Abstract
Salmonella enterica subsp. enterica serovar Enteritidis is a leading cause of human food-borne illness that is mainly associated with the consumption of contaminated poultry meat and eggs. To cause infection, S. Enteritidis is known to use two type III secretion systems, which are encoded on two salmonella pathogenicity islands, SPI-1 and SPI-2, the first of which is thought to play a major role in invasion and bacterial uptake. In order to study the role of SPI-1 in the colonization of chicken, we constructed deletion mutants affecting the complete SPI-1 region (40 kb) and the invG gene. Both ΔSPI-1 and ΔinvG mutant strains were impaired in the secretion of SipD, a SPI-1 effector protein. In vitro analysis using polarized human intestinal epithelial cells (Caco-2) revealed that both mutant strains were less invasive than the wild-type strain. A similar observation was made when chicken cecal and small intestinal explants were coinfected with the wild-type and ΔSPI-1 mutant strains. Oral challenge of 1-week-old chicken with the wild-type or ΔSPI-1 strains demonstrated that there was no difference in chicken cecal colonization. However, systemic infection of the liver and spleen was delayed in birds that were challenged with the ΔSPI-1 strain. These data demonstrate that SPI-1 facilitates systemic infection but is not essential for invasion and systemic spread of the organism in chickens.
Salmonella enterica is a gram-negative enteropathogenic bacterium. Within the S. enterica species, more than 2,300 serovars have been identified, of which the serovars Enteritidis and Typhimurium have been the most frequently associated with human infections (49). S. Enteritidis is a well-known zoonotic pathogen (30), and infected poultry are among the most common reservoir of salmonellae that can be transmitted through the food chain to humans (18). In young chicks, S. Enteritidis infection can lead to increased incidence of illness, while older birds are less susceptible to the effects of this pathogen, often experiencing intestinal colonization and even systemic dissemination without significant morbidity or mortality. Hence, a better understanding of the pathogenesis of S. Enteritidis in chickens may subsequently lower the rates of human disease caused by this pathogen.
S. Enteritidis requires a substantial number of genes for virulence, which are clustered in large chromosomal regions known as Salmonella pathogenicity islands (SPI) (40). Two of these pathogenicity islands encode two functionally distinct type III secretion systems (T3SS) that are utilized as “molecular syringes” to translocate virulence determinants, called effector proteins, from the bacterial cytoplasm into (29, 30) or in the vicinity of the target cell (52). Effector proteins delivered by the SPI-1 T3SS are mainly involved in host cell invasion by inducing membrane ruffling and disrupting actin polymerization to facilitate bacterial uptake (17). The SPI-2 T3SS plays a major role in systemic virulence and in facilitating intracellular survival, especially within macrophages. However, recent evidence suggests that the SPI-2 T3SS also plays a role in intestinal colonization (10, 11) and is expressed prior to bacterial uptake (6).
Invasion of epithelial cells by Salmonella enterica subspecies enterica serovar Typhimurium has been shown to disrupt tight junctions (15, 31, 54) which, along with other components, form important intercellular junctions found in polarized epithelial cells. Tight junctions regulate the paracellular flow of ions and solutes across the intestinal epithelium. They also maintain distinct apical and basolateral domains with well-defined plasma membrane components (42). SPI-1 effector proteins have been associated with the disruption of tight junctions (5, 31, 54) by activating Cdc42 and Rac-1 (Rho family GTPases), which subsequently activate signal transduction pathways that lead to the reorganization of actin, resulting in the uptake of Salmonella (45). Further, the S. Typhimurium SPI-1 effectors SopB, SopE, SopE2, and SipA have been identified as major contributors in the disruption of tight junctions (5).
The contribution of S. Typhimurium SPI-1 to intestinal cell invasion has been studied in cell culture using different SPI-1 deletion mutants. SPI-1 regulatory gene hilA and sipB (SPI-1 effector gene) mutants have been shown to be attenuated in invasion relative to the wild-type strain in porcine intestinal epithelioid (IPI-21) cells. However, the use of polarized porcine intestinal epithelial cells (IPEC-J2) has revealed that, in addition to hilA and sipB mutant strains, a sipA (SPI-1 effector gene) mutant strain was also recovered at a significantly lower rate than the wild-type strain (4), suggesting that SPI-1 is important for efficient invasion. In another study, it was demonstrated that the S. Typhimurium SPI-1 effectors SipA, SopA, SopB, SopD, and SopE2 all contributed to invasion of polarized human colon carcinoma cells (T84). Mutants lacking the genes for the aforementioned effector proteins were less invasive than the wild-type strain (51). Moreover, an S. Typhimurium strain lacking the invG gene (encoding an outer membrane component of the SPI-1 T3SS apparatus) was also impaired in invasion of COS-7 cells (24). Taken together, the results from these invasion studies suggest that SPI-1 plays a role in invasion in cell culture. However, most of the data available is from research that has been conducted using S. Typhimurium.
Several in vivo experiments have been performed to investigate the role of SPI-1 during the course of a Salmonella infection in different animal species using deletion mutants in SPI-1 genes. In the murine model of infection, it was observed that S. Typhimurium strains containing mutations in hilA and invG were recovered from intestinal contents and systemic sites at a lower frequency than the wild-type strain (43). Other groups have reported that a functional SPI-1 T3SS is required to induce intestinal inflammation and cause significant histopathological changes in the streptomycin-treated mouse model of infectious enterocolitis (1, 10, 24). Similarly, in the bovine model of enteritis, SPI-1 has been shown to be important for intestinal colonization (20). In addition, it has been demonstrated that S. Typhimurium SPI-1 gene (sipA, sipB, and hilA) mutants were impaired in their ability to colonize the porcine gut in a ligated intestinal loop model (4). However, a recent study has shown that a SPI-1 functional mutant induces intestinal pathology that is very similar to the wild-type strain when studied 5 days postchallenge in a novel bovine ileal loop model (11).
Studies investigating the contribution of SPI-1 in chicken during the course of an S. Enteritidis infection are limited in number and have mostly observed colonization and systemic infection over a short time frame. Infection of 1-day-old birds with S. Enteritidis strains containing mutations in the invA, invB, and invC genes (SPI-1 genes) has shown that these organisms are attenuated in the colonization of the gastrointestinal tract, as well as in the systemic spread of the organism over a period of 6 days postinfection (48). In another study, it was observed that S. Enteritidis sipD mutants were unable to colonize spleens compared to the wild-type strain 3 days postinfection in 1-day-old chicks (44). However, the colonization of the spleen was only measured at one time point, making it difficult to predict the effect of the mutant strain prior to and after, the third day after infection. A similar trend was seen in the ceca of 1-day-old chicks challenged with a S. Enteritidis hilA mutant strain over a period of 28 days postinfection (3). However, the hilA mutant strain did not have a significant impact on the infection of the livers and spleens. Recently, the impact of SPI-1 was examined using a S. Typhimurium spaS mutant strain in 1-day-old and 1-week-old birds (32). Inactivation of spaS (SPI-1 structural protein) did not affect colonization of the livers or ceca of 1-day-old birds over a period of 72 h postinfection. In 1-week-old birds, the same strain was recovered at lower levels from the ceca over a period of 14 days postinfection, while the recovery from the liver was lower at 3 days postinfection (32).
