Abstract
Borrelia burgdorferi, the Lyme disease-causing spirochete, can persistently infect its vertebrate hosts for years. B. burgdorferi is often found associated with host connective tissue, where it interacts with components of the extracellular matrix, including fibronectin. Some years ago, a borrelial surface protein, named BBK32, was identified as a fibronectin-binding protein. However, B. burgdorferi BBK32 mutants are still able to bind fibronectin, indicating that the spirochete possesses additional mechanisms for adherence to fibronectin. We now demonstrate that RevA, an unrelated B. burgdorferi outer surface protein, binds mammalian fibronectin in a saturable manner. Site-directed mutagenesis studies identified the amino terminus of the RevA protein as being required for adhesion to fibronectin. RevA bound to the amino-terminal region of fibronectin. RevA binding to fibronectin was not inhibited by salt or heparin, suggesting that adhesin-ligand interactions are primarily nonionic and occur through the non-heparin-binding regions of the fibronectin amino-terminal domains. revA genes are widely distributed among Lyme disease spirochetes, and the present studies determined that all RevA alleles tested bound fibronectin. In addition, RevB, a paralogous protein found in a subset of B. burgdorferi strains, also bound fibronectin. We also confirmed that RevA is produced during mammalian infection but not during colonization of vector ticks and determined that revA transcription is controlled through a mechanism distinct from that of BBK32.
Borrelia burgdorferi, the infectious agent of Lyme disease, is capable of infecting humans and other vertebrates for extensive periods, even for the host's lifetime (17, 54, 70). B. burgdorferi is an extracellular organism, frequently found associated with host connective tissues (8, 37, 44, 49, 57). In vitro, the Lyme disease spirochete shows affinity for various host extracellular matrix (ECM) components, such as fibronectin (6, 8, 15, 16, 31, 63). The hosts’ ECM may provide a protective niche for B. burgdorferi, allowing the spirochete to persist in the host despite vigorous humoral and cellular immune responses. The collective evidence suggests that B. burgdorferi's interactions with components of the ECM are important in both the pathogenesis of Lyme disease and the maintenance of B. burgdorferi infection in mammals.
The interactions between B. burgdorferi and fibronectin may be especially important, as soluble fibronectin or antifibronectin antibodies inhibit spirochetal interactions with cultured endothelial cells and their secreted matrices (43, 74). One B. burgdorferi fibronectin-binding protein, BBK32, has previously been identified and characterized (42, 63-65). That outer surface protein is not essential for mammalian infection, although 15-fold more BBK32 mutant bacteria were required for 50% infection of mice (46, 67). Importantly, BBK32 mutant bacteria still bind fibronectin (67). Those results indicate that the fibronectin-binding activity of BBK32 is redundant to another, previously unidentified borrelial fibronectin-binding activity.
RevA is a surface-exposed 17-kDa outer membrane protein of B. burgdorferi with no significant homology to any bacterial proteins outside Borrelia spp. (13, 61). RevA is expressed during mammalian infection and repressed in the tick vector (29, 30, 69; this work). The production of RevA can be regulated in vitro, with the protein being produced under temperature and pH conditions that mimic those found in its warm-blooded host (13). The function of RevA was previously unknown, yet its surface location and expression during mammalian infection suggested to us that this protein may be involved in interactions with the Lyme disease spirochete's host. To that end, we investigated the potential of RevA to adhere to mammalian tissue components, including fibronectin, and subsequently investigated RevA-fibronectin interactions in detail.
MATERIALS AND METHODS
Bacteria.
