Abstract
Exotoxin A is a major virulence factor of Pseudomonas aeruginosa. This toxin binds to a specific receptor on animal cells, allowing endocytosis of the toxin. Once in endosomes, the exotoxin can be processed by furin to generate a C-terminal toxin fragment that lacks the receptor binding domain and is retrogradely transported to the endoplasmic reticulum for retrotranslocation to the cytosol through the Sec61 channel. The toxin then blocks protein synthesis by ADP ribosylation of elongation factor 2, thereby triggering cell death. A shorter intracellular route has also been described for this toxin. It involves direct translocation of the entire toxin from endosomes to the cytosol and therefore does not rely on furin-mediated cleavage. To examine the implications of endosomal translocation in the intoxication process, we investigated whether the toxin required furin-mediated processing in order to kill cells. We used three different approaches. We first fused to the N terminus of the toxin proteins with different unfolding abilities so that they inhibited or did not inhibit endosomal translocation of the chimera. We then assayed the amount of toxin fragments delivered to the cytosol during cell intoxication. Finally we used furin inhibitors and examined the fate and intracellular localization of the toxin and its receptor. The results showed that exotoxin cytotoxicity results largely from endosomal translocation of the entire toxin. We found that the C-terminal fragment was unstable in the cytosol.
Several bacterial toxins are cleaved by cell proteases upon uptake by mammalian cells. Site-specific processing by furin-like endoproteases is often associated with toxin activation and is therefore a key step of the intoxication process. Indeed, toxins such as anthrax protective antigen, aerolysin, Shiga toxin, and diphtheria toxin (DT) require furin-mediated activation in order to intoxicate cells (47). Exotoxin A (PE) that is secreted by Pseudomonas aeruginosa can also be cleaved by furin in vivo and in vitro (19). PE is produced as a single polypeptide chain composed of three structural and functional domains (domains I, II, and III) that are successively involved in the intoxication process (3). This process starts with the binding of domain I to the low-density receptor-related protein (LRP) (25), a huge protein synthesized in a 600-kDa precursor form that is cleaved by furin into 515- and 85-kDa subunits (16). The LRP is the only functional receptor for PE since LRP-deficient cells are resistant to PE (53). This receptor enables endocytosis of PE and various other ligands (17). Once PE is internalized, exposure to a low endosomal pH triggers major conformational changes within its structure and leads to domain II-mediated insertion into the endosome membrane (32). After this step, two pathways have been documented for continuation of the PE intoxication process. Either the entire toxin could cross the endosome membrane (2, 32), or the toxin could be processed by furin after Arg279 within an exposed loop between the two first helices of the translocation domain (35). Then reduction of the Cys265-Cys287 disulfide generates two fragments, a 28-kDa fragment corresponding to the N-terminal part of PE and the 37-kDa carboxyl-terminal fragment of PE, which includes most of domain II and the entire enzymatic domain (domain III).
A number of studies suggested that the 37-kDa fragment can be retrogradely transported to the endoplasmic reticulum (ER) for retrotranslocation to the cytosol through the Sec61 complex. It was first observed that a PE carboxyl-terminal sequence (REDLK), which resembles the ER retrieval motif KDEL, is required for toxicity (8) and that it is possible to enhance PE toxicity in some types of cells by replacing its carboxyl-terminal sequence by KDEL (43). Moreover, brefeldin A, a drug that blocks transport between the Golgi apparatus and the ER, protects cells from PE (43, 56), and perturbation of the KDEL-mediated ER retrieval system by overexpression of lysozyme-KDEL prevents PE toxicity (20). It should nevertheless be noted that LRP requires a receptor-associated protein that ensures proper folding and export of the LRP molecules from the ER (55) and that receptor-associated protein retention in the ER relies on the KDEL retrieval system (7). Whatever the pathway enabling cytosolic delivery of PE, once PE is in the cytosol, the catalytic domain rapidly inactivates by ADP ribosylation elongation factor 2 (EF2), a key elongation factor of protein synthesis whose inactivation quickly induces cell death (18).
It is still not clear whether PE requires furin-mediated processing in order to kill cells. This possible requirement was first studied by introducing mutations within or close to the PE cleavage site. Depending on the cell type, from 0.5 to 20% of PE molecules are processed by furin, and mutations within the processed loop have produced contrasting results. Mutant toxins were either as toxic as or less toxic than PE. Some attenuated mutant proteins, such as PE-Gly276, were not cleaved by furin (35), while others, such as PE-Ala281 and mutant proteins in which the processed loop was engineered to resemble the Arg-rich DT loop, were more efficiently processed than native PE (9, 57). Hence, there was a lack of correlation between PE processing efficiency and cytotoxicity, and the mutagenesis approach did not allow workers to determine clearly whether PE required furin-mediated processing in order to kill cells.
A second approach for studying this requirement made use of furin-deficient cells, such as CHO RPE.40 or FD11 cells, that were found to be resistant to PE (14, 33). Nevertheless, the LRP also requires furin processing for proper functioning (15, 54). The lack of LRP processing within furin-deficient CHO cells was reported to produce a receptor deficient in endocytic function (54) and/or export from the ER to the cell surface (15, 24). It was not clear, therefore, whether protection from PE was due to the lack of PE processing or to the absence of ectopic LRP or whether both effects were involved in protection (15). The molecular bases for the PE resistance of furin-deficient cell lines remain to be determined.
Because of the presence of the receptor binding domain, entire PE remains bound to the LRP, even at low endosomal pH (48). Furin processing is therefore required to generate the active fragment that lacks the receptor binding domain and can be retrogradely transported to the ER (37), while unprocessed PE is thought to translocate only across the endosome membrane (2). Hence, a strict requirement for PE processing in order to kill cells should indicate that PE cytotoxicity relies only on retrograde transport, while, conversely, implication of unprocessed PE molecules in toxicity should show that endosomal translocation is also involved in cytotoxicity.
In this study, we used three different complementary approaches to examine the role of PE processing in the intoxication process.
MATERIALS AND METHODS
Reagents and cells.
Anti-actin and chemicals were obtained from Sigma, molecular biology enzymes were obtained from New England Biolabs, rabbit anti-goat immunoglobulin G (IgG) was obtained from Nordic, protein A-Sepharose was obtained from GE Healthcare, and furin inhibitors were obtained from Calbiochem. Rabbit anti-PE serum was obtained from Sigma; goat anti-PE antibodies were obtained from List Biological Laboratories and were affinity purified (2). These polyclonal antibodies detect both entire PE and the 55 kDa- and 37-kDa fragments and were used for immunofluorescence and Western blotting. Anti-cytochrome c was obtained from BD Biosciences, and anti-RNase A was obtained from Chemicon. Rabbit anti-dihydrofolate reductase (DHFR) and anti-LRP (Ab377) antibodies were provided by Karin Becker (Germany) and Joachim Herz (Southwestern Medical Center, Dallas, TX), respectively. Rat anti-mouse transferrin (Tf) receptor monoclonal antibody R17.217 was used as ascites fluid, while anti-LRP monoclonal antibody 11H4 (a gift from Emmanuelle Liaudet-Coopman, INSERM, Montpellier, France) was used as cell culture supernatant. The cell lines used for this study were L929 mouse fibroblasts (ATCC), which are highly sensitive to PE and were used for cytotoxicity assays and PE processing studies (34), BW mouse lymphocytes, which were used for cell fractionation (2), and furin-deficient CHO FD11 cells, which have been described previously (14). Cytosol from mouse BW lymphocytes was purified as described previously for HeLa S3 cells (49). Tf and ricin were labeled with fluorescein and colloidal gold, respectively, as indicated previously (5). Toxins were radiolabeled with 125I using the Iodogen method (2).