Taken together, the experimental evidence from the in vitro studies suggests that S. Typhimurium SPI-1 has an impact on invasion. However, studies investigating the role of S. Enteritidis SPI-1 in vivo are limited. Moreover, research from the murine, bovine, and porcine models of salmonellosis indicates that SPI-1 may play a role in breaching the intestinal epithelial layer during the course of an infection. Nevertheless, little is known about the virulence properties of the S. Enteritidis SPI-1 T3SS in the colonization of chickens. In addition, recent evidence suggests that S. enterica is capable of establishing infection without the presence of SPI-1 (11, 25, 28). The objective of the present study was to investigate the contribution of the S. Enteritidis SPI-1 T3SS in the invasion of polarized Caco-2 cells and chicken intestinal explants and in the colonization of chickens over a period of 4 days postchallenge. Our data indicate that S. Enteritidis SPI-1 is important for invasion in polarized Caco-2 cells and intestinal epithelial cells in vitro. We also show that a ΔSPI-1 mutant strain is not impaired in the cecal colonization of 1-week-old chickens. However, the deletion of the SPI-1 region causes a delay in systemic infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in the present study are listed in Table 1. S. Enteritidis Sal 18 was used as the wild type and parent strain for all gene disruptions. Escherichia coli K-12 DH5α strain was used as a negative control for invasion assays. Unless otherwise stated, all strains were grown in Luria-Bertani (LB) broth at 37°C, and bacterial cultures were agitated in an orbital shaker. SOC medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 20 mM Mg, 20 mM glucose) was used after transformation of the lambda red PCR products as described below.
TABLE 1.
Bacterial strains used in this study
| Bacterial strain | Description and/or genotype | Source or reference |
|---|---|---|
| S. enterica | ||
| Sal 18 | S. enterica serovar Enteritidis wild type | 47 |
| LS 21 | Sal 18, ΔSPI-1::cat | This study |
| LS 22 | Sal 18, ΔinvG::cat | This study |
| LS 29 | Sal 18, att Tn7::tet | Unpublished data |
| LS 25 | Sal 18, att Tn7::cat | Unpublished data |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
Construction of mutants.
Deletion mutants were made in the entire SPI-1 region (∼40 kb), as well as in invG (∼1.7 kb), using the phage lambda red one-step inactivation method (12). Briefly, PCR primers (Table 2) were designed for the flanking regions of SPI-1 (SPI-1F and SPI-1R) and invG (invGF and invGR), based on the S. enterica serovar Enteritidis PT4 NCTC 13349 sequence provided by the Welcome Trust Sanger Institute (United Kingdom) and the chloramphenicol resistance gene sequence on a template plasmid (pKD3). PCR products were transformed by electroporation into competent wild-type S. Enteritidis expressing the lambda red recombinase (pKD46) under the control of an arabinose inducible promoter. Transformants were incubated in SOC (23) for 3 h at 37°C in a shaker, plated initially on LB agar plates with 9 μg of chloramphenicol/ml, and then transferred to plates with 34 μg of chloramphenicol/ml to select for antibiotic-resistant transformants. The ΔSPI-1 and ΔinvG deletion mutants were confirmed by PCR using a 2720 thermal cycler (Applied Biosystems) and sequencing (Plant Biotechnology Institute) using a 3730 XL DNA analyzer (Applied Biosystems).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| SPI-1F | GCT GTC GCG TAT GAA GCG ATT GGG TAT TGA TAA AGA CGC GTT AGC GTA AGT GTA GGC TGG AGC TGC TTC |
| SPI-1R | ATA TGG TCT TAA TTA TAT CAT GAT GAG TTC AGC CAA CGG TGA TAT GGC CCA TAT GAA TAT CCT CCT TA |
| InvGF | TCG GCG TTT CGC CGC GGA AAT TAT CAA ATA TTA TTC AAT TGG CAG ACA AGT GTA GGC TTG GAG CTG CTT C |
| InvGR | GCC GGG GAC AAT ATT CTG GAA AAT GAA ATA CCG GAG GTT GAG CCA GGA ACA TAT GAA TAT CCT CCT TA |
Precipitation of SPI-1 secreted proteins.
Overnight cultures of S. Enteritidis Sal 18 wild-type, ΔSPI-1, and ΔinvG strains grown in LB broth containing 0.3 M NaCl were subcultured in fresh medium at a 1:50 dilution and grown for 4 h at 37°C with low aeration until the optical density at 600 nm reached ∼1.2. The cultures were centrifuged in Eppendorf tubes at 6,000 × g for 10 min (all centrifugation steps were at 4°C, unless stated otherwise). The supernatant was filtered into fresh tubes using a 0.2-μm-pore-size filter, while the sediment fraction was dissolved in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer until it was ready for use. Chilled 100% trichloroacetic acid was added to the filtered supernatant to 20% (vol/vol), and the mixtures were incubated on ice for 1 h. The precipitated proteins were centrifuged at 13,000 × g for 30 min. The supernatant was discarded, and 50 μl of phosphate-buffered saline (PBS; 136 mM NaCl, 2.6 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4), 20 μl of 1.5 M Tris (pH 8.8), and 1 ml of chilled (−20°C) acetone were added. The tubes were centrifuged at 13,000 × g for 15 min. The supernatant was discarded, and the sediment was washed with 300 μl of chilled acetone, followed by centrifugation at 13,000 × g for 10 min. The acetone was discarded, and the dried pellet was dissolved in 100 μl of SDS-PAGE sample buffer and stored until it was ready for use.
Cloning of sipD and purification of His tag SipD.
The sipD gene was amplified by using primers that were designed based on the S. Enteritidis PT4 NCTC 13349 sequence provided by the Welcome Trust Sanger Institute. The gene was cloned downstream of a phage T5 promoter into a His-tag pQE-30 vector (Qiagen). SipD was overexpressed in E. coli M15 (Qiagen) and purified by using a nickel-charged resin (Qiagen). Polyclonal antisera against SipD were raised in New Zealand White rabbits obtained from Charles River Canada.
Western immunoblots.
Proteins secreted under SPI-1 inducing conditions were separated by SDS-PAGE and transferred to a nitrocellulose membrane using a semidry transfer apparatus (Bio-Rad) according to the manufacturer's instructions. The membranes were probed by using rabbit polyclonal anti-His tag SipD serum.
Cell culture.
