holmes, and A. D. O'Brien. 1989. In vivo formation of hybrid toxins comprising Shiga toxin and the Shiga-like toxins and role of the B subunit in localization and cytotoxic activity. Infect. Immun. 573743-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, Y., K. Shima, K. Okazaki, H. Ito, Y. Oyamada, N. Sugimoto, M. Asakura, K. Nishimura, and S. Yamasaki. 2005. Abstr. 40th U.S.-Japan Cholera Other Bacterial Enteric Infect. Joint Panel Meet., abstr. 115-117.
- 50.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284611-615. [DOI] [PubMed] [Google Scholar]
- 51.Yoh, M., E. K. Frimpong, and T. Honda. 1997. Effect of antimicrobial agents, especially fosfomycin, on the production and release of Vero toxin by enterohaemorrhagic Escherichia coli O157:H7. FEMS. Immunol. Med. Microbiol. 1957-64. [DOI] [PubMed] [Google Scholar]
- 52.Yutsudo, T., N. Nakabayashi, T. Hirayama, and Y. Takeda. 1987. Purification and some properties of a Vero toxin from Escherichia coli O157:H7 that is immunologically unrelated to Shiga toxin. Microb. Pathog. 321-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.