Production of PE fusion proteins.
The pET3d expression vector containing the PE coding sequence after the proOmpA signal sequence (32) was used to construct expression plasmids. The coding sequences of mouse DHFR (6), bovine pancreatic RNase A (22), and rat apocytochrome c (45) were fused to the N terminus of PE. The chimera precursors consisted of the OmpA signal sequence followed by the first five residues from mature OmpA to enable efficient cleavage by the signal peptidase (13), the fused protein, a spacer peptide (Ala-Ser-Ala-Ser-Thr-Pro-Glu-Pro-Asp-Pro-Glu-Lys-Leu [6]), and PE. Each PCR-amplified DNA was sequenced.
For production, Escherichia coli BL21(λDE3) cells carrying the recombinant plasmid were grown at 37°C in L broth containing ampicillin (100 μg/ml). Isopropyl-β-d-thiogalactoside (1 mM) was added when the optical density at 600 nm reached 0.6, and bacteria were harvested 2 h later. The periplasmic fraction was prepared, dialyzed against 20 mM Tris-HCl (pH 7.6), and purified by anion-exchange chromatography, first on a Q-Sepharose column and then on a Mono-P column (GE Healthcare), except for DHFR-PE, which was purified on methotrexate (MTX)-agarose (6). The identity of the conjugates was confirmed by Western blotting against PE and the fused protein. Chimeras were at least 90% pure.
Assay for chimera catalytic activity.
A fusion protein (2 to 0.5 pmol; 10 μl) was added to 50 μl buffer (10 mM dithiothreitol [DTT], 4 mM EDTA, 100 mM Tris-HCl; pH 8.2), 20 μl purified EF2 (2), and 20 μl [32P]NAD (50 ng; 100,000 cpm). PE and the buffer were used as positive and negative controls, respectively. After 2 h of incubation at 30°C, the reaction was stopped by adding 1 ml of ice-cold 10% trichloroacetic acid before filtration onto glass fiber filters and scintillation counting.
PE-processing studies.
The processing efficiency was measured by quantification of PE or chimeras and their cleavage products after toxin internalization and translocation, as described previously (32). Briefly, L929 cells were incubated with 30 nM PE or a fusion protein for 4 h and then washed before a 30-min chase. Where indicated below, furin inhibitors (60 μM) were added during cell labeling and the chase, while an equivalent volume of solvent (dimethyl sulfoxide) was added to control cells. This inhibitor concentration is within the efficient range (10, 42), and higher concentrations were not more effective (data not shown). Cells were then lysed for PE immunoprecipitation before sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and anti-PE Western blotting. Membrane-bound native IgGs were labeled using recombinant protein G-peroxidase before detection using ECL+ (GE Healthcare). Films were scanned, and band quantification was performed using ImageQuant (GE Healthcare).
Toxicity measurement.
L929 cells were seeded in 96-well plates, and toxin solutions were added 4 h later. After 24 h of incubation at 37°C, [35S]methionine plus [35S]cysteine (Trans Label; 8 kBq) was added, and the preparations were incubated for 24 h. Medium was then removed, and cells were solubilized in 0.1 N NaOH before precipitation of the proteins with trichloroacetic acid. Proteins were collected on glass fiber filters, washed with 5% trichloroacetic acid, and then dried for determination of the radioactivity. Background incorporation was determined by using cells treated with 1 mM cycloheximide (32).
Kinetics of protein synthesis inhibition.
L929 cells were treated with 100 pM toxin, and 1-h pulse incorporation of [35S]methionine was performed at various times after the beginning of intoxication. Where indicated below, furin inhibitors were used at a concentration of 60 μM. Translocation activity (expressed in h−1) was calculated by determining the inverse of the time required for 100 pM toxin to inhibit protein synthesis by 50% (32).
Cell-free translocation assay.
The cell-free translocation assay has been described elsewhere (2). Briefly, BW mouse lymphocytes were labeled for 40 min at 37°C with 100 nM 125I-chimera, with 125I-PE, or with 125I-Tf as a control. Cells were then washed and cooled on ice, ricin-gold was added, and the preparations were incubated for 30 min at 4°C before cell homogenization and separation of crude membranes from the postnuclear supernatant using a sucrose step gradient. Endosomes were collected from the 30% sucrose-20% sucrose interface and washed before resuspension in translocation buffer. Cytosol and ATP-MgCl2 were then added at final concentrations of 3 mg protein/ml and 10 mM, respectively. After incubation for 1 h at 37°C, endosomes were separated from the medium by ultracentrifugation before counting. Chimera translocation activity was calculated by determining the (supernatant radioactivity)/(supernatant radioactivity + pellet radioactivity) ratio (expressed as a percentage) and was linear with time for up to 2 h. For analysis of the translocated proteins, the proteins were precipitated (51) before separation by SDS-PAGE and autoradiography.
Preparation of cytosol from PE-treated cells.
Subconfluent L929 cells were incubated at 37°C with 20 nM PE (in the presence or absence of 50 μM monensin) for the periods of time indicated below. Cells were washed in ice-cold phosphate-buffered saline (PBS) before a 30-min chase at 37°C in prewarmed culture medium was performed. They were then scraped, washed, and resuspended in a streptolysin O (SLO) solution (20 U/106 cells) in PBS containing 0.01% bovine serum albumin, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (Complete; Roche). After 1 h of incubation at 0°C, 200 μl of ice-cold RPMI containing 3% fetal calf serum was added to neutralize unbound SLO, and cells were incubated at 37°C for 15 min to permeabilize the plasma membrane. Samples were centrifuged, and each supernatant was frozen at −80°C for assay of PE fragments or EF2-ADP ribosylation activity.
Analysis of cytosolic PE fragments.
Cytosol (350 μl) was mixed with RPMI containing 10 mM phenylmethylsulfonyl fluoride, 10% sodium deoxycholate, and 10% Nonidet P-40. Then 2 μl of goat anti-PE antibody was added to the mixture, and after 1 h of incubation on a rotating wheel, 20 μl of protein A-Sepharose saturated with rabbit anti-goat IgG was added. After 1 h of incubation on the wheel, the immune complexes were recovered by centrifugation (250 × g for 1 min), washed twice with immunoprecipitation buffer, twice with PBS-0.05% Tween, and once with PBS and then eluted by 5 min of boiling in SDS-PAGE reducing sample buffer. Proteins were separated by SDS-PAGE and then transferred to nitrocellulose, which was incubated with goat anti-PE antibodies (0.8 μg/ml blocking buffer). Membrane-bound native IgGs were labeled using recombinant protein G-peroxidase (1 μg/ml blocking buffer; Sigma), and after 1 h of incubation detection was performed using an ECL+ kit (GE Healthcare). Films were exposed in their linear range of detection and scanned for band quantification using ImageQuant (GE Healthcare).
Assays for cytosolic EF2 ADP ribosylation activity.
Cytosol (40 μl) was mixed with 40 μl buffer (10 mM DTT, 4 mM EDTA, 100 mM Tris-HCl; pH 8.2), 15 μl purified EF2 (2), and 5 μl [32P]NAD (50 ng; 1.7 kBq). After 2 h of incubation at 30°C, the reaction was stopped by adding SDS-PAGE reducing sample buffer and boiling. Gels were dried for autoradiography. Films were scanned for quantification of EF2 labeling using ImageQuant (GE Healthcare).
LRP immunoprecipitation.
