Abstract
Delivery of bacterial proteins into mammalian cells by type III secretion systems (TTSS) is thought to require the intimate association of bacteria with target cells. The molecular bases of this intimate association appear to be different in different bacteria involving TTSS components, as well as surface determinants not associated with TTSS. We show here that the protein translocases SipB, SipC, and SipD of the Salmonella enterica serovar Typhimurium pathogenicity island 1 (SPI-1)-encoded TTSS are required for the intimate association of these bacteria with mammalian cells. S. Typhimurium mutant strains lacking any of the translocases were defective for intimate attachment. Immunofluorescence microscopy showed that SipD is present on the bacterial surface prior to bacterial contact with host cells. In contrast, SipB and SipC were detected on the bacterial surface only subsequent to bacterial contact with the target cell. We conclude that the coordinated deployment and interaction between the protein translocases mediate the SPI-1 TTSS-dependent intimate association of S. Typhimurium with host cells.
Type III secretion systems (TTSS) are central to the interactions of many pathogenic or symbiotic bacteria with their hosts (12). They exert their function by delivering bacterial proteins, so-called “effectors,” into host cells (8, 32). These proteins have the capacity to modulate a variety of cellular functions, including cytoskeleton dynamics, vesicular trafficking, cell cycle progression, programmed cell death, and transcription. Salmonella enterica serovar Typhimurium encodes two TTSS in discrete pathogenicity islands (9). One, encoded in the Salmonella pathogenicity island 1 (SPI-1), modulates a variety of cellular processes including actin cytoskeleton dynamics, which is required to mediate bacterial uptake into nonphagocytic cells (28). The other, whose expression is induced upon bacterial internalization, is encoded within SPI-2 and modulates vesicular trafficking to secure bacterial replication within cells (38).
A central component of TTSS is the needle complex, a multiring supramolecular structure that is thought to serve as a conduit through which the bacterial proteins transiting this secretion pathway traverse the bacterial envelope (21, 24). Other essential components include a set of inner membrane proteins, an associated ATPase (1), and a family of customized chaperones that facilitate the targeting of cognate effector proteins to the secretion machine (7, 23, 33). In addition, there is a group of proteins known as “translocases,” which themselves are secreted through the TTS pathway and mediate the passage of effectors through the target host cell membrane (4, 14). Although significant advances have been made in the understanding of the assembly of the needle complex and the mechanisms of substrate recognition, relatively little is known about other essential aspects of the function of these machines. A characteristic feature of at least some TTSS is that their activity is stimulated upon contact with target cells (26, 37, 41). However, little is known about the events that lead to the deployment of the translocases at the plasma membrane of the host cells and the subsequent events that lead to protein translocation through the host-cell plasma membrane. The intimate association of bacteria with host cells is thought to be essential for efficient protein translocation (13). However, the mechanisms of intimate attachment leading to TTSS-mediated protein translocation appear to be different in different bacteria. In Yersinia enterocolitica and Yersinia pseudotuberculosis, for example, two surface proteins, YadA (39) and invasin (16), have been proposed to redundantly mediate the intimate attachment necessary for TTSS function. In enteropathogenic E. coli, another surface protein, intimin, mediates intimate attachment by binding tir, a protein that itself is translocated to the host cell membrane by the TTSS (20, 25). S. enterica encodes multiple adhesins that can potentially mediate its association to host cells (36). However, the mechanisms that result in the intimate association necessary for TTS function have not been investigated. We show here that the SPI-1 TTSS translocases SipB, SipC, and SipD are coordinately deployed to mediate intimate attachment of S. Typhimurium to target host cells.
MATERIALS AND METHODS
Bacterial strains.
All of the strains used in the present study are derivatives of S. enterica serovar Typhimurium strain SL1344 and are listed in Table S1 in the supplemental material. The construction of nonpolar mutations in invA (11), invJ (5), sipB and sipC (19), sipD (18), and the triple mutant ΔsopE ΔsopE2 ΔsopB (40) have been described elsewhere. To introduce a 3×Flag (3×F)tag at amino acid 20 of the translocases SipB, SipC, and SipD, an R6K suicide vector was constructed with regions of homology placed at each side of the tag by standard recombinant DNA techniques. The first 50 amino acids of the resulting tagged proteins are shown in Fig. S1 in the supplemental material. The 3×Flag-tagged alleles were introduced into the chromosome of wild-type S. Typhimurium by allelic exchange (17), and its expression was confirmed by anti-Flag Western blot. When required sipB::aphT, sipC::aphT, and sipD::aphT alleles were moved into the 3×Flag-tagged translocase background by P22 HT-mediated transduction (31). All strains were grown in LB agar or in LB supplemented with 0.3 M sodium chloride to induce optimal in vitro expression of SPI-1.