Caco-2 cells were grown in HyClone Dulbecco modified Eagle medium (DMEM; Fischer) with 10% fetal bovine serum (Seracare) and 1% nonessential amino acids (Invitrogen) at 37°C and 5% CO2. To obtain polarized monolayers, Caco-2 cells were seeded onto Transwell inserts (24 mm in diameter, 0.4-μm pore size; Corning) for about 21 days. The cells were used for gentamicin protection assays once the transepithelial resistance (TER) was determined to be between 250 and 300 Ω cm−2, as described elsewhere (5). All assays were performed in triplicate and were repeated twice.
Invasion assay using polarized Caco-2 cells.
Invasion was assessed by using the gentamicin protection assay as described elsewhere (38). Briefly, polarized Caco-2 cells were apically infected with either Sal 18 wild-type (LS 29), ΔSPI-1 (LS 21), ΔinvG (LS 22), or E. coli DH5α strains grown in LB broth at an approximate multiplicity of infection (MOI) of 100 for 1 h. For competition experiments, the cells were apically infected with a 1:1 ratio of wild-type and ΔSPI-1 strains or wild-type and ΔinvG strains (total MOI of 100). The Caco-2 cells were washed three times with 200 μl of PBS to remove excess bacteria. The cells were then incubated for 2 h with DMEM containing gentamicin (400 μg/ml) to kill the extracellular bacteria. The cells were washed two times with 200 μl of PBS and lysed with 1% Triton. Serial dilutions were plated on LB agar containing either tetracycline (5 μg/ml) for the wild-type strain or chloramphenicol (34 μg/ml) for the ΔSPI-1 and ΔinvG strains. For competition experiments, serial dilutions were plated in duplicate on LB agar containing tetracycline and LB agar containing chloramphenicol. The invasiveness of the wild-type strain (∼105 CFU/ml) was accepted to be 100%, while the invasiveness of the mutant strains was calculated as a percentage of the wild type by using a similar approach as published previously (41).
Measurement of TER.
The TER was measured by using a Millicell electrical resistance system (Millipore) to determine cell monolayer health and polarity as described elsewhere (5). For bacterial invasion assays, the TER was measured prior to infection of the polarized Caco-2 cells with S. Enteritidis, as well as after gentamicin treatment, to determine the effect of the bacterial strains on the TER.
Chicken intestinal tissue explants.
Small-intestinal and cecal tissue samples were obtained from chickens to assess S. Enteritidis invasion as described elsewhere (51). Briefly, the tissue samples were placed on biopsy disks (25.4 by 2 mm; Fisher) with the mucosal side facing up in six-well tissue culture plates (Corning) containing DMEM (Sigma) supplemented with 10% fetal bovine serum. The tissues were coinfected with 107 CFU of S. Enteritidis wild-type and ΔSPI-1 strains/well at a 1:1 ratio for 1 h to allow for bacterial invasion. The plates were incubated at 41°C with gentle shaking (90 rpm) in an air-tight container under 95% O2. The samples were washed in DMEM and then incubated with DMEM containing gentamicin (400 μg/ml) for 1 h, washed in DMEM, and homogenized in PBS. Serial dilutions were plated on brilliant green agar (BGA) containing either tetracycline (5 μg/ml) or chloramphenicol (9 μg/ml) to enumerate the bacteria. All assays were performed in triplicate and were repeated at least three times. The invasiveness of the wild-type strain was accepted to be 100% (∼105 CFU/ml), as described above, while the invasiveness of the mutant strain was calculated as a percentage of the wild-type strain. Samples of the explants in all experiments were subjected to histopathological analysis, and the integrity of the intestinal epithelial layers was confirmed.
Passage of strains.
S. Enteritidis wild-type (LS 25) and ΔSPI-1 (LS 21) strains were passaged through 1-week-old specific-pathogen-free chickens prior to the colonization experiments. Isolates were obtained and processed from the livers or spleens as described below and confirmed by PCR and antibiotic selection.
Infection of 1-week-old chickens.
Specific-pathogen-free eggs were obtained from Charles River Laboratories and incubated for 21 days until hatch at the Department of Poultry Science (University of Saskatchewan). The chicks were transferred and housed in isolation rooms at the Vaccine and Infectious Disease Organization (VIDO) for the duration of the experiment. The birds were screened by using fecal swabs that were plated on BGA to test for the presence of Salmonella species. At 1 week of age the birds were randomly divided into two groups containing 40 birds each. The birds were orally challenged with 0.5 ml (1010 CFU) of either wild-type S. Enteritidis or the ΔSPI-1 mutant grown in LB broth that was administered using an oral gavage needle (18 gauge by 1.5 in.) down the throat. Ten birds from each group were euthanized on days 1, 2, 3, and 4 postchallenge. Samples from the liver, spleen, and cecal contents were weighed and homogenized in saline (0.85% sodium chloride), and serial dilutions were plated on BGA to determine bacterial counts. In addition, homogenates from the livers and spleens were enriched by incubation in selenite broth overnight at 37°C. Enriched samples were streaked on BGA to determine whether they contained Salmonella species.
Statistical analysis.
Statistical analysis was performed by using GraphPad Prism 5.0. The recovery of S. Enteritidis from the cecal contents, livers, and spleens based on direct plating was analyzed by a nonparametric analysis using the Kruskal-Wallis test. Dunn's multiple comparison test was used to compare the different groups. Enrichment data from the livers and spleens were analyzed by using the chi-squared test. The Fisher exact test was used to compare the different groups. A P value of <0.05 was considered significant.
RESULTS
The ΔSPI-1 and ΔinvG strains are impaired in the secretion of SipD.
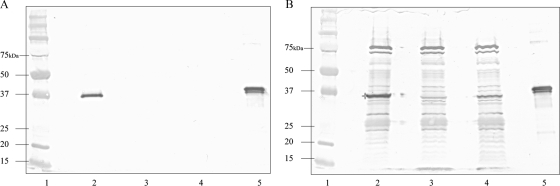
In order to determine whether the S. Enteritidis mutants constructed were impaired in their ability to secrete SPI-1 T3SS proteins, Western blots of bacterial cell lysates and culture supernatants were probed with serum against the His tag derivative (38.5 kDa) of the SipD protein, the gene of which is encoded by SPI-1 (39). As expected, SipD (37 kDa) was expressed in the wild-type strain and was found in both the supernatant and the cellular fractions (Fig. 1A and B, lane 2). Likewise, SipD was not detected in either the supernatant or cellular fractions of the ΔSPI-1 strain (Fig. 1A and B, lane 3). The ΔinvG strain was unable to secrete SipD into the culture supernatant (Fig. 1A, lane 4), although the protein was present in the cellular fraction (Fig. 1B, lane 4).
FIG. 1.