B. burgdorferi strain B31 MI-16 is an infectious clone of the sequenced type strain (14, 26) which contains all parental plasmids (53). Strain B31-A3 is a distinct clonal derivative of strain B31 (21). Strains B31-A3ntrA and B31-A3rpoS are rpoN (ntrA) and rpoS mutants, respectively, of strain B31-A3 and were kind gifts of Patricia Rosa (21, 25). Strain ML23, a B31 derivative lacking lp25, and strain JS315, a BBK32 mutant of ML23, were kind gifts from Jonathan Skare (67). Strain N40 is a wild-type isolate originally obtained from a tick (1), and strain 297 was isolated from human spinal fluid (71). Borrelia garinii strain PBi, a European isolate from human cerebrospinal fluid (62), was a kind gift from Peter Kraiczy. All Borrelia strains were grown at 34°C to cell densities of approximately 1 × 107 bacteria/ml in modified Barbour-Stoenner-Kelly (BSK-II) medium supplemented with 6% rabbit serum (78). Total DNA (genomic and plasmids) was isolated using a DNEasy blood and tissue kit (Qiagen, Valencia, CA).
Recombinant proteins.
Recombinant proteins contained amino-terminal polyhistidine tags, with the RevA or RevB segment beginning with that protein's first amino acid following the cysteine lipidation site. revA genes were PCR amplified from total genomic DNA of B. burgdorferi strains B31 MI-16, N40, and 297 and of B. garinii strain PBi and revB was amplified from B. burgdorferi strain B31 MI-16 using oligonucleotides listed in Table 1. Amplicons were cloned into pET200 (Invitrogen, Carlsbad, CA). Recombinant ErpA has been described previously (72). The resultant plasmid inserts were entirely sequenced on both strands to ensure that no undesired mutations had occurred during PCR or cloning procedures. Recombinant proteins were expressed in Escherichia coli strain Rosetta (DE3) pLysS (Novagen, Madison, WI) upon induction with isopropyl thiogalactopyranoside. Induced E. coli cultures were harvested and lysed by sonication, and debris cleared by centrifugation. Recombinant proteins were purified from cleared lysates by using either MagneHis nickel-conjugated magnetic beads (Promega, Madison, WI) or His-Trap HP columns and an ÄKTA-FPLC system equipped with a UPC-900 UV absorbance monitor and a Frac920 fraction collector (GE Healthcare, Piscataway, NJ). Proteins were eluted from fast protein liquid chromatography columns by a constantly increasing gradient between the lysis buffer (30 mM imidazole, 0.5 M NaCl, 20 mM NaPO4, pH 7.4) and the elution buffer (0.75 M imidazole, 5 M NaCl, 20 mM NaPO4, pH 7.4). All recombinant proteins were dialyzed at 4°C overnight against phosphate-buffered saline (PBS) using 3500 MWCO Slide-A-Lyzer cassettes (Pierce, Rockford, IL). Protein purity was assessed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue (data not shown). Protein concentrations were determined by bicinchoninic acid protein assays (Pierce).
TABLE 1.
Oligonucleotide primer sequences used in this study
| Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| B. burgdorferi B31 N40 revA forward | TGTAAAGCATATGTAGAAGAAAAG | Cloning |
| B. burgdorferi B31 revA reverse | TTAATTAGTGCCCTCTTCGAGGAA | Cloning |
| B. burgdorferi N40 revA reverse | TTAATTGGTGCCCTCTTCGAGGAA | Cloning |