Imaging and quantification of bacterial attachment to eukaryotic cells by immunofluorescence microscopy.
The levels of S. Typhimurium entry or attachment to cultured epithelial cells were determined by differential immunofluorescence staining of inside versus outside bacteria as previously described (2), in which internalized bacteria stain exclusively in red, whereas outside bacteria stain both green and red. To quantify the levels of attachment, at least 20 random low-magnification (×20) images of this assay were acquired (with an average of ∼20 cells per image), and both the number of cells and bacteria in each field were counted and the average number of bacteria per cell were determined.
Quantification of bacterial attachment by a CFU-based assay.
Almost-confluent monolayers of Henle-407 embryonic intestinal epithelial cells were infected at a multiplicity of infection of 10 with different strains of S. Typhimurium grown under SPI-1 TTSS-inducing conditions as described above. Infections were performed in triplicates in 500 μl of Hanks balanced salt solution in 24-well plates. After 30 min of incubation, the infection medium was aspirated, and the cells were washed six times with 1 ml of Hanks balanced salt solution and 5 min of horizontal shaking (250 rpm) after each medium exchange. After the last wash, medium was aspirated, and the cells were lysed in 500 μl of 0.05% sodium deoxycholate in water. Dilutions were plated on LB plates, and attachment was represented as the percentage of the inoculum that remained in the well after the washes.
Immunoprecipitation of 3×Flag epitope-tagged SipB and SipC from culture supernatants.
Overnight cultures of wild-type S. Typhimurium and derivatives expressing 3×Flag epitope-tagged versions of SipB or SipC were diluted 1:20 in 20 ml of 0.3 M NaCl LB and grown to optical density at 600 nm of 0.9. Bacterial cells were pelleted, and the culture supernatants were filtered through a 0.45-μm-pore-size syringe filter. Portions (14 ml) of the filtered supernatants were transferred to 15-ml conical tubes, and 50 μl of anti-FLAG M2 affinity gel (Sigma) was added to each supernatant. A cocktail of protein inhibitors was added (Complete mini EDTA-free proteinase inhibitors mix; Roche), and the reactions were incubated overnight at 4°C in a rotating wheel. Beads were pelleted at 1,000 × g for 5 min, the supernatants were transferred to a fresh tube, and the beads were washed three times with 10 ml of TBST (20 mM Tris [pH 7.6], 150 mM NaCl, 0.05% Tween 20). Bound proteins were then eluted from the beads in 25 μl of 0.1 M glycine (pH 3.5) after incubation for 5 min at room temperature with gentle shaking. The resin was pelleted by centrifugation, and the supernatant containing the eluted proteins was transferred to a fresh tube containing 2 μl of 1 M Tris (pH 8). The elution step was performed twice, and the eluates were pooled, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western immunoblotting with monoclonal antibodies directed to SipB and SipC according to standard procedures.
Evaluation of SPI-1 TTSS-dependent phenotypes.
The functionality of the SPI-TTSS in strains carrying 3×Flag-tagged versions of SipB, SipC, or SipD was determined by examining the ability of these strains to secrete TTSS-dependent proteins and their ability to enter into cultured epithelial cells. These assays were carried out as previously described (3, 10).
Immunofluorescence staining of the TTSS translocases prior to cell contact.
Overnight cultures of the strains to be tested were diluted 1:20 in 0.3 M NaCl LB and grown under aeration to an optical density at 600 nm of 0.9. One milliliter of each culture was spun down at 6,000 rpm for 5 min. The supernatant was discarded, and the pellet was resuspended in 200 μl of a 4% paraformaldehyde solution in phosphate-buffered saline (PBS). Bacterial cells were fixed for 30 min and then washed with PBS once and resuspended in 1 ml of PBS. Then, 10 μl of this suspension was applied on top of poly-l-d-lysine-treated coverslips and allowed to settle by gravity for 5 h or overnight. The coverslips were washed with PBS, treated with 3% bovine serum albumin-0.1% Triton X-100 in PBS, and labeled with the following primary antibodies: mouse monoclonal anti-Flag M2 (Sigma) at a 1:10,000 dilution and rabbit polyclonal anti-S. Typhimurium lipopolysaccharide (LPS; Difco Laboratories) at a 1:2,000 dilution. Secondary anti-rabbit antibody conjugated to Alexa 594 and biotinylated anti-mouse antibody were used at 1:2,000, followed by fluorescein isothiocyanate-conjugated anti-biotin antibody. Confocal images were captured by using a Improvision spinning disc confocal microscope and processed by using Volocity software (version 4.0; Improvision).