Western blot analysis of bacterial culture supernatants (A) and pellet fractions (B) of wild-type Sal 18 and the mutant strains constructed, using rabbit anti-His tag SipD polyclonal serum. Lane 1, prestained protein marker; lane 2, wild-type Sal 18; lane 3, Sal 18 ΔSPI-1; lane 4, Sal 18 ΔinvG; lane 5, purified His tag SipD (38.5 kDa). The SipD protein (37 kDa) was present in the supernatant of the wild-type strain (A, lane 2) but not in the Sal 18 ΔSPI-1 and Sal 18 ΔinvG strains (A, lanes 3 and 4, respectively). In the pellet fractions, SipD was present (*) in the wild-type strain pellet (B, lane 2), but not in the Sal 18 ΔSPI-1 pellet (B, lane 3). The Sal 18 ΔinvG pellet contained some SipD, but not as much as the wild type (B, lane 4).
SPI-1 is important for efficient invasion in polarized Caco-2 cells and causes a reduction in the TER.
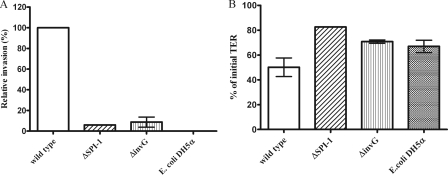
S. Enteritidis ΔSPI-1 and ΔinvG mutant strains were tested for their ability to invade polarized Caco-2 cells relative to the wild-type strain by using the gentamicin protection assay. The ΔSPI-1 and ΔinvG mutant strains were recovered at lower levels relative to the wild-type strain, while the E. coli strain (control) was unable to invade the Caco-2 cells. Invasion by the ΔSPI-1 and ΔinvG mutant strains was 6.02 and 8.79%, respectively, of the wild-type CFU (Fig. 2A). The TER (Fig. 2B), on the other hand, was significantly reduced in the wild type-infected group (50% of the initial value) compared to the ΔSPI-1 group (82.59% of the initial value), while the TER of the ΔinvG mutant-infected group (70.79% of the initial value) was also reduced, although not to wild-type levels, indicating that SPI-1 is important for tight junction disruption. The TER of the control group incubated with E. coli was reduced to 67% of the initial value.
FIG. 2.
Single infection of polarized Caco-2 monolayers by either S. Enteritidis wild-type, ΔSPI-1, or ΔinvG strains or E. coli DH5α at an MOI of 100. Values represent means ± the standard error of the mean of two independent experiments performed in triplicate. (A) Invasion is expressed as a percentage of the wild-type strain (B) The change in TER is expressed as a percentage of the initial TER value.
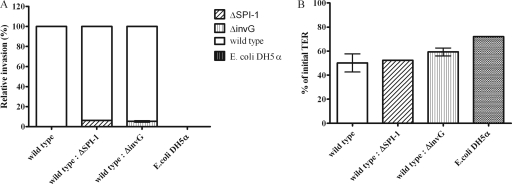
Using competition studies, polarized Caco-2 cells were infected with a 1:1 ratio of either S. Enteritidis wild-type and ΔSPI-1 strains or wild-type and ΔinvG strains to test the ability of the mutant strains to invade Caco-2 cells in the presence of the wild-type strain. ΔSPI-1 and ΔinvG strains were impaired in invasion compared to the wild-type strain, confirming what was seen in Fig. 2A (single strain infection). The ΔSPI-1 and ΔinvG strains showed reduced invasion (6.28 and 5.31%, respectively) relative to the wild-type strain CFU (Fig. 3A). The TERs (Fig. 3B) of the coinfected groups (wild-type and ΔSPI-1 strains or wild-type and ΔinvG strains) were similar (54 and 58% of the initial TER, respectively) to that of the wild type-infected group (50% of the initial TER). This was expected since the coinfected groups contained the wild-type strain.
FIG. 3.
Mixed infection of polarized Caco-2 monolayers by S. Enteritidis wild-type and ΔSPI-1 strains or wild-type and ΔinvG strains at a 1:1 ratio with an MOI of 100. Values represent the means of two independent experiments ± the standard error of the mean performed in triplicate. (A) Invasion is expressed as a percentage of the wild-type strain. (B) The change in TER is expressed as a percentage of the initial TER value.
The SPI-1-deficient strain is less invasive than the wild-type strain in chicken intestinal tissue explants.
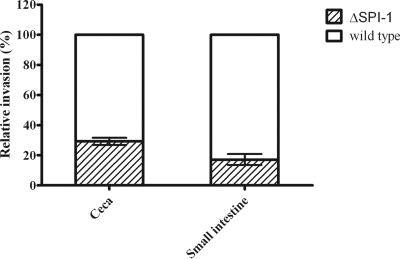
Chicken intestinal tissue samples were used to test the invasion of the S. Enteritidis wild-type and ΔSPI-1 strains using the gentamicin protection assay as described above. In this competition experiment, the SPI-1-deficient strain was recovered at a lower rate from both the cecal and small-intestinal tissue explants (29.3 and 17.2%, respectively) compared to the wild-type strain (Fig. 4), while the E. coli strain (control) was not recovered (data not shown). This provided more evidence to suggest that SPI-1 plays a role in breaching the chicken intestinal epithelial barrier. However, the invasion defect of the ΔSPI-1 strain was relatively smaller in comparison with the invasion defect observed in polarized Caco-2 cells.
FIG. 4.
Invasion of chicken cecal and small-intestinal explants by S. Enteritidis wild-type and ΔSPI-1 strains at a 1:1 ratio. Values represent means of at least three independent experiments ± the standard error of the mean performed in triplicate. Invasion is expressed as a percentage of the wild-type strain.
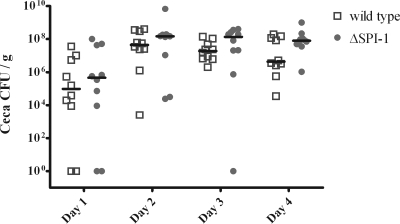
Cecal colonization levels were similar in both wild-type strain- and ΔSPI-1 strain-challenged groups.
One-week-old birds were orally challenged with 1010 CFU of either the wild-type strain or the ΔSPI-1 strain as described in Materials and Methods. Birds were euthanized on days 1, 2, 3, and 4 postchallenge. We did not see any statistically significant difference between the cecal colonization (Fig. 5) of the wild-type strain- and the ΔSPI-1 strain-challenged groups over the duration of the experiment.
FIG. 5.
Colonization of ceca from 1-week-old chicken by either wild-type S. Enteritidis or ΔSPI-1. The chickens were orally challenged with 1010 CFU of either wild type or mutant. Ten birds per group were euthanized on days 1, 2, 3, and 4 postinfection.
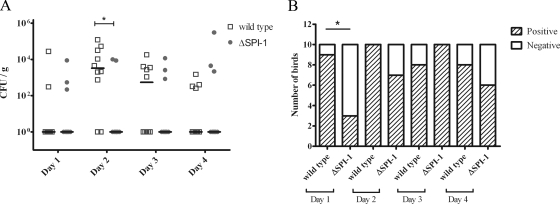
The deletion of SPI-1 results in delayed systemic infection in chickens.