| B. burgdorferi 297 cp32-4 revA forward | TGTAAAGCATATGTAGAAGAAAAG | Cloning |
| B. burgdorferi 297 cp32-4 revA reverse | TTAATTAGTACCTTCTTCGAGAAA | Cloning |
| B. burgdorferi 297 cp32-12 revA forward | TGTAAAGCATATGTGGAAGAGAAG | Cloning |
| B. burgdorferi 297 cp32-12 revA reverse | TTAATTAGTACCCTCTTCAAGAAAC | Cloning |
| B. garinii PBi revA forward | TGTAAAACATATGTAAAAGAAAAAGAAGAG | Cloning |
| B. garinii PBi revA reverse | TCAATTAGTACCTTCTTCGAGAAACTTTA | Cloning |
| B. burgdorferi B31 revB forward | GAACTATTTATAATAAAAAGGAG | Cloning |
| B. burgdorferi B31 revB reverse | TTAATCTTCTTCAAGATATTTTATTAT | Cloning |
| B. burgdorferi B31 revAC1 F | ATGTCTAAAAACTAAAAGGGCGAGCTCAACGATC | Overlap PCR |
| B. burgdorferi B31 revAC1 R | CGCCCTTTTAGTTTTTAGACATTTTATCCCCATTACC | Overlap PCR |
| B. burgdorferi B31 revAC2 F | GACAAGGATCAGGATATTTCAAAAAGATAAAAGGGC | Overlap PCR |
| B. burgdorferi B31 revAC2 R | ATCGTTGAGCTCGCCCTTTTATCTTTTTGAAAT | Overlap PCR |
| B. burgdorferi B31 revAN1 F | GATGACGATAAGAATCATCCCTTCACCGATGTTTTA | Overlap PCR |
| B. burgdorferi B31 revAN1 R | AGAAGAATCATTTACAAGAGCTAAAACATCGGTGAAGGGATG | Overlap PCR |
| B. burgdorferi revA1 | GAA ATA GAT TCA TTA ATG GAG GA | qRT-PCR |
| B. burgdorferi revA2 | CTT GTC AGT AAT GTT TTC AAT GG | qRT-PCR |
| B. burgdorferi flaB3 | GGG TCT CAA GCG TCT TGG | qRT-PCR |
| B. burgdorferi flaB4 | GAA CCG GTG CAG CCT GAG | qRT-PCR |
For protein identification by mass spectrometry, proteins were separated by SDS-PAGE followed by staining with Sypro Ruby (Invitrogen) and excision of the band of interest. The mass spectrometric analysis was performed at the University of Kentucky's Center for Structural Biology Protein Core Facility.
Amino- and carboxy-terminal truncated RevA proteins were produced from mutated plasmids created by overlap extension PCR (40) of the B31 MI-16 RevA pET200 plasmid, using primers listed in Table 1.
Polyclonal antiserum directed against RevA was produced by inoculation of purified recombinant protein into a New Zealand White rabbit at Animal Pharm (Healdsburg, CA), using one round of their standard protocol. Antiserum was adsorbed against sonicated Escherichia coli Rosetta (DE3) pLysS (Novagen) and then affinity purified using HiTrap protein A columns (GE Healthcare) according to the manufacturer's instructions. The specificity of the purified antibody for RevA was tested by Western blotting against recombinant RevA proteins, B. burgdorferi lysates, and control proteins (bovine serum albumin [BSA] and human plasma fibronectin) (data not shown). Briefly, proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Membranes were blocked overnight at 4°C with 5% (wt/vol) BSA in Tris-buffered saline-Tween 20 (TBS-T; 20 mM Tris [pH 7.5], 150 mM NaCl, 0.05% [vol/vol] Tween 20). Membranes were next washed with TBS-T and incubated for 2 h at room temperature with purified anti-RevA antibody diluted 1:500 in TBS-T. After extensive washing with TBS-T, membranes were incubated for 1 h at room temperature with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (IgG) antibody (GE Healthcare) diluted 1:5,000 in TBS-T. After a final series of washes with TBS-T, bound antibodies were detected by using SuperSignal West Pico enhanced chemiluminescence substrate (Pierce).