Immunofluorescence staining of the TTSS translocases after cell contact.
Cultured Henle-407 cells were infected with the different Salmonella strains at a multiplicity of infection of 50. Infected cells were fixed 30 min postinfection with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, and then treated with an avidin/biotin blocking kit (Vector Laboratories). After these treatments staining was performed as described above.
RESULTS
A Salmonella Typhimurium ΔsopE ΔsopE2 ΔsopB mutant strain can intimately attach to but cannot enter into nonphagocytic cells.
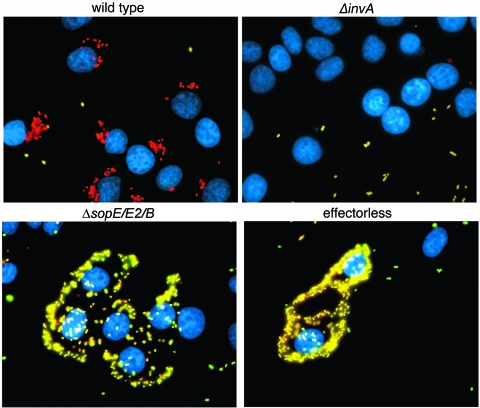
S. Typhimurium induces its own actin-cytoskeleton-dependent internalization into nonphagocytic cells by delivering the SPI-1 TTSS effector proteins SopE, SopE2, and SopB which, in a functionally redundant manner, activate Rho-family GTPases. SopE and SopE2 are nucleotide exchange factors that target Cdc42, Rac, and RhoG (15), while SopB is a phosphoinositide phosphatase that stimulates entry by activating the endogenous RhoG exchange factor SGEF (27). Although introduction of a loss-of-function mutation in any of these effectors only marginally affects entry, an S. Typhimurium strain simultaneously lacking these three effectors is unable to enter nonphagocytic cells (40). During the course of experiments analyzing this mutant strain, we observed that, although unable to enter into cells, the ΔsopE ΔsopE2 ΔsopB mutant readily associated with cultured epithelial cells (Fig. 1). Such association was very robust as it resisted the extensive handling associated with the immunofluorescence staining protocol (see Materials and Methods). Intimate attachment of bacteria encoding TTSS has been reported to be essential for the function of this protein delivery system. However, TTSS-dependent intimate attachment has not been previously reported in S. Typhimurium most likely because the rapid internalization of these bacteria has prevented its observation. Although in some cases the intimate attachment of bacteria encoding TTSS has been shown to be mediated by proteins associated or delivered by this system (e.g., tir in enteropathogenic E. coli) (20, 25), in other cases this intimate attachment has been shown to be mediated by unrelated proteins (e.g., invasin or YadA in Yersinia spp.) (16, 39). We therefore investigated whether the intimate attachment of the ΔsopE ΔsopE2 ΔsopB S. Typhimurium mutant strain was dependent on a functional SPI-1 TTSS. Using an immunofluorescence assay, we compared the association of the ΔsopE ΔsopE2 ΔsopB S. Typhimurium mutant with the association of a ΔinvA strain, a mutant that lacks a functional SPI-1 TTSS. This assay is more stringent than a standard, CFU-based assay, and therefore a tighter association is required for bacteria to remain associated to host cells after sample processing (for comparison, see the results in Fig. S2 in the supplemental material showing the attachment of the different strains assayed by a standard CFU-based assay). As shown in Fig. 1, the S. Typhimurium invA mutant strain showed a much reduced ability to associate with cells compared to the ΔsopE ΔsopE2 ΔsopB S. Typhimurium strain. These results indicate that the presence of a functional TTSS confers on S. Typhimurium the ability to associate intimately with cells. Although this intimate association must precede bacterial entry, most likely it has been previously overlooked in wild-type S. Typhimurium because these bacteria undergo uptake into cells very rapidly, thereby effectively preventing the observation of this transient early stage of the internalization process.
FIG. 1.
The invasion-defective but type III secretion-proficient ΔsopE ΔsopE2 ΔsopB S. Typhimurium strain intimately attaches to cultured epithelial cells in a type III secretion-dependent manner. Cultured Henle-407 cells were infected with wild-type S. Typhimurium, an ΔinvA mutant, which is defective in the SPI-1 TTSS; a ΔsopE ΔsopE2 ΔsopB mutant, which is TTS proficient but lacks the effectors that mediate entry into cells; or a mutant that lacks all effectors of the SPI-1 TTSS (effectorless), which is also defective for entry. Infected cells were stained according to a protocol that can distinguish extracellular (appearing yellow in these images) from intracellular (appearing red in these images) bacteria and stained with DAPI to visualize the nucleus of epithelial cells (blue) (see Materials and Methods for experimental details).