Analysis of the bacterial load in the liver (Fig. 6A) revealed that on the first day postchallenge, there was very little detectable S. Enteritidis wild-type and ΔSPI-1 strains, suggesting that the systemic spread of the bacteria was still in its early stages. This idea is supported by data from enriched samples (Fig. 6B), which revealed that on day 1 postchallenge, 9 of 10 livers were infected in the wild-type-challenged group, while only 3 of 10 livers (P < 0.05) were infected in the ΔSPI-1 strain-challenged group. At 2 days postchallenge, the recovery of the ΔSPI-1 strain (Fig. 6A), as evident from direct plating, was significantly lower (P < 0.05) in comparison to the wild-type strain (3.3 × 103 CFU/g). However, liver enrichment data (Fig. 6B) demonstrated that the number of livers that were positive in the ΔSPI-1 strain-challenged group (7/10) was only marginally lower than in the wild-type-challenged group (10/10). On day 3 postchallenge the levels of both the wild-type and ΔSPI-1 strains (Fig. 6A), based on direct counts, followed a similar trend. The data from enriched samples (Fig. 6B) suggested that the infection was starting to clear in the wild-type-challenged group on the third day (8 of 10 livers were positive), while 10 of 10 livers were positive for Salmonella in the ΔSPI-1 strain-inoculated group. After 4 days, the bacterial levels (Fig. 6A) in both the wild-type- and ΔSPI-1 strain-challenged groups was mostly below the detection limit using direct plating, which demonstrated that a clearing of the infection was in progress. As for the enriched liver samples (Fig. 6b), 8 of 10 livers were still positive for the Salmonella wild-type strain versus 6 of 10 livers in the ΔSPI-1 strain-challenged group. This suggests that the ΔSPI-1 strain was initially slower in spreading systemically but also started to clear faster relative to the wild-type strain.
FIG. 6.
Systemic infection of the liver in 1-week-old chickens challenged with 1010 CFU of either wild-type S. Enteritidis or ΔSPI-1. Ten birds were euthanized per day in each group. (A) Viable counts of Salmonella from direct plating represented as the median CFU/g. (B) Number of birds positive for Salmonella after enrichment. *, P < 0.05.
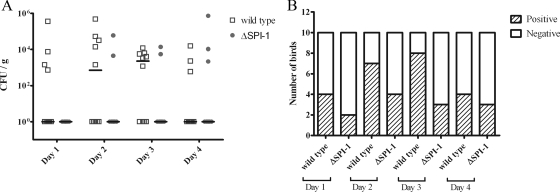
The recovery of S. Enteritidis wild-type and ΔSPI-1 strains from chicken spleens was similar to what was observed in the liver. On the first day postchallenge (Fig. 7A) the infection was still at its early stage, and therefore the median group CFU recovery, as determined by direct plating, was below the detection limit in the wild-type- and ΔSPI-1 strain-challenged birds. Similarly, the data from enriched spleen samples (Fig. 7B) suggested that there was little difference in the number of spleens that were positive for Salmonella in both groups. On day 2 postchallenge, the recovery of bacteria in the wild-type-challenged group was 7.2 × 102 CFU/g, while that of the ΔSPI-1-challenged group remained unchanged (Fig. 7A). Likewise, data from enriched samples showed that the number of spleens that were positive for wild-type Salmonella were slightly higher (7/10) than that of the ΔSPI-1 strain-challenged group (4/10). On the third day after challenge, bacterial levels in the spleen, measured by direct plating (Fig. 7A) and after enrichment (Fig. 7B), by both wild-type and ΔSPI-1 strains followed a similar pattern as on day 2. On the last day after challenge (Fig. 7A), only a few birds were found with countable Salmonella on BGA in either the wild-type- or ΔSPI-1 strain-challenged groups. In accordance with this finding, data from enriched samples showed a marginal difference between the number of spleens that were positive in both wild-type- and ΔSPI-1 strain-challenged groups, suggesting that the infection was clearing. The values described above represent median group numbers after statistical processing. For CFU recovered from individual birds, see Fig. 5, 6, and 7.
FIG. 7.
Systemic infection of the spleen in 1-week-old chickens challenged with 1010 CFU of either wild-type S. Enteritidis or ΔSPI-1. Ten birds were euthanized per day in each group. (A) Viable counts of Salmonella from direct plating represented as the median CFU/g. (B) Number of birds positive for Salmonella after enrichment.
DISCUSSION
The role of SPI-1 in the pathogenesis of S. enterica has been studied in detail in both the mouse enterocolitis and the bovine enteritis models of infection (10, 11, 21, 25, 50). However, the contribution of SPI-1 is still not fully understood in the chicken colonization model. The goal of the present study was to determine the role of S. Enteritidis SPI-1 in invasion using polarized Caco-2 cells, as well as chicken intestinal explants, and to test the effects of a SPI-1 deletion mutant on cecal colonization and systemic spread of the organism in chickens over a period of 4 days postchallenge.
We chose to construct deletion mutants in SPI-1 since it has been well documented that the SPI-1 T3SS is essential for the secretion of effector proteins and plays an important role in infection in different animal models (3, 4, 43, 52). Thus, it would be expected that deletion of the pathogenicity island in its entire form or the knockout of certain individual genes would not allow the bacterium to form a fully functional T3SS apparatus or to secrete any of the SPI-1 related effectors in order to cause intestinal invasion and facilitate bacterial uptake. The ΔSPI-1 and ΔinvG mutants constructed were tested for their ability to secrete effector proteins in vitro by using Western blot analysis, which has been previously used in several studies as a functional assay to determine the effect of SPI-1 deletions on secretion (35, 37, 56). We chose to monitor the secretion of SipD in the supernatant and pellet fractions of the mutant strains, since SipD is secreted using this system and forms part of the SipB/C/D translocase complex at the tip of the SPI-1 T3SS apparatus (52). The SipD protein was not detected in the supernatant or pellet fractions of the ΔSPI-1 mutant strain, confirming that SipD was not expressed in the cell, since the gene encoding SipD had been deleted. In addition, SipD was not detected in the supernatant fraction of the ΔinvG strain, as suggested by reports that show that ΔinvG strains are unable to secrete effector proteins (46). The presence of SipD in the cellular fraction of the ΔinvG strain is also expected since the sipD gene was not deleted in this strain. Interestingly, the level of SipD present in the cellular fraction of the ΔinvG mutant strain was much lower than the amount present in the wild-type fraction. We speculate that this may be due to the fact that the regulation of SPI-1 involves several regulators, including the main regulator, HilA (encoded on SPI-1). The HilA protein is known to bind directly to promoters on the inv-spa and prg-org operons on SPI-1, thus activating the expression of several genes encoding components of the SPI-1 T3SS. Activation of the inv-spa operon by HilA eventually leads to the activation of the sic-sip operon, which encodes SipD. Further, activation of the inv-spa operon results in the expression of InvF (a positive transcriptional activator) which, along with the chaperone SicA, also induces the expression of SPI-1 secreted proteins on the sic-sip operon and those located outside SPI-1 (14). Therefore, it is possible that the deletion of invG affects the expression of genes in the inv-spa operon, which ultimately affects downstream genes such as sipD in the sic-sip operon.