Where indicated below, cells were first treated with 40 nM PE for 4 h in the presence or absence of 60 μM furin inhibitor. They were then washed in PBS and lysed in 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 50 mM HEPES (pH 7.5) containing a protease inhibitor cocktail (Complete; Roche Molecular Biochemicals) for 20 min on ice. The lysate was centrifuged for 15 min at 10,000 × g before rabbit anti-LRP and protein G-Sepharose were added to the supernatant. After 2 h of incubation on a rotating wheel at 4°C, the beads were washed five times in lysis buffer and finally resuspended in SDS-PAGE reducing sample buffer. Proteins were separated on 4 to 8% acrylamide gradient gels (RunBlue; Euromedex), and Western blotting was performed using rabbit anti-LRP.
Confocal microscopy.
To examine the intracellular routing of PE fusion proteins, L929 cells were grown on coverslips and labeled for 45 min at 37°C with 100 nM Tf-fluorescein and 15 nM PE or fusion protein in Dulbecco modified Eagle medium supplemented with 0.2 mg/ml bovine serum albumin. Cells were then washed, fixed in 3.7% paraformaldehyde, permeabilized with 0.015% saponin, and then labeled with affinity-purified goat anti-PE antibodies that were revealed using tetramethylrhodamine-labeled donkey anti-goat IgG (Nordic). To monitor the effect of furin inhibitors on PE and LRP endocytosis, cells were treated with 8 nM PE for 4 h before fixation and permeabilization. The Tf receptor, PE, and the LRP were then labeled using the R17.217 rat antibody, rabbit anti-PE antiserum, and the 11H4 mouse antibody, respectively. These antibodies were revealed using swine anti-rabbit (Nordic), goat anti-rat (Sigma), and donkey anti-mouse (Jackson Immunoresearch) antibodies labeled with fluorescein, tetramethylrhodamine, and Cy5, respectively. Cells were then mounted for examination with a Leica confocal microscope.
RESULTS
Fusion protein approach.
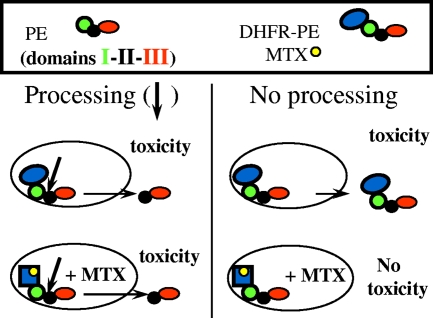
Small proteins whose molecular masses are in the 11- to 23-kDa range and which have various unfolding abilities (Table 1) were fused to the PE N terminus. We previously found that exogenous disulfide introduction inhibited PE translocation, indicating that this transmembrane transport requires toxin unfolding (32). The rationale for the fusion protein approach was therefore that a tightly folded protein should keep PE within endosomes to some extent, thereby protecting cells from entire molecules, while processing and generation of the active 37-kDa fragment should not be hampered by the small fused protein. We used (i) RNase A, a tightly folded protein with four intramolecular disulfides (50), to make RNase-PE; (ii) a control protein able to unfold easily, PE domain III, to prepare III-PE; (iii) apocytochrome c, a poorly folded protein that has some affinity for membranes (11), to obtain ApoC-PE; and (iv) DHFR, which is stabilized upon tight binding of its specific inhibitor MTX, which was thus expected to inhibit DHFR-PE transport to the cytosol. Indeed, when DHFR was fused to the enzymatic subunit of another toxin targeting the protein synthesis system, such as a DT or ricin A chain, it was transported to the cytosol in a manner that could be inhibited by MTX, showing that unfolding is a prerequisite for DHFR transport through membranes when this type of toxin is used as a vehicle (6, 23). Figure 1 shows the fusion protein approach as exemplified by using DHFR-PE.
TABLE 1.
Main features of the PE fusion proteins used in this studya
| N-terminal protein | Mol wt (103) | No. of disulfides | Fusion protein |
|---|---|---|---|
| None | PE | ||
| PE domain III | 22.8 | 0 | III-PE |
| DHFR | 20 | 0 | DHFR-PE |
| Apocytochrome c | 11.6 | 0 | ApoC-PE |
| RNase A | 13.7 | 4 | RNase-PE |
A protein was fused to the N terminus of PE between the signal sequence and the mature toxin. Short linkers were used at the protein-signal sequence junction to preserve correct processing of the signal sequence once the protein was in the periplasm and between the protein and the toxin to enable correct folding of the two parts of the chimera.
FIG. 1.
Schematic diagram of the fusion protein approach using the DHFR-MTX system. Arrows indicate the site of furin cleavage at the beginning of PE domain II. DHFR-PE efficiently kills cells in the absence of MTX (see Fig. 4). The presence of this drug stabilizes DHFR and inhibits its transmembrane transport in most biological systems. Therefore, toxicity that can be inhibited by MTX should correspond to endosome-trapped entire chimeras.
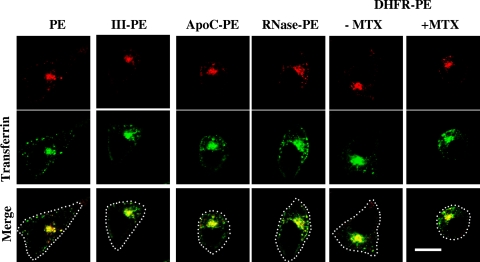
The fusion proteins were purified to homogeneity from E. coli periplasm, and we first checked whether the fused proteins altered PE intracellular routing. Upon uptake, PE is known to be concentrated within the endosome recycling compartment together with cellubrevin (36) or fluorescent Tf (2). In agreement with these findings, PE, Tf, and fusion proteins accumulated in this structure in L929 fibroblasts (Fig. 2). This was also the case for DHFR-PE in the presence of MTX, and the labeling pattern remained unchanged for up to ∼1.5 h, after which cells started to detach from the coverslips due to PE toxicity. These data indicated that the fused protein did not alter PE intracellular routing.
FIG. 2.
PE and fusion proteins are routed to the endosome recycling compartment. After labeling for 45 min with Tf-fluorescein and PE or the indicated fusion protein, L929 cells were processed for detection of PE by immunofluorescence. Where indicated, 200 nM MTX was present during cell labeling. Median optical sections were obtained using a confocal microscope. Both Tf and PE accumulated in the recycling compartment at the center of the cell. The dotted lines indicate the outlines of the cells in the merged images. Bar, 10 μm.
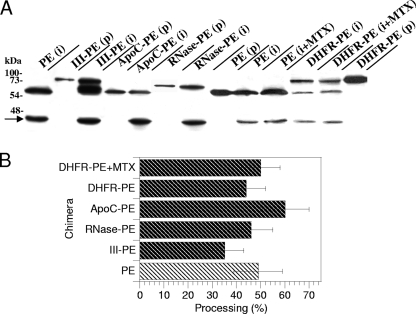
We then used purified EF2 to assess the catalytic activities of the fusion proteins. All of these proteins showed the same catalytic activity as PE, and there was no increase for III-PE (data not shown). The latter result might indicate that the additional catalytic site of III-PE is inactive. The presence of MTX did not affect DHFR-PE catalytic activity (data not shown). A key control experiment involved examining whether the fusion proteins could be efficiently cleaved by furin intracellularly. In this assay, ∼50% of PE molecules were processed to generate the active 37-kDa fragment. The fusion proteins all produced similar amounts of active fragment. The presence of MTX did not impair DHFR-PE processing by furin (Fig. 3). Together, these control experiments showed that a fused protein did not significantly affect PE routing to recycling endosomes, processing, or catalytic activity.
FIG. 3.