The SPI-1 TTSS-associated translocon is required for intimate attachment.
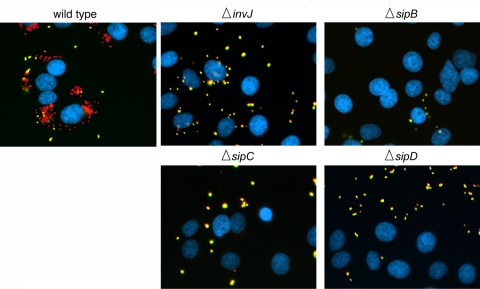
The observation that a functional SPI-1 TTSS is required for the intimate attachment of S. Typhimurium to host cells indicated that a protein(s) secreted via this pathway must be required for this association. Since a strain lacking all effectors secreted by this pathway was still able to intimately associate with cells (Fig. 1), some components of the needle complex or the associated translocases must be responsible for this intimate association. The ΔinvA strain, which was defective for intimate attachment, has a fully assembled base of the needle complex, but it lacks the needle substructure since secretion and assembly of its main subunit, PrgI, requires a functional TTSS (35). Indeed, it is an attractive hypothesis that the needle of the TTSS could recognize a yet-unknown receptor on the cell surface, and this interaction could be responsible for the intimate attachment observed in the absence of entry but in the presence of a functional SPI-1 TTSS. Alternatively, the deployment of the translocases to form the translocon through which the effectors are translocated could be responsible for the close association. To distinguish between these possibilities, we assayed the attachment of the S. Typhimurium ΔinvJ mutant strain. The TTSS of this mutant strain is unable to undergo “substrate switching” and therefore is “locked” in a secretion mode only compatible with the secretion of the needle protein PrgI and the inner rod protein PrgJ (22). As a result, this strain exhibits longer needles on its surface but is unable to secrete any other protein normally secreted by the SPI-1 TTSS including the protein translocases (i.e., SipB, SipC, or SipD). As shown in Fig. 2 and 3, the ΔinvJ mutant strain was unable to intimately attach to cultured intestinal epithelial cells, suggesting that the needle per se is unable to mediate this association. We then tested the intimate association of S. Typhimurium strains lacking the protein translocases SipB, SipC, or SipD. Each one of these mutant strains can assemble a fully functional needle complex and can secrete (but cannot translocate) all proteins traveling the SPI-1 TTS pathway. All of these mutant strain were equally defective for intimate attachment (Fig. 2 and 3), indicating that all of the translocases, and presumably the assembly of the translocon, are required for intimate attachment. In summary, these results indicate that the TTSS-dependent intimate association of S. Typhimurium with cultured mammalian cells is mediated by the protein translocases SipB, SipC, and SipD.
FIG. 2.
The S. Typhimurium type III secretion-dependent intimate attachment to cultured epithelial cells requires the protein translocases SipB, SipC, and SipD. Cultured Henle-407 cells were infected with wild-type S. Typhimurium or the indicated isogenic strains, stained according to a protocol that can distinguish extracellular (appearing yellow in these images) from intracellular (appearing red in these images) bacteria, and stained with DAPI to visualize the nucleus of epithelial cells (blue) (see Materials and Methods for experimental details). The ΔinvJ strain can assemble the needle substructure of the needle complex but is unable to secrete the protein translocases SipB, SipC, and SipD.
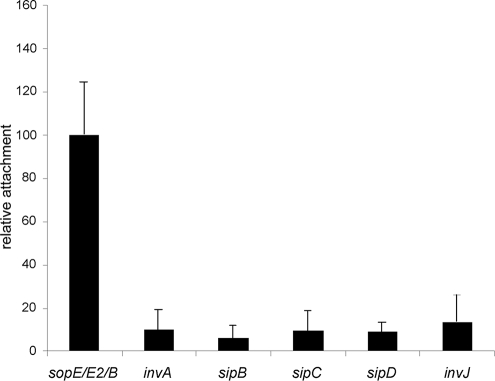
FIG. 3.