In order to examine the contribution of S. Enteritidis SPI-1 to invasion, we used polarized Caco-2 cells. This cell line was chosen due to the absence of well-characterized chicken epithelial cell lines. Moreover, Caco-2 cells are capable of polarizing on transwells, resulting in well-defined apical and basolateral compartments, making them a good model for studying host cell invasion. Caco-2 cells also form well-defined brush borders and express several markers that are present on small-intestinal villus cells (15). Further, it has been demonstrated that polarized epithelial cells reveal additional information, with respect to invasion, which would not have been possible with nonpolarized cells (51). The choice of this cell line was also relevant to the present study since S. Enteritidis is also a potential human pathogen (19). Our results from the invasion assays using Caco-2 cells implied that ΔSPI-1 and ΔinvG mutant strains were impaired in invasion relative to the wild-type strain in both the single infection experiments and competition experiments. This was in line with other reports that demonstrated that SPI-1 was important for invasion in cell culture (24, 33, 34, 36). The difference in invasion between the wild-type and SPI-1 mutant strains was ∼10-fold based on our invasion assays, which is comparable to what has been observed by others in similar experiments with S. Typhimurium and S. enterica subspecies enterica serovar Typhi (2, 4, 16, 41, 51, 56). These data also suggested that the role of S. Enteritidis SPI-1 in tissue culture models was similar to that observed for S. Typhimurium. To our knowledge, this is the first time S. Enteritidis ΔSPI-1 and ΔinvG mutants have been tested (both single and mixed infections) using polarized Caco-2 cells.
The disruption of tight junctions is a strategy used by different bacterial pathogens, including Salmonella, to cause damage to the integrity of the epithelial cell layer (53). We used the TER as an indicator of cell monolayer health, which has been widely used by others to assess the effect of bacterial pathogens on tight junctions (7, 8, 15). Our results from the invasion assays (single infection) suggest that both S. Enteritidis ΔSPI-1 and ΔinvG mutant strains caused a smaller reduction of TER compared to the wild-type strain. This was expected since the SPI-1 mutants used in our study were impaired in the secretion of SPI-1 effector proteins. On the other hand, there was no difference in the TER between the wild type alone and coinfected groups, which contained the wild-type and either ΔSPI-1 or ΔinvG strains. This can be explained by the fact that the coinfected groups all contained the wild-type strain, which is capable of secreting SPI-1 effector proteins. The E. coli strain also caused a reduction in the TER, which was similar to that caused by the SPI-1 mutant strains. This finding was not expected because the E. coli strain used was a nonpathogenic strain that was not able to invade Caco-2 cells in our assays. A possible reason for this could be that E. coli K-12 may produce unidentified factors that reduce the TER.
Intestinal tissue explants represent a valid model for studying invasion since they bridge the gap between tissue culture-based assays and animal experiments. Such explants are commonly used to provide more insights into host-pathogen interactions (22, 26, 51). Our data from the chicken cecal and small intestinal explants suggest that SPI-1 mediated invasion is important for breaching the chicken intestinal epithelial barrier. This is in agreement with data obtained from bovine intestinal explants that were infected with a SPI-1 mutant of S. Typhimurium (51). However, we observed that the difference in invasiveness between the wild type and the ΔSPI-1 strain was smaller than the difference observed in polarized Caco-2 cells. A similar finding reported in bovine intestinal explants (51) implies that other mechanisms besides SPI-1 may also be involved in the process of invasion. We have shown for the first time that SPI-1 mutants are impaired in invasion of chicken cecal and small-intestinal tissue explants.
To extend our observations from the in vitro studies using SPI-1 mutants, we tested the effect of the ΔSPI-1 mutant on the colonization and systemic spread of the organism in chickens. Only the ΔSPI-1 mutant was tested in chickens, since the ΔinvG mutant strain was similar to the ΔSPI-1, in that both strains were impaired in the secretion of SipD and showed reduced invasion in polarized Caco-2 cells. In addition, to get a better understanding of the contribution of SPI-1 in chickens, we examined the effect of a SPI-1 deletion over a period of 4 days after oral challenge with either wild-type S. Enteritidis or ΔSPI-1 mutant strains. The extended time frame is crucial since it is possible that the absence of SPI-1 may have a pronounced effect on pathogenesis at a certain stage of infection, while at other stages the effect may not be obvious.
Our data indicate that the absence of SPI-1 does not significantly affect cecal colonization on all 4 days after oral challenge. This is in agreement with what has been observed in day-old chicks orally challenged with a S. Typhimurium ΔspaS (SPI-1 gene deletion) mutant strain. A similar finding was reported when 1-week-old chicks were used in the same study. However, cecal colonization levels of the ΔspaS strain-challenged group were lower than for the wild-type strain on day 14 postchallenge (32). Likewise, lower levels of cecal colonization were observed in day-old chicks challenged with an S. Enteritidis ΔhilA strain than in chicks challenged with the wild type only after day 6 postchallenge (3). As well, reduced levels of S. Typhimurium ΔSPI-1 were reported in cecal tissue of 1-week-old chickens over a 14-day period (13). Hence, it is likely that differences between cecal colonization levels of wild-type- and SPI-1 mutant-challenged groups are evident when the colonization is monitored over several weeks. We did not extend the chicken experiments beyond 4 days after oral inoculation since we have observed that the chickens start to clear the infection.
Our systemic infection data imply that the absence of SPI-1 affects systemic infection, as detected on days 2 and 3 after oral challenge. However, since the detection limit of viable Salmonella is 102 CFU/g in our sample processing regime (direct plating without enrichment), it is highly likely that many liver and spleen samples from infected birds appear as false negatives. The systemic infection data after enrichment of the liver and spleen samples provide a better insight into the effect of a SPI-1 deletion on the systemic spread of S. Enteritidis. It is apparent from the enrichment data that the ΔSPI-1 strain causes a delay in infection of the livers (P < 0.05) and spleens on the first day postchallenge. Subsequently, the ΔSPI-1 strain was recovered from the liver and spleen at levels that were similar to those for the wild-type strain. This is in line with what has been observed when chickens were challenged with an S. Enteritidis ΔhilA strain (3), S. Typhimurium ΔspaS strain (32), or S. Typhimurium ΔSPI-1 strain (13). However, our results from the systemic infection data are in contrast to what has been seen when 3-week-old chicks were challenged with an S. Enteritidis ΔsipD strain, where none of the spleens were infected on day 3 postchallenge (44). A likely explanation for this might be that the birds were sampled at one time point and were only based on direct plating. Hence, it would be difficult to determine how the sipD mutant would have affected colonization over a longer time period, given the fact that our data indicate that there is a delay in systemic infection initially by the ΔSPI-1 mutant strain. As well, our results are not in agreement with the findings from a study in which the S. Typhimurium ΔspaS strain was not detected on days 1, 7, and 14 postchallenge in the livers of 1-week-old chickens, relative to the wild-type strain, based on direct plating (32). One possible explanation for this difference is that S. Typhimurium and S. Enteritidis are two different organisms, and thus their method of establishing infection may be different in chickens. Alternatively, the ΔspaS strain may have been present, but below the detection limit, which is what we observed in our systemic infection data.