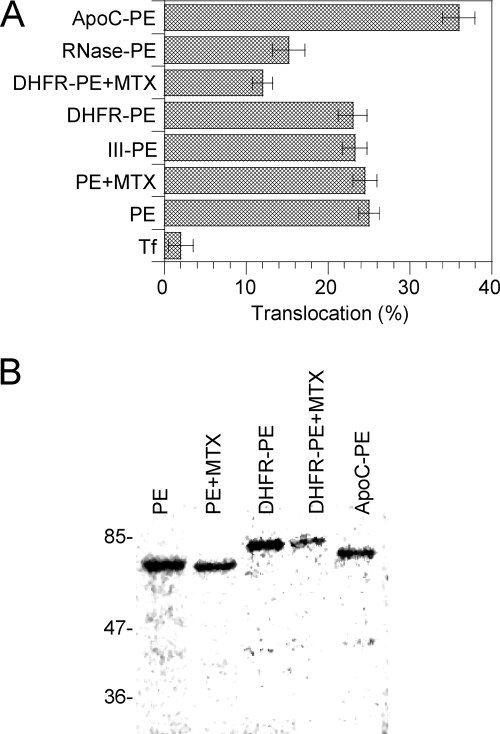
Processing efficiency of PE and fusion proteins. L929 cells were labeled for 4 h with PE or the indicated fusion protein before a 30-min chase. Where indicated, 150 nM MTX was present throughout the experiment. Cells were then lysed, and PE was immunoprecipitated. (A) Purified proteins (p) together with immmunoprecipitates (i) were separated by SDS-PAGE before immunoblotting and ECL detection. PE (66 kDa) is processed by furin into a 37-kDa fragment, whose position is indicated by an arrow, and a 28-kDa fragment. The latter fragment is quickly degraded (34, 57) and is not visible on films. (B) Quantification. Films from three independent experiments were scanned before band quantification. The furin processing efficiency (expressed as a percentage) was calculated as follows: 37-kDa fragment/(37-kDa fragment + entire protein).
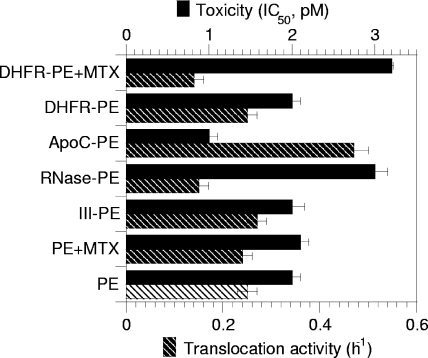
Nevertheless, when cytotoxicity was examined, significant differences were observed. While III-PE and DHFR-PE were as toxic as PE, RNase-PE was twofold less toxic than PE and ApoC-PE was twofold more toxic than PE (Fig. 4). Since toxicity is the result of a multistep process, and to obtain more specific insight into the translocation efficiency of the chimeras, we monitored the kinetics of protein synthesis inactivation using intact cells. Indeed, since translocation is the rate-limiting step of the intoxication process, the slope of the protein synthesis inactivation curve is directly proportional to the translocation activity (18, 32). Again, while III-PE and DHFR-PE translocated as efficiently as PE, the translocation of RNase-PE and the translocation of ApoC-PE were inhibited 50% and enhanced 80%, respectively, compared to the translocation of PE (Fig. 4). The enhanced translocation activity (and cytotoxicity) of ApoC-PE was probably due to the affinity of apocytochrome c for membranes (11). The decrease in translocation activity of RNase-PE compared to that of PE is likely explained by the fact that the tight folding of the protein (50) prevented its efficient transport across the endosome membrane. To directly examine these issues and confirm the previous findings, we tested the endosomal translocation activities of the chimeras using a cell-free assay that we developed previously to characterize this process (2, 32, 46). As observed for intact cells, III-PE and DHFR-PE behaved like PE, while ApoC-PE was translocated ∼50% more efficiently than PE and RNase-PE was translocated 40% less actively than PE (Fig. 5A). Analysis of translocated material showed that fusion proteins remained essentially intact during this process (Fig. 5B).
FIG. 4.
Toxicities and translocation activities of PE and fusion proteins measured using intact cells. To assay cytotoxicity, L929 cells were treated for 24 h with different concentrations of a toxin or chimera, and protein synthesis was examined using the 24-h incorporation of [35S]methionine. Toxicity is expressed as the 50% inhibitory concentration (IC50), which is the toxin concentration that inhibits protein synthesis by 50%. To assay translocation activity with entire cells, the cells were treated with 100 pM PE or chimera for different periods of time, before protein synthesis was assayed using pulse-labeling (1 h) of [35S]methionine. Protein synthesis activity was then plotted against time, and the slopes of protein inactivation curves were then measured (h−1). These slopes are directly proportional to the translocation activity (18, 32). MTX was used at concentrations of 50 nM and 150 nM for toxicity and translocation assays, respectively.
FIG. 5.
Cell-free endosomal translocation of PE and fusion proteins. (A) Translocation activity. Mouse lymphocytes were labeled with 125I-Tf (as a negative control), 125I-PE, or 125I-labeled fusion protein for 45 min before endosomes were purified and translocation was assayed for 1 h in the presence of ATP and cytosol as described in Materials and Methods. Where indicated, translocation assays were performed in the presence of 1 μM MTX. (B) Translocated proteins (at 1 h) were concentrated before SDS-PAGE and autoradiography. The molecular masses of standards (in kDa) are indicated on the left. RNase-PE is poorly translocated and did not produce a significant band.
Although the RNase-PE data clearly indicated that endosomal translocation of full-length PE is responsible for more than one-half of the toxicity, it could be argued that the RNase moiety modified PE intracellular routing in a subtle manner that we were unable to visualize microscopically and that this modification was responsible for the differences between PE and RNase-PE that we observed. The use of DHFR-PE enabled us to overcome this type of limitation. We first used intact cells. The 50% inhibitory concentration of DHFR-PE increased from 2 to 3.1 pM in the presence of 50 nM MTX, indicating that MTX inhibited DHFR-PE toxicity by ∼50%. Moreover, the DHFR-PE-mediated protein synthesis inactivation rate decreased from 0.25 to 0.14 h−1 in the presence of 150 nM MTX (Fig. 4). These results indicated that MTX caused retention of 45 to 50% of DHFR-PE molecules within endosomes. The MTX-resistant protein synthesis inactivation activity could result either from 37-kDa fragments or from entire DHFR-PE molecules that were not efficiently trapped within endosomes because the MTX concentration was not sufficient to block all DHFR-PE molecules. To discriminate between these possibilities, and because MTX toxicity prevented the use of concentrations greater than 200 nM with entire cells, we used purified endosomes and the cell-free translocation assay. In this experimental system, 1 μM MTX inhibited DHFR-PE translocation by 50% (Fig. 5A). We concluded that, whatever its concentration, MTX could not block translocation of more than one-half of the DHFR-PE molecules. Hence, the MTX-resistant protein synthesis inactivation activity likely resulted from the cytosolic delivery of entire DHFR-PE molecules that were not blocked by MTX.
Collectively, data obtained with this first approach using fusion proteins indicated that at least one-half of PE toxicity results from endosomal translocation of entire molecules. The results also indicated that PE passenger proteins have to unfold so that they can be transported into the cytosol.
Quantification of cytosolic PE fragments.
The nature of PE fragments within the cytosol of intoxicated cells should provide information regarding the importance of furin processing in PE toxicity. A previous study used conventional cell homogenization methods to obtain a cytosolic fraction and found that there was some enrichment of the 37-kDa fragment, although entire PE was also detected. Cytosolic delivery of the molecules was not studied kinetically (34). Here we quantified PE and its processed fragments within the cytosol as a function of time and compared these kinetics of cytosolic accumulation with those of EF2 ADP-ribosylating material. Cytosol was obtained by SLO permeabilization, a mild method that enables specific permeabilization of the plasma membrane with no effect on the integrity of endocytic elements (28). In these experiments, monensin, an ionophore that does not inhibit PE endocytosis (2) but neutralizes endosomal acidic pH and prevents the PE membrane insertion required for toxicity (32), was used for control purposes. EF2 ADP-ribosylating activity accumulated with time in the cytosol of PE-treated cells (Fig. 6A). This accumulation was largely inhibited (by 70%) in the presence of monensin, showing that most EF2 ADP-ribosylating material in cytosol preparations actually originated from the cell cytosol and not from damaged organelles or the plasma membrane.