Relative attachment levels of different isogenic strains of S. Typhimurium. Cultured Henle-407 cells were infected with the indicated isogenic strains of S. Typhimurium and stained according to a protocol that can distinguish extracellular from intracellular bacteria, and the numbers of attached bacteria per cell were counted after the acquisition of several images (see Materials and Methods for experimental details). The values have been standardized relative to the ΔsopE ΔsopE2 ΔsopB mutant strain (considered 100%) and represent the mean ± the standard deviation of at least three independent experiments in which the numbers of bacteria attached to a minimum of 100 epithelial cells were determined. The difference between the values corresponding to the ΔsopE ΔsopE2 ΔsopB strain compared to any of the other strains in the assay were statistically significant (P < 0.0001). The differences between the values corresponding to the invA, sipB, sipC, sipD, and invJ mutants were not statistically significant (P ≥ 0.250) compared to each other.
SipD, but not SipB or SipC, is located on the surface before interaction with cells.
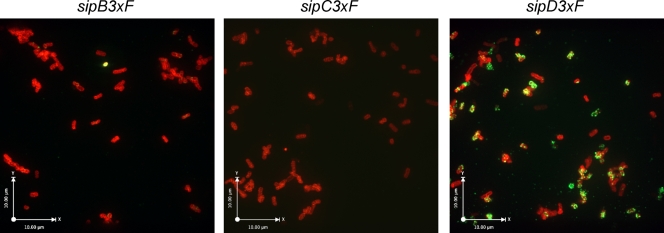
The fact that S. Typhimurium ΔsipB, ΔsipC, or ΔsipD mutant strains cannot intimately attach to cultured cells coupled with the observation that no effector proteins are involved in this process, suggested at least two possibilities: (i) the three translocases form a complex that mediates this attachment, or (ii) one or more translocases are required for the proper deployment of the translocase(s) that mediates attachment. To facilitate the study of the translocases in this process, we generated epitope-tagged versions of SipB, SipC, and SipD. Since the addition of a tag at the carboxyl terminus interfered with the function of these proteins (data not shown), and their extreme amino terminus is required for their secretion, we placed the epitope tag immediately after the first 20 amino acids and introduced them into the wild-type S. Typhimurium chromosome by allelic exchange. The epitope placed at that position did not interfere with the secretion or the function of any of the translocases, as demonstrated by their ability to mediate the translocation of effectors requires for bacterial uptake into cells (Fig. 4). We then examined the localization of the translocases on the bacterial surface by immunofluorescence confocal microscopy of nonpermeabilized bacterial cells after growth under conditions that result in the stimulation of expression of the SPI-1 TTSS. SipD was readily observed as puncta distributed over the bacterial surface (Fig. 5 and see Video S1 in the supplemental material). The pattern of the observed staining suggests that SipD may be localized at the tip of the needle complex prior to contact with target cells. In contrast, no SipB or SipC staining was observed on the bacterial surface after identical growth conditions and staining protocol. Therefore, the absence of staining suggests that SipB or SipC are not exposed on the bacterial surface prior to contact with the target cell. Alternatively, it is also possible that the conformation of SipB or SipC on the bacterial surface prior to bacterial contact is such that the epitope tag is not exposed on the surface. However, this is unlikely since (i) the amino termini of TTSS secreted proteins are thought to be unstructured and (ii) soluble SipB and SipC can be immunoprecipitated under nondenaturing conditions using the antibody directed to the epitope tag (see Fig. S3 in the supplemental material), suggesting that the epitope is not buried on the protein structure. In summary, these observations indicate that SipD is surface exposed prior to contact with target host cells and that, as previously suggested for its close homolog IpaD (6), it may be localized at the tip of the needle complex and therefore potentially positioned to mediate intimate attachment.
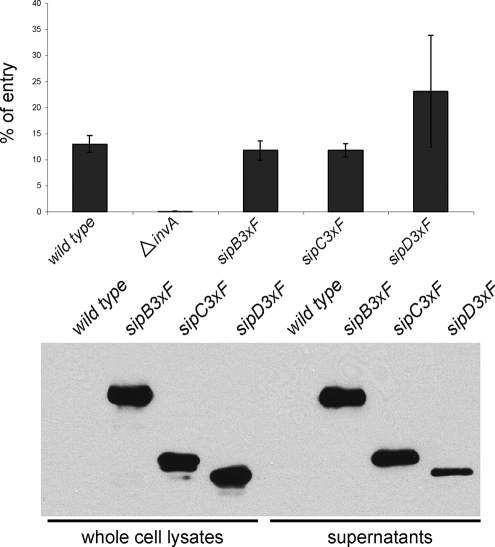
FIG. 4.