The results from our in vivo study imply that the SPI-1 virulence determinant does not play a major part in S. Enteritidis infection in chicken. A possible hypothesis for this finding could be that S. Enteritidis SPI-1 genes are not expressed in vivo in chicken. However, data from our lab indicate that sera from chickens challenged with wild-type S. Enteritidis react with purified recombinant components of the SPI-1 T3SS (data not shown), suggesting that the SPI-1 T3SS is expressed during infection of chickens. Our systemic infection results (enrichment) illustrate that the absence of SPI-1 results in a delay in infection relative to the wild-type strain. This further confirms the concept that the SPI-1 T3SS comes into play at some point during the process of infection in chickens. Recently, SipA, SopA, SopB, SopD, and SopE2 (SPI-1 effector proteins) have been detected in the spleens of mice challenged with S. Typhimurium (21). Although the murine model of salmonellosis is different from the chicken model, this finding not only confirms that SPI-1 is expressed in vivo but also provides support for the role of SPI-1 in systemic infection. A recent report has shown that SPI-1 genes are highly expressed at early and late stages of infection in cultured epithelial cells (27). This finding, despite being from an in vitro study, is significant since it provides further evidence to suggest that the role of SPI-1 can be extended to the systemic phase of infection and requires further investigation.
The prevailing view has been that SPI-1 is essential for intestinal pathogenesis in different animal models. However, recent evidence from the bovine and murine models of infection suggests that S. Typhimurium can cause infection in an SPI-1-independent manner (11, 25, 55). Further, it has been reported that SPI-1 mutants retain their ability to invade M cells in a murine gut loop model (9), implying that intestinal invasion may involve other factors than SPI-1. In addition, it has been shown that human isolates of S. enterica subspecies enterica serovar Senftenberg cause intestinal inflammation despite the fact that they are SPI-1 deficient (28). Taken together, this, along with our data from the chicken colonization study, suggests that the SPI-1 T3SS is not a critical factor for crossing the intestinal epithelial barrier in some animals.
Although there are differences between Salmonella infections that occur naturally under field conditions and those that occur under experimental conditions, the results from our chicken colonization study, as well as those obtained by others, provide insights into the mechanisms by which S. enterica serovars Enteritidis and Typhimurium colonize chickens. We chose to use a high dose for oral challenge of chickens, unlike what the birds would encounter in a natural setting, since we observed very low levels of colonization at lower doses (data not shown), making it difficult to interpret the data. In addition, other groups have also used high challenge doses in chicken experiments (3, 13, 32), thus validating our findings.
In summary, we have shown that S. Enteritidis ΔSPI-1 and ΔinvG strains were unable to secrete effector proteins using SipD as an indicator of SPI-1 effector protein secretion. The mutants showed reduced invasion of both polarized Caco-2 cells and tissue explants obtained from chicken small intestine and ceca. Our data further suggest that the deletion of SPI-1 does not affect cecal colonization in one-week-old chicken but causes a milder and delayed systemic infection, as revealed by samples from the liver and spleen. Additional work needs to be done to determine the individual SPI-1 effectors responsible for systemic infections in chicken.
Acknowledgments
We thank Aaron P. White and Michael G. Surrette (University of Calgary) for providing the lambda red plasmids. We also thank Erin Boyle and B. Brett Finlay (University of British Columbia) for providing the Caco-2 cells and for assistance in establishing polarized monolayers. We are indebted to Brenda Allan (VIDO) for stimulating discussions. We especially thank VIDO Animal Care for help with the animal experiments and GMP for providing reagents.
This project was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and Bioniche Life Sciences, Inc. A.A.P. holds an NSERC Senior Industrial Research Chair in food and water safety vaccines. W.K. holds an NSERC Associate Industrial Research Chair in food and water safety vaccines. W.K. received a New Investigator Establishment Grant from the Saskatchewan Health Research Foundation.
This report was published with the permission of the Director of VIDO as journal series number 517.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 13 April 2009.
REFERENCES
- 1.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Rüssmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, A., D. House, T. Perkins, S. Baker, R. A. Kingsley, and R. Dougan. 2008. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: role of surface structures in adhesion and invasion. Microbiology 1541914-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohez, L., R. Ducatelle, F. Pasmans, N. Botteldoorn, F. Haesebrouck, and F. Van Immerseel. 2006. Salmonella enterica serovar Enteritidis colonization of the chicken caecum requires the HilA regulatory protein. Vet. Microbiol. 116202-210. [DOI] [PubMed] [Google Scholar]
- 4.Boyen, F., F. Pasmans, F. Van Immerseel, E. Morgan, C. Adriaensen, J. Hernalsteens, A. Decostere, R. Ducatelle, and F. Haesebrouck. 2006. Salmonella Typhimurium SPI-1 genes promote intestinal but not tonsillar colonization in pigs. Microbes Infect. 11335-44. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, E. C., N. F. Brown, and B. B. Finlay. 2006. Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2, and SipA disrupt tight junction structure and function. Cell. Microbiol. 81946-1957. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. F., B. A. Vallance, B. K. Coombes, Y. Valdez, B. A. Coburn, and B. B. Finlay. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog. 10001-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canil, C., I. Rosenshine, S. Ruschkowski, M. S. Donnenberg, J. B. Kaper, and B. B. Finlay. 1993. Enteropathogenic Escherichia coli decreases the transepithelial electrical resistance of polarized epithelial monolayers. Infect. Immun. 612755-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M. L., Z. Ge, J. G. Fox, and D. B. Schauer. 2006. Disruption of tight junctions and induction of proinflammatory cytokine responses in colonic epithelial cells by Campylobacter jejuni. Infect. Immun. 746581-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, M. A., K. A. Reed, J. Lodge, J. Stephen, B. H. Hirst, and M. A. Jepson. 1996. Invasion of murine intestinal M cells by Salmonella typhimurium inv mutants severely deficient for invasion of cultured cells. Infect. Immun. 644363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coburn, B., Y. Li, D. Owen, B. A. Vallance, and B. B. Finlay. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of Infectious enterocolitis. Infect. Immun. 733219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombes, B. K., B. A. Coburn, A. A. Potter, S. Gomis, K. Mirakhur, Y. Li, and B. B. Finlay. 2005. Analysis of the contribution of Salmonella pathogenicity island 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect. Immun. 737161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieye, Y., K. Ameiss, M. Mellata, and R. R. Curtiss. 2009. The Salmonella pathogenicity island (SPI) 1 contributes more than SPI2 to the colonization of the chicken by Salmonella enterica serovar Typhimurium. BMC Microbiol. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 1024-29. [DOI] [PubMed] [Google Scholar]
- 15.Finlay, B. B., and S. Falkow. 1990. Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J. Infect. Dis. 1621096-1106. [DOI] [PubMed] [Google Scholar]
- 16.Forbes, S. J., M. Eschmann, and N. J. Mantis. 2008. Inhibition of Salmonella enterica serovar Typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun. 764137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 1753-86. [DOI] [PubMed] [Google Scholar]
- 18.Gast, R. K. 2003. Salmonella infections, p. 567. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, and L. R. McDougald (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames, IA.