FIG. 6.
Accumulation of EF2-ADP ribosylation activity and PE fragments in the cytosol of intoxicated cells. L929 cells were treated with 18 nM PE for the indicated times. Where indicated, 50 μM monensin was added to cells 20 min before PE was added. Cytosol preparations were then obtained using SLO permeabilization and divided in two for the EF2-ADP ribosylation activity assay and immunoprecipitation of PE fragments detected by immunoblotting and ECL detection. (A) EF2-ADP ribosylation by cytosol preparations from cells that were treated with PE for the indicated times. Assays were performed using purified EF2 and [32P]NAD, and 32P-labeled EF2 was detected by autoradiography. Films were scanned, and the 32P-labeled EF2 band was quantified using ImageQuant (GE Healthcare). The results are expressed in arbitrary units (a.u.). (B) Quantification of PE cytosolic fragments. PE fragments were immunoprecipitated from cytosol samples and then detected by immunoblotting and ECL detection before band quantification. □, entire PE from control cells; ▪, entire PE from monensin-treated cells; ○, 37-kDa fragment from control cells; •, 37-kDa fragment from monensin-treated cells; ▴, 37-kDa fragment from lactacystin-treated cells. The results are expressed as percentages of the intensity of the entire PE band in control cells at time zero. The lines show linear regression of the data. The data are the results of a typical experiment that was repeated three times.
When we monitored the presence of PE in these cytosol preparations, we found that entire molecules accumulated with kinetics similar to those of EF2 ADP-ribosylating material (Fig. 6B). Moreover, PE cytosolic accumulation was inhibited by ∼80% when monensin was present during cell labeling. The curve for the 37-kDa fragment was not expected since the cytosolic concentration of this fragment decreased with time. We thought that cell protein degradation activity was likely responsible for the disappearance of the 37-kDa fragment and suspected the proteasome that is known to modulate the stability and toxicity of DT mutants (12). Indeed, when cytosol was prepared from cells treated with PE in the presence of lactacystin, an efficient proteasome inhibitor (30), the levels of the 37-kDa fragment became stable over time, although no increase in the concentration was observed (Fig. 6B). It should be noted that lactacystin did not affect PE toxicity (data not shown). Together, the results obtained with this second approach indicated that entire PE molecules accumulate in the cytosol of intoxicated cells and are likely responsible for EF2 ADP-ribosylating activity and cell death.
Furin-deficient cells can process PE.
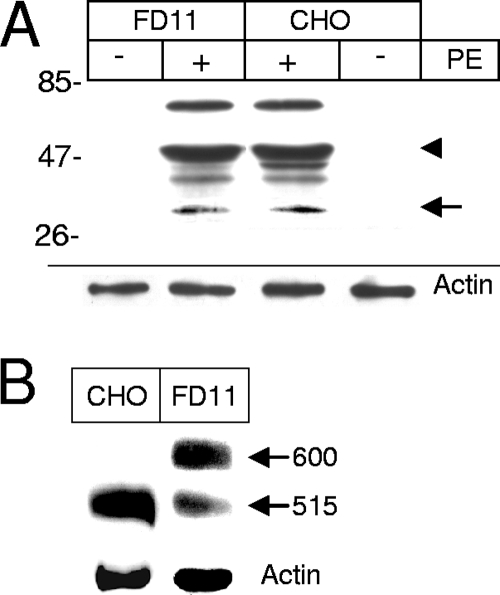
Furin-deficient cells (CHO FD11 cells) are considerably more resistant than the parent cell line (CHO), indicating that furin processing is required at some stage for PE toxicity (14). Nevertheless, it was not known whether PE processing is actually blocked in these cell lines. When we monitored PE processing by FD11 cells, we found that a large proportion of internalized PE was proteolysed by CHO cells to a 55-kDa protein (Fig. 7A), which is presumably the N-terminal fragment generated by trypsin-mediated cleavage after Arg-490 (34). Although the parent CHO cell line generated a ∼45-kDa fragment that was not generated by FD11 cells, both cell lines produced similar amounts of the 37-kDa PE fragment. Hence, FD11 resistance to PE is not due to a defect in toxin processing. It should be noted that, in addition to furin, other proprotein convertases, such as PC1/3, PC2, PC4, and PC7, can cleave PE at the same site at a low pH (39). These proprotein convertases are most likely responsible for PE processing in furin-deficient cells.
FIG. 7.
Furin-deficient cells can efficiently proteolyse PE, but they poorly process the LRP. (A) PE processing. Where indicated, PE (40 nM) was added to CHO (wild type) or CHO FD11 (furin-deficient) cells for 4 h before immunoprecipitation and anti-PE Western blotting. The molecular masses of markers (in kDa) are indicated on the left. The positions of the 55-kDa and 37-kDa fragments are indicated by an arrowhead and an arrow, respectively. (B) LRP processing. LRP was immunoprecipitated before anti-LRP Western blotting. The positions of the precursor (600 kDa) and the large subunit of the mature LRP (515 kDa) are indicated on the right. Lysate blots were probed for actin as a loading control.
Because a deficiency in LRP function could explain the resistance of FD11 cells to PE, we determined whether the LRP was processed by these cells. While in CHO cells the LRP is quantitatively converted from the precursor form (600 kDa) to the mature form (515 kDa), in FD11 cells only 22% ± 12% (n = 3) of the protein was processed to the mature form (Fig. 7B). The presence of PE did not affect LRP processing (data not shown). A similar defect in LRP processing was observed in another furin-deficient CHO cell line (RPE.40); the impaired maturation was associated with a slight but significant reduction in the endocytosis efficiency of plasminogen activator and α2-macroglobulin, which are LRP-specific ligands (54). Hence, the cell line FD11 resistance to PE does not result from impaired toxin processing but could be related to a defect in LRP maturation or to a deficiency in other furin-dependent events required for PE intoxication.
Furin inhibition protects cells from PE by impairing receptor processing and/or endocytosis.
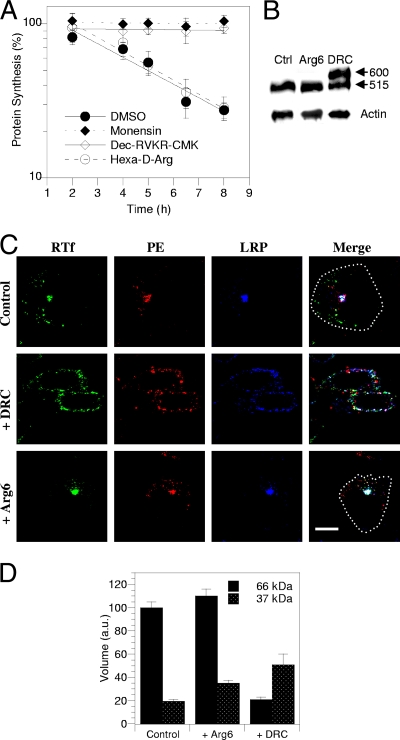
We thought that furin inhibitors applied for a limited period of time would affect the furin processing of incoming toxin more specifically than they would affect the furin processing of the LRP, especially when cell-impermeable inhibitors were used. We used a furin inhibitor that is reversible and poorly cell permeable, hexa-d-Arg, and an irreversible, readily cell-permeable inhibitor, decanoyl-RVKR-chloromethylketone (Dec-RVKR-CMK) (4). We treated cells with these inhibitors for up to 8 h and determined the toxicity and PE and LRP processing together with PE and LRP intracellular localization. When such a short exposure time was used, hexa-d-Arg (60 μM) did not significantly affect PE toxicity (Fig. 8A), LRP processing (Fig. 8B), or PE or LRP intracellular routing (Fig. 8C). Nevertheless, this inhibitor significantly stabilized (by 84%) the 37-kDa PE fragment intracellularly (Fig. 8D). This surprising effect was likely related to the intracellular instability of this fragment observed in previous experiments (Fig. 6B), and hexa-d-Arg apparently inhibited its degradation. Hence, increasing the amount of the intracellular 37-kDa fragment did not favor toxicity.