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (73). Maxisorp 96-well plates (Nalge-Nunc, Rochester, NY) were coated overnight at 4°C with 10 μg/ml of the following proteins: BSA (Millipore, Kankakee, IL); recombinant RevA or RevB protein; human plasma fibronectin (Sigma-Aldrich, St. Louis, MO); Engelbreth-Holm-Swarm mouse sarcoma laminin (Sigma-Aldrich); and three human fibronectin proteolytic fragments, the amino-terminal human fibronectin 70-kDa fragment prepared by cathepsin D digestion and the 30-kDa amino-terminal domain and the 45-kDa gelatin-binding domain generated by trypsin digestion of the 70-kDa fragment (Sigma-Aldrich), each in PBS. The plates were brought to room temperature and washed once with PBS plus PBS-T with 0.5% (vol/vol) Tween-20. The wells were blocked for 2 h at room temperature with SEABLOCK (Pierce) and then washed three times with PBS-T. One hundred microliters per well of either recombinant RevA/RevB protein or fibronectin (see below for concentrations and details) was added, and then the plates were incubated for 2 h at 37°C. Wells were washed three times with PBS-T and then incubated for 1 h at room temperature with affinity-purified rabbit antibodies raised against recombinant B. burgdorferi B31 MI-16 RevA diluted 1:500 in PBS or rabbit anti-human fibronectin diluted 1:500 in PBS (Sigma-Aldrich). Plates were washed three times with PBS-T and then incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit IgG diluted 1:5,000 (GE Healthcare). Wells were again washed three times with PBS-T. One hundred microliters per well of tetramethylbenzidine substrate (Pierce) was added, and then reactions stopped by the addition of 100 μl/well 2N H2SO4. Absorbance was read at 450 nm using a Spectramax plate reader and SoftMax Pro software (Molecular Devices, Sunnyvale, CA). For experiments examining the role of ionic interactions in RevA protein binding to fibronectin, additional NaCl was added to the PBS-based binding buffer. For experiments analyzing the role of heparin-binding domains in the interactions between RevA and fibronectin, porcine heparin (Sigma-Aldrich) was added to the RevA-coated wells for 1 h prior to the addition of tested protein and was included in the binding buffer along with fibronectin (73).
Protein binding affinities (Kd [dissociation constant]) were calculated as the concentration of ligand required for half-maximal binding activity.
Ligand affinity blot.
Recombinant RevA or control protein BSA was suspended in SDS gel loading buffer (125 mM Tris [pH 6.8], 20% [vol/vol] glycerol, 4% [wt/vol] SDS, 10% [vol/vol] β-mercaptoethanol) and heated in a boiling water bath for approximately 1 min. An aliquot (approximately 1 mg) of each protein was subjected to electrophoresis. Proteins were electrotransferred to nitrocellulose membranes and then blocked overnight at 4°C with 5% (wt/vol) BSA in TBS-T. Membranes were next washed with TBS-T and incubated for 2 h at room temperature in 20 μg/ml human plasma fibronectin (Sigma-Aldrich) in TBS-T. After extensive washing with TBS-T, membranes were incubated for 1 h at room temperature in rabbit antifibronectin polyclonal antibodies (Sigma-Aldrich) diluted 1:2,500 in TBS-T. Membranes were again washed with TBS-T and then incubated for 1 h at room temperature with horseradish peroxidase-conjugated donkey anti-rabbit IgG antibody (GE Healthcare), diluted 1:5,000 in TBS-T. After a final series of washes with TBS-T, bound antibodies were detected by using SuperSignal West Pico enhanced chemiluminescence substrate (Pierce). See reference 73 for additional methodological details.
Interactions between intact B. burgdorferi bacteria and fibronectin.
Glass microscope slides were washed with deionized water and then coated by overnight incubation with 10 μg/ml human plasma fibronectin (Sigma-Aldrich) in PBS. Control slides were coated similarly with BSA. The following day, slides were washed three times with PBS and then blocked by incubation with 3% (wt/vol) BSA for 2 h at room temperature. Bacteria from mid-exponential-phase cultures of B31 MI-16, ML23, or BBK32 mutant JS315 were harvested by centrifugation, washed three times with PBS, and resuspended in PBS to 2 × 106 bacteria/ml. For inhibition of binding, affinity-purified anti-RevA antibody (1:100 dilution in PBS) or recombinant RevA protein (50 μg/ml) was added to slides 30 min prior to the addition of bacteria. Slides were covered with suspended bacteria, incubated at 37°C for 2 h, and then gently washed 10 times with PBS. Bacteria were visualized by dark-field microscopy. The numbers of adherent bacteria observed at ×200 magnification in 10 fields per slide were counted. Each assay was repeated three times.