- 19.Gast, R. K. 2007. Serotype-specific and serotype-independent strategies for preharvest control of food-borne Salmonella in poultry. Avian Dis. 51817-828. [DOI] [PubMed] [Google Scholar]
- 20.Gaylov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Waston, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25903-912. [DOI] [PubMed] [Google Scholar]
- 21.Giacomodonato, M. N., S. Uzzau, D. Bacciu, R. Caccuri, S. H. Sarnacki, S. Rubino, and M. C. Cerquetti. 2007. SipA, SopA, SopB, SopD, and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology 1531221-1228. [DOI] [PubMed] [Google Scholar]
- 22.Grant, A. J., J. Woodward, and D. J. Maskell. 2006. Development of an ex vivo organ culture model using human gastro-intestinal tissue and Campylobacter jejuni. FEMS Microbiol. Lett. 263240-243. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hapfelmeier, S., K. Ehrbar, B. Stecher, M. Barthel, M. Kremer, and W. Hardt. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72795-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hapfelmeier, S., B. Stecher, M. Barthel, M. Kremer, A. J. Müller, M. Heikenwalder, T. Stallmach, M. Hensel, K. Pfeffer, S. Akira, and W.-D. Hardt. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 1741675-1685. [DOI] [PubMed] [Google Scholar]
- 26.Haque, A., F. Bowe, R. J. Fitzhenry, G. Frankel, M. Thomson, R. Heuschkel, S. Murch, M. P. Stevens, T. S. Wallis, A. D. Philips, and G. Dougan. 2004. Early interactions of Salmonella enterica serovar Typhimurium with human small intestinal epithelial explants. Gut 531424-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hautefort, I., A. Thompson, S. Eriksson-Ygberg, M. L. Parker, S. Lucchini, V. Danino, R. J. M. Bongaerts, N. Ahmad, M. Rhen, and J. C. D. Hinton. 2008. During infection of epithelials cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10958-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, Q., B. Coburn, W. Deng, Y. Li, X. Shi, Q. Lan, B. Wang, B. K. Coombes, and B. B. Finlay. 2008. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J. Clin. Microbiol. 461330-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imke Hansen-Wester, M. H. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3549-559. [DOI] [PubMed] [Google Scholar]
- 31.Jepson, M. A., C. B. Collares-Busto, M. A. Clark, B. H. Hirst, and N. L. Simmons. 1995. Rapid disruption of epithelial barrier function by Salmonella typhimurium is associated with structural modification of intercellular junctions. Infect. Immun. 63356-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, M. A., S. D. Hulme, P. A. Barrow, and P. Wigley. 2007. The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol. 36199-203. [DOI] [PubMed] [Google Scholar]
- 33.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1777078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 1773965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 9711008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubori, T., and J. E. Galan. 2002. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J. Bacteriol. 1844699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubori, T., A. Sukhan, S.-I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 9710225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth rate. Infect. Immun. 874304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lostron, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 31281-1291. [DOI] [PubMed] [Google Scholar]
- 40.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2145-156. [DOI] [PubMed] [Google Scholar]
- 41.Mirold, S., K. Ehrbar, A. Weissmüller, R. Prager, H. Tschäpe, H. Rüssmann, and W.-D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 1832348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi, J., and Y. Takai. 2005. Molecular perspective on tight-junction assembly and epithelial polarity. Adv. Drug Deliv. Rev. 57815-855. [DOI] [PubMed] [Google Scholar]
- 43.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 685050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker, C. T., and J. Guard-Petter. 2001. Contribution of flagella and invasion proteins to pathogenesis of Salmonella enterica serovar Enteritidis in chicks. FEMS Microbiol. Lett. 204287-291. [DOI] [PubMed] [Google Scholar]
- 45.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host cytoskeleton by Salmonella: all in the name of entry. Curr. Opin. Microbiol. 810-15. [DOI] [PubMed] [Google Scholar]
- 46.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24697-709. [DOI] [PubMed] [Google Scholar]
- 47.Poppe, C. 2000. Salmonella infections in the domestic fowl, p. 107-132. In C. Wray and A. Wray (ed.), Salmonella in domestic animals. Oxford University Press, New York, NY.
- 48.Porter, S. B., and R. Curtiss III. 1997. Effect of inv mutations on Salmonella virulence and colonization in 1-day-old White Leghorn chicks. Avian Dis. 4145-57. [PubMed] [Google Scholar]
- 49.Porwollik, S., F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 1865883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pullinger, G. D., S. M. Paulin, B. Charleston, P. R. Watson, A. J. Bowen, F. Dziva, E. Morgan, B. Villarreal-Ramos, T. S. Wallis, and M. P. Stevens. 2007. Systemic translocation of Salmonella enterica serovar Dublin in cattle occurs predominantly via efferent lymphatics in a cell-free niche and requires type III secretion system 1 (T3SS-1) but not T3SS-2. Infect. Immun. 755191-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raffatellu, M., R. P. Wilson, D. Chessa, H. Andrews-Polymenis, Q. T. Tran, S. Lawhon, S. Khare, L. G. Adams, and A. J. Baümler. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73146-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlumberger, M. C., and W. D. Hardt. 2005. Salmonella type III secretion effectors: pulling the host cell's strings. Curr. Opin. Microbiol. 91-9. [DOI] [PubMed] [Google Scholar]
- 53.Sousa, S., M. Lecuit, and P. Cossart. 2005. Microbial strategies to target, cross or disrupt epithelia. Curr. Opin. Microbiol. 17489-498. [DOI] [PubMed] [Google Scholar]
- 54.Tafazoli, F., K. Magnusson, and L. Zheng. 2003. Disruption of epithelial barrier integrity by Salmonella enterica serovar Typhimurium requires geranylgeranylated proteins. Infect. Immun. 71872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vazquez-Torres, A., J. Jones-Carson, A. J. Baümler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401804-808. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, J. W., and C. A. Nickerson. 2006. Cloning of a functional Salmonella SPI-1 type III secretion system and development of a method to create mutations and epitope fusions in the cloned genes. J. Biotechnol. 122147-160. [DOI] [PubMed] [Google Scholar]