FIG. 8.
A furin inhibitor protects cells from PE toxicity and prevents LRP processing and endocytosis. (A) Intoxication kinetics in the presence of inhibitors. L929 cells were treated for up to 8 h with 100 pM PE and, where indicated, 50 μM monensin, 60 μM hexa-d-Arg, 60 μM Dec-RVKR-CMK, or solvent (dimethyl sulfoxide [DMSO]) before protein synthesis was assayed for 1 h. (B) Effects on LRP processing. Cells were treated with 60 μM hexa-d-Arg (Arg6), Dec-RVKR-CMK (DRC), or solvent (Ctrl) for 4 h before LRP immunoprecipitation and anti-LRP Western blotting. The positions of the precursor (600 kDa) and the large subunit of the mature LRP (515 kDa) are indicated on the right. Lysate blots were probed for actin as a loading control. (C) Effects on PE, LRP, and Tf receptor intracellular localization. Cells were treated with 60 μM inhibitor or solvent for 4 h and then fixed and permeabilized for detection of PE, LRP, and the Tf receptor (RTf) by immunofluorescence. The images show median confocal sections. Where necessary, a dotted line indicates the outline of the cell in the merged image. Bar, 10 μm. (D) Effects of inhibitors on furin-mediated PE processing efficiency. L929 cells were labeled for 4 h with PE before a 30-min chase. Where indicated, the inhibitor was present throughout the experiment. Cells were then lysed for PE immunoprecipitation. Films from three independent experiments were scanned for band quantitation. Volumes were normalized using the 66-kDa control band.
The second inhibitor, Dec-RVKR-CMK (60 μM), completely blocked PE toxicity (Fig. 8A). It was also more effective than hexa-d-Arg in stabilizing the 37-kDa fragment (by 160%); nevertheless, in the presence of Dec-RVKR-CMK, less PE was found intracellularly (Fig. 8D). This was likely due to general inhibition of endocytosis by this drug that caused retention at the plasma membrane level of PE, the LRP, and the Tf receptor, together with significant PE aggregation at this level (Fig. 8C). After 4 h of exposure to Dec-RVKR-CMK, only 50% ± 5% (n = 3) of LRP molecules were processed into the 515-kDa mature form (Fig. 8B). This result is consistent with measurements of the LRP half-life (∼3 h) (30) and showed that Dec-RVKR-CMK is an efficient furin inhibitor. Although it is not clear whether Dec-RVKR-CMK protected cells from PE by inhibiting endocytosis or preventing LRP processing, this protection did not result from inhibition of PE processing.
Two conclusions can be drawn from these furin inhibition studies. First, increasing 37-kDa intracellular stability does not affect PE toxicity, and second, efficient furin inhibition protects cells from PE by impairing LRP cleavage and/or affecting other furin-dependent steps that enable PE cytosolic delivery, but it does not inhibit PE processing.
DISCUSSION
An important biological issue regarding the PE intoxication process is evaluation of the implications of the ER and recycling endosomes as doorsteps for PE cytosolic delivery. The PE retrograde route has not been characterized yet at the molecular level, and no molecular tool to specifically interfere with this transport route is available (44). Thus, to evaluate the roles of the ER and endosomes in PE access to the cytosol, we studied the requirement of this toxin for processing. Indeed, entire PE strongly binds to the LRP (in the absence of membranes), even at low pH (48), and it is acknowledged that furin processing is strictly required to ensure production of the active fragment that lacks a receptor binding domain and can be retrogradely transported to the ER (37). It therefore seems reasonable to assume that the lack of a processing requirement indicates that intoxication is due to entire PE molecules reaching the cytosol from endosomes, whereas, conversely, if PE requires furin processing to kill cells, it is likely because the active fragment triggers toxicity after retrograde transport to the ER.
To study the PE requirement for furin processing to kill cells, we used techniques ranging from conventional assays for determining cytotoxicity and intoxication kinetics to PE and LRP processing analyses, together with intracellular localization of the toxin and its receptor. The three approaches were independent. They were also complementary since, for instance, the fusion protein approach was drug free, while in the furin inhibitor studies native PE was used. These different experimental systems provided similar information, which indicated that PE intoxication relies largely on endosomal translocation of entire PE molecules.
Previous evidence for implication of the endosome as a doorstep for PE cytosolic delivery was essentially based on cell fractionation experiments (2), and it was important to confirm these findings using entire cells and toxicity assays. The use of RNase-PE and DHFR-PE provided confirmation. The fusion protein approach also showed that cargos attached to PE have to unfold during transport to the cytosol. This was previously observed for other toxins, such as DT (23) and ricin (6), using DHFR chimeras, like those used here. An interesting PE fusion protein was ApoC-PE, which translocated more efficiently than PE and was more toxic. These results were most likely due to the apocytochrome c affinity for membranes (11). They further indicate the key role of membrane interaction and insertion for PE translocation (32).
The last two approaches that we used to study PE processing involved quantification of cytosolic toxin fragments. The cytosolic levels of entire PE were correlated with the accumulation of EF2 ADP ribosylation activity with time (Fig. 6A and 6B). Moreover, treatment of cells with the furin inhibitor Dec-RVKR-CMK resulted in inhibition of PE toxicity (Fig. 8A), together with a decrease in the cytosolic PE concentration (Fig. 8D). Toxicity was therefore associated with cytosolic accumulation of entire PE molecules. Conversely, the 37-kDa fragment was unstable in the cytosol. The fact that degradation of the PE 37-kDa fragment was prevented by lactacystin (Fig. 6B) indicated that, despite the fact that this fragment has very few lysine residues (only three residues close to its C terminus), the proteasome was responsible for its degradation. Surprisingly, the cytosolic concentration of this fragment was increased upon treatment of cells by furin inhibitors (by 80 to 160%). Nevertheless, neither furin nor proteasome inhibitors protected cells from PE. These observations are consistent with previous results showing that increasing the production efficiency of the 37-kDa fragment intracellularly by replacing the PE processed loop with a DT Arg-rich loop did not favor toxicity (9). Similarly, W281A and W281K mutations enhanced processing and decreased toxicity, while L284M and L284W mutations inhibited processing and decreased toxicity (57). Moreover, we found that, although furin-deficient cells are resistant to PE, they can process this toxin properly (Fig. 7A). All these results provide compelling evidence that there is no correlation between the amount of the 37-kDa fragment produced intracellularly and PE toxicity.
According to the data presented here and elsewhere (2, 32) and data obtained by other groups that documented PE insertion into membranes upon acidification (21, 38, 41), endosomal translocation of entire PE is involved in cytotoxicity. This result is consistent with data for the intracellular routing of other bacterial toxins and their ability to undergo conformational modifications and membrane insertion upon exposure to endosomal pH. Indeed, the ER-targeted toxins Shiga toxin and cholera toxin are closely related AB5 toxins that recognize glycolipids. Once in the endosome lumen, low pH induces in these toxins only subtle conformational changes that are unrelated to membrane penetration (29, 31). These toxins eventually reach the ER, where they can be visualized (27, 52). The characteristics of endosomes translocating toxins, such as DT, anthrax toxin protective antigen, and PE, are different. The endocytic routing of the proteinic receptor is restricted to the endocytic pathway (1, 26, 30). Once these toxins are in early endosomes, low pH induces major conformational changes in them, leading to membrane insertion and translocation (1, 32, 40).