Infection of mice and ticks.
Eight female BALB/c mice were infected by subcutaneous injection of 104 B31 MI-16 bacteria from a mid-exponential-phase culture grown at 34°C. Seven days later, the infection status of the mice was assessed by inoculation of a 1-mm2 ear biopsy specimen from each animal into BSK-II medium. The medium contained antibiotics (phosphomycin and rifampin[rifampicin]) and antifungal agents (amphotericin B) that allow growth of B. burgdorferi but inhibit contaminants. Biopsy cultures were examined 10 days later by dark-field microscopy. These mice then served to infect Ixodes scapularis larvae as follows. Egg masses laid by pathogen-free I. scapularis ticks were obtained from the Department of Entomology, Oklahoma State University—Stillwater, and held in a humidified chamber until they hatched. For B. burgdorferi acquisition studies, approximately 200 naïve larvae were placed on each of the above-described B. burgdorferi-infected mice. After 96 h, the ticks had fully engorged and naturally dropped off the mice. These ticks were returned to the humidified chamber and were allowed to molt to the nymphal stage. Two weeks after ecdysis, three independent pools of 20 to 30 ticks were frozen in liquid nitrogen for later real-time quantitative reverse transcription-PCR (qRT-PCR) analysis and the remainder were fed upon uninfected female BALB/c mice. Nymphs were forcibly removed after only 72 h of feeding. Pools of 20 to 30 ticks from three independent experiments were frozen for analysis by qRT-PCR. Infection of mice was confirmed by analysis of serum samples by immunoblot for antibodies directed against the B. burgdorferi BmpA (P39) protein, which is indicative of mammalian infection (68). Eight mice infected through feeding by infected nymphs were killed 2 weeks after completion of tick feeding, and their ear pinnae, hearts, and tibiotarsal joints were collected and frozen for RNA extraction and qRT-PCR.
All infection studies were performed under protocols approved by the University of Kentucky Institutional Animal Care and Use Committee and the University of Kentucky Institutional Biosafety Committee.
Analysis of B. burgdorferi mRNA levels.
Total RNA was extracted from cultured bacteria or tissue samples by using TRIzol reagent (Invitrogen). Frozen mouse tissue samples were first ground with a mortar and pestle, followed by homogenization in TRIzol reagent at 4°C with a Tissue Tearor (Biospec Products, Bartlesville, OK). RNA was resuspended in RNAsecure reagent (Ambion, Austin, TX) and treated with DNase I (Ambion) to remove contaminating DNA. The DNase was inactivated by using DNase inactivation reagent (Ambion). Aliquots (1 μg) of each DNA-free RNA preparation were reverse transcribed by using FirstStrand cDNA synthesis kits (Roche Applied Science, Indianapolis, IN) with random hexamers and avian myeloblastosis virus reverse transcriptase enzyme (RTase). As controls, mixtures containing all components except RTase were prepared and treated similarly. Primers and templates were annealed for 10 min at room temperature, followed by cDNA synthesis at 42°C for 1 h. RTase was inactivated by heating for 5 min at 99°C, followed by 10 min at 4°C. All cDNA and control reaction mixtures were diluted 10-fold with water before being used as templates for qRT-PCR.
qPCR was performed by using a LightCycler thermal cycler (Roche Applied Science) as previously described (7, 51). All cDNA samples were analyzed in triplicate. Each LightCycler run included RNA processed without RTase (see above) as a negative control to test for DNA contamination of each RNA preparation and a sample that lacked template to test for DNA contamination of reagents.