The epitope-tagged type III secretion translocases SipB, SipC, and SipD retain wild-type activity. (A) S. Typhimurium strains encoding epitope-tagged SipB, SipC, or SipD are fully competent for entry into cultured epithelial cells. Henle-407 cells were infected with the indicated strains, and the internalization levels were determined by the gentamicin protection assay (see Materials and Methods). Values represent the percentage of the inoculum that survived the gentamicin treatment as a consequence of internalization and are the mean ± the standard deviation of three independent determinations. The differences between the values corresponding to the wild-type, sipB3×F, sipC3×F, and sipD3×F strains were not statistically significant compared to each other (P ≥ 0.150), although the difference between the values of any of these strains and the ΔinvA strain were statistically significant (P < 0.001). (B) The epitope-tagged protein translocases are efficiently secreted into cultured supernatants. Whole-cell lysates and cultured supernatants of the S. Typhimurium strains expressing epitope-tagged SipB, SipC, or SipD (as indicated) were analyzed by Western immunoblotting as indicated in Materials and Methods.
FIG. 5.
SipD, but not SipB or SipC, is surface exposed before bacterial contact with host cells. After growth in LB broth under conditions that stimulate the expression of the SPI-1 TTSS, S. Typhimurium strains expressing the indicated epitope-tagged translocases were immobilized on poly-lysine-coated slides and stained with an antibody directed to the epitope (green) under conditions that do not permeabilize the bacterial envelope and counterstained with an antibody directed to the LPS (red). Bacteria were then visualized under a fluorescence confocal microscope.
SipB and SipC become surface exposed upon contact with mammalian cells.
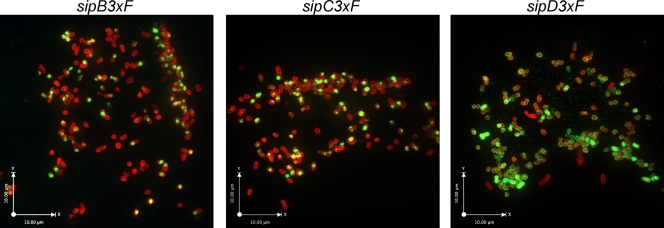
We have previously shown that contact with epithelial cells stimulates secretion through the SPI-1 TTSS (4, 41). We therefore examined the localization of the translocases upon contact with epithelial cells. Since the rapid SPI-1 TTSS-mediated bacterial internalization significantly interferes with the examination of the fate of the translocases upon their deployment, we introduced the epitope-tagged versions of these proteins into the S. Typhimurium ΔsopE ΔsopE2 ΔsopB triple-mutant background. As shown above, this mutant strain can intimately attach to but is unable to enter cultured cells. Cultured intestinal epithelial cells were infected with the different strains, and at different times after infection the localization of the translocases was examined by immunofluorescence microscopy. As early as 10 min after infection, not only SipD but also SipB and SipC were observed on the bacterial surface (data not shown). Later in infection, more prominent staining of the three translocases was observed not only on the bacterial surface but also on the epithelial cell surface (Fig. 6). These results suggest that upon contact with host cells, SipB and SipC become surface exposed where they could be available to mediate intimate attachment.
FIG. 6.
SipB and SipC become surface exposed upon S. Typhimurium contact with epithelial cells. Cultured Henle-407 cells were infected with different strains of S. Typhimurium expressing epitope-tagged SipB, SipC, or SipD, as indicated. At 30 min after infection, the cells were fixed and stained with an antibody directed to the epitope (green) under conditions that do not permeabilize the bacterial envelope and counterstained with an antibody directed to the LPS (red). Bacteria were then visualized under a fluorescence confocal microscope.
Intimate attachment is mediated by SipD together with the translocases.
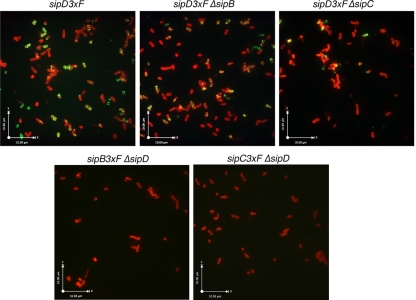
The fact that SipD is localized at the bacterial surface, presumably at the tip of the needle complex before contact with host cells, raises the possibility that SipD itself mediates intimate attachment. However, strains lacking either SipB or SipC are also defective for intimate attachment (Fig. 2), suggesting the possibility that these proteins may be required for the surface localization of SipD. We investigated this possibility by examining by immunofluorescence microscopy the localization of SipD in strains lacking either SipB or SipC. We found that strains lacking either SipB or SipC are also capable of displaying SipD at the surface (Fig. 7), indicating that the surface-localized SipD by itself is unable to mediate bacterial attachment. In addition, we observed no bacterial surface staining of SipB and SipC in the absence of SipD (Fig. 7), even though SipD is not required for the secretion of SipB or SipC. Taken together, these results suggest that a complex of SipD, SipB, and SipC mediates the intimate attachment of S. Typhimurium to host cells.