Nevertheless, several lines of evidence, such as the two- to threefold increase in PE toxicity obtained by replacing its C-terminal end with the ER retrieval motif KDEL (43, 44), support a model in which delivery to the ER of the 37-kDa PE fragment favors toxicity. PE intoxication models based on endosomal translocation or retrotranslocation from the ER are not mutually exclusive. Depending on the cell type for instance (44), PE could enter cells directly from endosomes or be subjected to processing and retrograde transport to the ER before it reaches the cytosol.
Acknowledgments
We are indebted to Harald Sheraga (Cornell University) and Gottfried Schatz (University of Basel) for RNase and apocytochrome c expression vectors, to Joachim Herz, Karin Becker, and Emma Coopman for antibodies, and to Laurence Abrami and Stephen Leppla for FD11 cells.
This work was supported by the Association pour le Recherche sur le Cancer.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 20 April 2009.
REFERENCES
- 1.Abrami, L., N. Reig, and F. G. van der Goot. 2005. Anthrax toxin: the long and winding road that leads to the kill. Trends Microbiol. 1372-78. [DOI] [PubMed] [Google Scholar]
- 2.Alami, M., M. P. Taupiac, H. Reggio, A. Bienvenue, and B. Beaumelle. 1998. Involvement of ATP-dependent Pseudomonas exotoxin translocation from a late recycling compartment in lymphocyte intoxication procedure. Mol. Biol. Cell 9387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allured, V. S., R. J. Collier, S. Carroll, and D. B. McKay. 1986. Structure of exotoxin A of Pseudomonas aeruginosa at 3-Ångstrom resolution. Proc. Natl. Acad. Sci. USA 831320-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak, A. 2005. Inhibitors of proprotein convertases. J. Mol. Med. 83844-855. [DOI] [PubMed] [Google Scholar]
- 5.Beaumelle, B., and C. R. Hopkins. 1989. High-yield isolation of functionally competent endosomes from mouse lymphocytes. Biochem. J. 264137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaumelle, B., M. P. Taupiac, J. M. Lord, and L. M. Roberts. 1997. Ricin A chain can transport unfolded dihydrofolate reductase into the cytosol. J. Biol. Chem. 27222097-22102. [DOI] [PubMed] [Google Scholar]
- 7.Bu, G., H. J. Geuze, G. J. Strous, and A. L. Schwartz. 1995. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 142269-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhary, V. K., Y. Jinno, D. J. FitzGerald, and I. Pastan. 1990. Pseudomonas exotoxin contains a specific sequence at the carboxyl terminus that is required for cytotoxicity. Proc. Natl. Acad. Sci. USA 87308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiron, M. F., M. Ogata, and D. FitzGerald. 1996. Pseudomonas exotoxin exhibits increased sensitivity to furin when sequences at the cleavage site are mutated to resemble the arginine-rich loop of diphtheria toxin. Mol. Microbiol. 22769-778. [DOI] [PubMed] [Google Scholar]
- 10.de Haan, C. A., K. Stadler, G. J. Godeke, B. J. Bosch, and P. J. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 786048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demel, R., W. Jordi, H. Lambrechts, H. van Damme, R. Hovius, and B. de Kruijff. 1989. Differential interactions of apo- and holocytochrome c with acidic membrane lipids in model systems and the implications for their import into mitochondria. J. Biol. Chem. 2643988-3997. [PubMed] [Google Scholar]
- 12.Falnes, P. O., and S. Olsnes. 1998. Modulation of the intracellular stability and toxicity of diphtheria toxin through degradation by the N-end rule pathway. EMBO J. 17615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geller, B., H. Y. Zhu, S. Cheng, A. Kuhn, and R. E. Dalbey. 1993. Charged residues render pro-OmpA potential dependent for initiation of membrane translocation. J. Biol. Chem. 2689442-9447. [PubMed] [Google Scholar]
- 14.Gordon, V. M., K. R. Klimpel, N. Arora, M. A. Henderson, and S. H. Leppla. 1995. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect. Immun. 6382-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, M., V. M. Gordon, D. J. FitzGerald, and S. H. Leppla. 1996. Furin regulates both the activation of Pseudomonas exotoxin A and the quantity of the toxin receptor expressed on target cells. Infect. Immun. 64524-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herz, J., R. C. Kowal, J. L. Goldstein, and M. S. Brown. 1990. Proteolytic processing of the 600 kd low density lipoprotein receptor-related protein (LRP) occurs in a trans-Golgi compartment. EMBO J. 101769-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herz, J., and D. K. Strickland. 2001. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Investig. 108779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson, T. H., and D. M. Neville, Jr. 1987. Temporal separation of protein toxin translocation from processing events. J. Biol. Chem. 26216484-16494. [PubMed] [Google Scholar]
- 19.Inocencio, N. M., J. M. Moehring, and T. J. Moehring. 1994. Furin activates Pseudomonas exotoxin A by specific cleavage in vivo and in vitro. J. Biol. Chem. 26931831-31835. [PubMed] [Google Scholar]
- 20.Jackson, M. E., J. C. Simpson, A. Girod, R. Pepperkok, L. M. Roberts, and J. M. Lord. 1999. The KDEL retrieval system is exploited by Pseudomonas exotoxin A, but not by Shiga-like toxin-1, during retrograde transport from the Golgi complex to the endoplasmic reticulum. J. Cell Sci. 112467-475. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, J. X., and E. London. 1990. Involvement of denaturation-like changes in Pseudomonas exotoxin A hydrophobicity and membrane penetration determined by characterization of pH and thermal transitions. J. Biol. Chem. 2658636-8641. [PubMed] [Google Scholar]
- 22.Juminaga, D., W. J. Wedemeyer, R. Garduno-Juarez, M. A. McDonald, and H. A. Scheraga. 1997. Tyrosyl interactions in the folding and unfolding of bovine pancreatic ribonuclease A: a study of tyrosine-to-phenylalanine mutants. Biochemistry 3610131-10145. [DOI] [PubMed] [Google Scholar]
- 23.Klingenberg, O., and S. Olsnes. 1996. Ability of methotrexate to inhibit translocation to the cytosol of dihydrofolate reductase fused to diphtheria toxin. Biochem. J. 313647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko, K. W., R. S. McLeod, R. K. Avramoglu, J. Nimpf, D. J. FitzGerald, J. Vukmirica, and Z. Yao. 1998. Mutation at the processing site of chicken low density lipoprotein receptor-related protein impairs efficient endoplasmic reticulum exit, but proteolytic cleavage is not essential for its endocytic functions. J. Biol. Chem. 27327779-27785. [DOI] [PubMed] [Google Scholar]
- 25.Kounnas, M. Z., R. E. Morris, M. R. Thompson, D. J. FitzGerald, D. K. Strickland, and C. B. Sealinger. 1992. The a2 macroglobulin receptor /low density lipoprotein receptor binds and internalizes Pseudomonas exotoxin A. J. Biol. Chem. 26712420-12423. [PubMed] [Google Scholar]
- 26.Lemichez, E., M. Bomsel, G. Devilliers, J. vanderSpek, J. R. Murphy, E. V. Lukianov, S. Olsnes, and P. Boquet. 1997. Membrane translocation of diphtheria toxin fragment A exploits early to late endosome trafficking machinery. Mol. Microbiol. 23445-457. [DOI] [PubMed] [Google Scholar]