Oligonucleotide primers used for amplification are listed in Table 1. Reaction conditions consisted of a 2-min initial 94°C denaturation followed by 45 cycles of 94°C for 5 s, 50°C for 5 s, and 72° for 30 s. Tenfold serial dilutions of B31 MI-16 genomic DNA (100 ng to 100 fg) were included in every assay for each primer set. This enabled the generation of standard curves from which the amount of transcript present in each cDNA sample could be calculated, which was done using LightCycler software version 3.5.3 (Roche Applied Science). The same software package was also used for melting-curve analyses. To verify amplicon sizes and purities, all products were separated by agarose gel electrophoresis, and DNA was visualized with ethidium bromide. Average expression values obtained from triplicate runs of each cDNA sample for all the genes of interest were calculated relative to the average triplicate value for the B. burgdorferi housekeeping gene flaB from the same cDNA preparation (7). Statistical analyses of data were performed using Student's t test and assuming unequal variances.
Modeling analyses.
Predictions of disorder within proteins were determined by using Predictor of Naturally Disordered Regions (PONDR) using VL-XT. Access to PONDR was provided by Molecular Kinetics (Indianapolis, IN). Primary amino acid sequences of B. burgdorferi proteins and truncated protein fragments were compared with all known sequences in the nonredundant protein sequence database of GenBank by using BLAST-P and Psi-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Protein-folding probabilities were determined by using Protein Homology/analogY Recognition Engine (PHYRE) (http://www.sbg.bio.ic.ac.uk/∼phyre). Coiled-coil formation probability was assessed by using PCOILS (http://toolkit.tuebingen.mpg.de/pcoils).
RESULTS
RevA binds fibronectin.
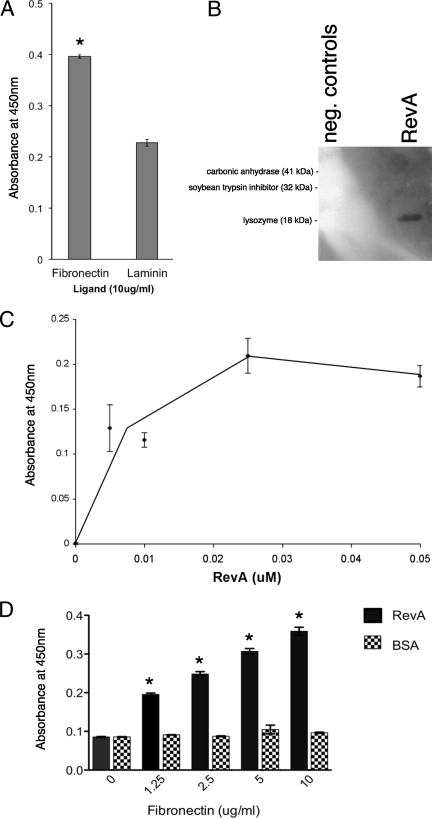
Since B. burgdorferi so frequently infects ECM-containing tissues and produces the RevA outer surface protein during mammalian infection, we hypothesized that RevA may be an ECM adhesin. Our laboratory recently characterized a laminin-binding protein of B. burgdorferi, as well as a family of proteins from the spirochete Leptospira interrogans that bind host fibronectin and laminin (6, 73), so we investigated whether RevA bound to these ligands. Initial studies used the RevA allele encoded by the genome of type strain B31, designated herein as RevAB31. Although strain B31 carries two distinct revA genes, both encode identical proteins. Microtiter plates were coated by overnight incubation with either 10 μg/ml human plasma fibronectin or mouse laminin, and RevAB31 binding to these ECM proteins was detected by using RevAB31-specific antibody. RevAB31 exhibited a significantly greater affinity for fibronectin than it did for laminin (Fig. 1A). The ability of RevAB31 to bind fibronectin was further evaluated by ligand affinity blot (Fig. 1B). Interactions between RevAB31 and fibronectin were then quantified by ELISA. Saturable, dose-dependent binding of RevAB31 to fibronectin was observed, with a calculated Kd of 12.5 nM (Fig. 1C), consistent with RevA binding fibronectin through a specific site. This value is comparable to the Kd of 10 nM previously determined for the other borrelial fibronectin-binding protein, BBK32 (42). Reverse ELISAs, in which the plate was coated with RevAB31, likewise demonstrated dose-dependent binding of fibronectin to RevA (Fig. 1D).