FIG. 7.
SipD is surface exposed in the absence of SipB or SipC. After growth in LB broth under conditions that stimulate the expression of the SPI-1 TTSS, S. Typhimurium strains (as indicated) expressing epitope-tagged SipD were immobilized in poly lysine coated slides and stained with an antibody directed to the epitope (green) under conditions that do not permeabilize the bacterial envelope and counterstained with an antibody directed to the LPS (red). Bacteria were then visualized under a fluorescence confocal microscope.
DISCUSSION
It is widely believed that translocation of bacterial proteins to mammalian cells via TTSS requires an intimate association between the bacteria and the host cell. Studies in Yersinia spp. have shown that such intimate association is promoted by surface proteins such as invasin (16) and YadA (39), which are not components or substrates of its TTSS. In contrast, in enteropathogenic E. coli, the intimate attachment is mediated by the joint activity of a TTSS-unrelated adhesin, intimin, which binds to a TTSS-translocated receptor, tir (20). Although several adhesins have been described in S. Typhimurium (36), the determinants involved in the intimate attachment presumably required for TTSS-mediated protein delivery have not been previously reported, most likely because these bacteria rapidly enter into host mammalian cells effectively hampering the study of such a phenotype. Using a strain of S. Typhimurium that is specifically defective in the TTSS effector proteins that mediate bacterial internalization (40), we have been able to detect the intimate association phenotype that has been observed in other bacteria encoding TTSS. We show here that the intimate association of S. Typhimurium with host cells required for TTSS-mediated protein delivery is mediated by the concerted activity of the TTSS protein translocases SipB, SipC, and SipD. Consistent with this conclusion, strains lacking any of these proteins were unable to intimately associate with host cells. Previous studies have shown that SipB and SipC are deployed to the host cell plasma membrane to mediate the passage of effector proteins through this membrane (4, 30). Although we have not specifically investigated its location, like its Shigella sp. close homolog IpaD (6), S. Typhimurium SipD is presumably located at the tip of the needle extension of the TTSS needle complex. Previous studies have shown that the S. Typhimurium SPI-1 TTSS, like other TTSS, is stimulated upon contact with host cells (41). However, it was unclear whether the translocases were deployed on the bacterial surface prior to bacterial contact with host cells. Our results indicate that SipD is present at the tip of the needle complex prior to contact with host cells since immunofluorescence staining with a protocol that does not permeabilize the bacterial envelope readily detected this protein on the bacterial surface after growth in vitro. In contrast, the same staining protocol did not detect the presence of SipB or SipC on the bacterial surface prior to cell infection, indicating that these translocases may not be exposed prior to cell contact. These observations suggest that SipD is properly positioned to play a central role in mediating the association of S. Typhimurium with host cells, which is consistent with the observation that a ΔsipD S. Typhimurium mutant strain is defective for intimate attachment. However, S. Typhimurium strains lacking SipB or SipC were also unable to associate intimately with host cells even though SipD could be readily detected on the surface of these mutant strains, suggesting that SipD plays an essential but not sufficient role in the intimate association of S. Typhimurium with host cells. In addition, with its location at the tip of the needle complex, SipD would be positioned to work as a sensor for host cell contact resulting in the “activation” of the TTS apparatus as previously suggested for its Shigella sp. homolog IpaD (34). Subsequent to contact with host cells, both SipB and SipC were detected on the bacterial surface as well as on the host cell membrane. These observations suggest the possibility that SipD mediates the association of S. Typhimurium to host cells by interacting with the membrane-deployed SipB and SipC. This would be analogous to what has been suggested for LcrV, the needle tip protein of Yersinia spp. that interacts with the translocases YopB and YopD (29). This hypothesis would also be consistent with the observation that SipB, SipC, and SipD can interact with one another and are equally required for bacterial attachment. However, more studies will be required to understand the details of the coordinated deployment of the TTSS-associated translocases and the nature of their interactions at the plasma membrane.
Supplementary Material
Acknowledgments
This study was supported by Public Health Service grant AI30492 to J.E.G.