- 27.Majoul, I. V., P. I. H. Bastiaens, and H. D. Sölling. 1996. Transport of an external Lys-Asp-Glu-Leu (KDEL) protein from the plasma membrane to the endoplasmic reticulum: studies with cholera toxin in vero cells. J. Cell Biol. 133777-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallard, F., B. L. Tang, T. Galli, D. Tenza, A. Saint-Pol, X. Yue, C. Antony, W. Hong, B. Goud, and L. Johannes. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCann, J. A., J. A. Mertz, J. Czworkowski, and W. D. Picking. 1997. Conformational changes in cholera toxin B subunit-ganglioside GM1 complexes are elicited by environmental pH and evoke changes in membrane structure. Biochemistry 369169-9178. [DOI] [PubMed] [Google Scholar]
- 30.Melman, L., H. J. Geuze, Y. Li, L. M. McCormick, P. Van Kerkhof, G. J. Strous, A. L. Schwartz, and G. Bu. 2002. Proteasome regulates the delivery of LDL receptor-related protein into the degradation pathway. Mol. Biol. Cell 133325-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menikh, A., M. T. Saleh, J. Gariepy, and J. M. Boggs. 1997. Orientation in lipid bilayers of a synthetic peptide representing the C-terminus of the A1 domain of shiga toxin. A polarized ATR-FTIR study. Biochemistry 3615865-15872. [DOI] [PubMed] [Google Scholar]
- 32.Méré, J., J. Morlon-Guyot, A. Bonhoure, L. Chiche, and B. Beaumelle. 2005. Acid-triggered membrane insertion of Pseudomonas exotoxin A involves an original mechanism based on pH-regulated tryptophan exposure. J. Biol. Chem. 28021194-21201. [DOI] [PubMed] [Google Scholar]
- 33.Moehring, J. M., N. M. Inocencio, B. J. Robertson, and T. J. Moehring. 1993. Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and virus complements the genetic lesion. J. Biol. Chem. 2682590-2594. [PubMed] [Google Scholar]
- 34.Ogata, M., V. K. Chaudhary, I. Pastan, and D. J. FitzGerald. 1990. Processing of Pseudomonas exotoxin by a cellular protease results in the generation of a 37,000-Da toxin fragment that is translocated to the cytosol. J. Biol. Chem. 26520678-20685. [PubMed] [Google Scholar]
- 35.Ogata, M., C. M. Fryling, I. Pastan, and D. J. FitzGerald. 1992. Cell-mediated cleavage of Pseudomonas aeruginosa exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. J. Biol. Chem. 26725396-25401. [PubMed] [Google Scholar]
- 36.Ornatowski, W., J. F. Poschet, E. Perkett, J. L. Taylor-Cousar, and V. Deretic. 2007. Elevated furin levels in human cystic fibrosis cells result in hypersusceptibility to exotoxin A-induced cytotoxicity. J. Clin. Investig. 1173489-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastan, I., V. Chaudhary, and D. FitzGerald. 1992. Recombinant toxins as novel therapeutic agents. Annu. Rev. Biochem. 61331-354. [DOI] [PubMed] [Google Scholar]
- 38.Rasper, D. M., and A. R. Merrill. 1994. Evidence for the modulation of Pseudomonas aeruginosa exotoxin A-induced pore formation by membrane surface charge density. Biochemistry 3312981-12989. [DOI] [PubMed] [Google Scholar]
- 39.Remacle, A. G., S. A. Shiryaev, E. S. Oh, P. Cieplak, A. Srinivasan, G. Wei, R. C. Liddington, B. I. Ratnikov, A. Parent, R. Desjardins, R. Day, J. W. Smith, M. Lebl, and A. Y. Strongin. 2008. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem. 28320897-20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosconi, M. P., and E. London. 2002. Topography of helices 5-7 in membrane-inserted diphtheria toxin T domain: identification and insertion boundaries of two hydrophobic sequences that do not form a stable transmembrane hairpin. J. Biol. Chem. 27716517-16527. [DOI] [PubMed] [Google Scholar]
- 41.Sandvig, K., and J. O. Moskaug. 1987. Pseudomonas toxin binds Triton X-114 at low pH. Biochem. J. 245899-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarac, M. S., A. Cameron, and I. Lindberg. 2002. The furin inhibitor hexa-d-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 707136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seetharam, S., V. K. Chaudhary, D. J. FitzGerald, and I. Pastan. 1991. Increased cytotoxic activity of Pseudomonas exotoxin and two chimeric toxins ending in KDEL. J. Biol. Chem. 26617376-17381. [PubMed] [Google Scholar]
- 44.Smith, D. C., R. A. Spooner, P. D. Watson, J. L. Murray, T. W. Hodge, M. Amessou, L. Johannes, J. M. Lord, and L. M. Roberts. 2006. Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic 7379-393. [DOI] [PubMed] [Google Scholar]
- 45.Sprinkle, J. R., T. B. Hakvoort, T. I. Koshy, D. D. Miller, and E. Margoliash. 1990. Amino acid sequence requirements for the association of apocytochrome c with mitochondria. Proc. Natl. Acad. Sci. USA 875729-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taupiac, M. P., M. Bebien, M. Alami, and B. Beaumelle. 1999. A deletion within the translocation domain of Pseudomonas exotoxin A enhances translocation efficiency and cytotoxicity concomitantly. Mol. Microbiol. 311385-1393. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, M. R., J. Forristal, P. Kauffman, T. Madden, K. Kozak, R. E. Morris, and C. B. Saelinger. 1991. Isolation and characterization of Pseudomonas aeruginosa exotoxin A binding glycoprotein from mouse LM cells. J. Biol. Chem. 2662390-2396. [PubMed] [Google Scholar]
- 49.Vendeville, A., F. Rayne, A. Bonhoure, N. Bettache, P. Montcourrier, and B. Beaumelle. 2004. HIV-1 Tat enters T-cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol. Biol. Cell 152347-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volles, M. J., X. Xu, and H. A. Scheraga. 1999. Distribution of disulfide bonds in the two-disulfide intermediates in the regeneration of bovine pancreatic ribonuclease A: further insights into the folding process. Biochemistry 387284-7293. [DOI] [PubMed] [Google Scholar]
- 51.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138141-143. [DOI] [PubMed] [Google Scholar]
- 52.White, J., L. Johannes, F. Mallard, A. Girod, S. Grill, S. Reinsch, P. Keller, B. Tzschaschel, A. Echard, B. Goud, and E. H. Stelzer. 1999. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 147743-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willnow, T. E., and J. Herz. 1994. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. J. Cell Sci. 107719-726. [PubMed] [Google Scholar]
- 54.Willnow, T. E., J. M. Moehring, N. M. Inocencio, T. J. Moehring, and J. Herz. 1996. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem. J. 313 (Pt 1)71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willnow, T. E., A. Rohlmann, J. Horton, H. Otani, J. R. Braun, R. E. Hammer, and J. Herz. 1996. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptor. EMBO J. 152632-2639. [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida, T., C. Chen, M. Zhang, and H. C. Wu. 1991. Disruption of the Golgi apparatus by brefeldin A inhibits the cytotoxicity of ricin, modeccin and Pseudomonas toxin. Exp. Cell Res. 192389-395. [DOI] [PubMed] [Google Scholar]
- 57.Zdanovsky, A. G., M. Chiron, I. Pastan, and D. J. FitzGerald. 1993. Mechanism of action of Pseudomonas exotoxin. J. Biol. Chem. 26821791-21799. [PubMed] [Google Scholar]