Editor: A. Camilli
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akeda, Y., and J. E. Galan. 2004. Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J. Bacteriol. 1862402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, L. M., S. Hobbie, and J. E. Galan. 1996. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 2742115-2118. [DOI] [PubMed] [Google Scholar]
- 3.Collazo, C., and J. E. Galán. 1996. Requirement of exported proteins for secretion through the invasion-associated type III system in Salmonella typhimurium. Infect. Immun. 643524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collazo, C., and J. E. Galán. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24747-756. [DOI] [PubMed] [Google Scholar]
- 5.Collazo, C. M., M. K. Zierler, and J. E. Galán. 1995. Functional analysis of the Salmonella typhimurium invasion genes invI and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol. Microbiol. 1525-38. [DOI] [PubMed] [Google Scholar]
- 6.Espina, M., A. Olive, R. Kenjale, D. Moore, S. Ausar, R. Kaminski, E. Oaks, C. Middaugh, W. Picking, and W. Picking. 2006. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 744391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman, M., and G. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219151-158. [DOI] [PubMed] [Google Scholar]
- 8.Galan, J. E. 2007. SnapShot: effector proteins of type III secretion systems. Cell 130192. [DOI] [PubMed] [Google Scholar]
- 9.Galán, J. E. 2001. Salmonella interaction with host cells: type III Secretion at Work. Annu. Rev. Cell Dev. Biol. 1753-86. [DOI] [PubMed] [Google Scholar]
- 10.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sc. USA 866383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galán, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444567-573. [DOI] [PubMed] [Google Scholar]
- 13.Grosdent, N., I. Maridonneau-Parini, M. Sory, and G. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 704165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 155812-5823. [PMC free article] [PubMed] [Google Scholar]
- 15.Hardt, W.-D., L.-M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galán. 1998. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93815-826. [DOI] [PubMed] [Google Scholar]
- 16.Isberg, R. R., and J. M. Leong. 1990. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60861-871. [DOI] [PubMed] [Google Scholar]
- 17.Kaniga, K., J. C. Bossio, and J. E. Galán. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol. Microbiol. 13555-568. [DOI] [PubMed] [Google Scholar]
- 18.Kaniga, K., D. Trollinger, and J. E. Galán. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1777078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga, K., S. C. Tucker, D. Trollinger, and J. E. Galán. 1995. Homologues of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 1773965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny, B., and B. B. Finlay. 1997. Intimin-dependent binding of enteropathogenic Escherichia coli to host cells triggers novel signaling events, including tyrosine phosphorylation of phospholipase C-γ1. Infect. Immun. 652528-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galán, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280602-605. [DOI] [PubMed] [Google Scholar]
- 22.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galán. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 9710225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. H., and J. E. Galan. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51483-495. [DOI] [PubMed] [Google Scholar]
- 24.Marlovits, T. C., T. Kubori, A. Sukhan, D. R. Thomas, J. E. Galan, and V. M. Unger. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 3061040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nougayrède, J., P. Fernandes, and M. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell Microbiol. 5359-372. [DOI] [PubMed] [Google Scholar]
- 26.Parsot, C., R. Ménard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16291-300. [DOI] [PubMed] [Google Scholar]
- 27.Patel, J. C., and J. E. Galan. 2006. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 175453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host actin cytoskeleton by Salmonella: all in the name of entry. Curr. Opin. Microbiol. 810-15. [DOI] [PubMed] [Google Scholar]
- 29.Sarker, M. R., C. Neyt, I. Stainier, and G. R. Cornelis. 1998. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J. Bacteriol. 1801207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer, C., E. Cooper, and S. Miller. 2000. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 371133-1145. [DOI] [PubMed] [Google Scholar]
- 31.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 11974-88. [DOI] [PubMed] [Google Scholar]
- 32.Stavrinides, J., H. McCann, and D. Guttman. 2008. Host-pathogen interplay and the evolution of bacterial effectors. Cell Microbiol. 10285-292. [DOI] [PubMed] [Google Scholar]
- 33.Stebbins, C. E., and J. E. Galan. 2003. Priming virulence factors for delivery into the host. Nat. Rev. Mol. Biol. 4738-743. [DOI] [PubMed] [Google Scholar]
- 34.Stensrud, K., P. Adam, C. La Mar, A. Olive, G. Lushington, R. Sudharsan, N. Shelton, R. Givens, W. Picking, and W. Picking. 2008. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J. Biol. Chem. 28318646-18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galán. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 1831159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Velden, A., A. Bäumler, R. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 662803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 142461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 5501-511. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Y., and R. R. Isberg. 1993. Cellular internalization in the absence of invasin expression is promoted by the Yersinia pseudotuberculosis yadA product. Infect. Immun. 613907-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galán. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39248-259. [DOI] [PubMed] [Google Scholar]
- 41.Zierler, M. K., and J. E. Galán. 1995. Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect. Immun. 634024-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.