FIG. 1.
RevA binds human fibronectin. (A) Binding of recombinant RevAB31 (10 μg/ml) to immobilized human plasma fibronectin or murine laminin was analyzed by ELISA, with bound RevAB31 detected by using purified specific antibodies. Data represent the means and standard errors of the results from two separate experiments with six replicates per ligand. The asterisk indicates a significantly different value (P = < 0.05, Student's t test assuming unequal variances). (B) Binding of human plasma fibronectin (20 μg/ml) to recombinant RevA, carbonic anhydrase, soybean trypsin inhibitor, or lysozyme as analyzed by ligand affinity blot with bound fibronectin detected by using specific antibodies. Note that all three control proteins exhibited relative affinities for fibronectin below that of even the blocking agent BSA, as demonstrated by the white areas on the blot which correspond to those proteins. (C) Dose-dependent, saturable binding of RevAB31 to human fibronectin. Binding of recombinant RevA (10 μg/ml) to immobilized human plasma fibronectin was analyzed by ELISA, with bound RevA detected by using purified specific antibodies. Control wells were coated with immobilized laminin, and values represent RevA binding to fibronectin minus background readings for laminin. Data represent the means and standard errors of the results from two separate experiments with six replicates per RevA concentration. Protein binding affinity (Kd) was calculated as the concentration of ligand required for half-maximal binding activity. (D) Fibronectin also binds to RevA in a dose-dependent manner. Binding of human plasma fibronectin (0 to 10 μg/ml) to recombinant RevA or BSA was analyzed by ELISA, with bound fibronectin detected by using specific antibodies. Data represent the means and standard errors of the results from two separate experiments with six replicates per RevA concentration. The asterisks indicate values significantly different from the level of fibronectin binding to BSA (P = < 0.05, Student's t test assuming unequal variances).
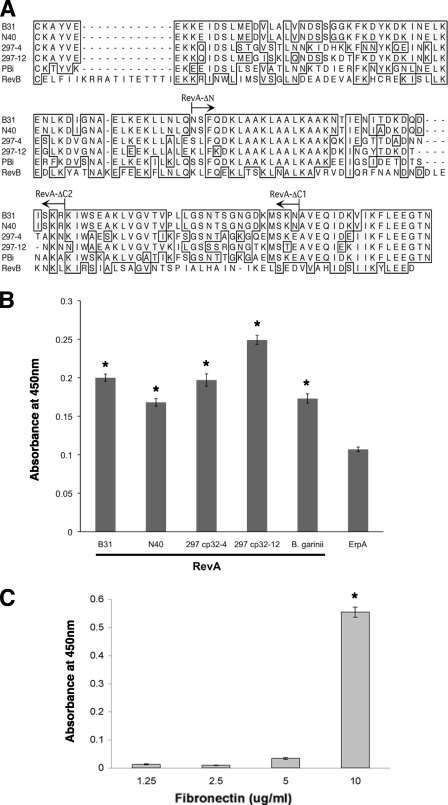
The predicted amino acid sequences of RevA proteins are moderately to highly conserved among Lyme disease spirochetes (Fig. 2A). The two revA genes of B. burgdorferi strain B31 are located on its native circular prophages cp32-1 and cp32-6. B. burgdorferi strain 297 has two copies of revA, which share only 72% identity with each other and are located on its naturally occurring cp32-4 and cp32-12 prophages. B. burgdorferi strain N40 and B. garinii strain PBi each carry one revA locus. The revA genes of B. burgdorferi strain 297 cp32-4 and cp32-12 and of strain N40 and B. garinii strain PBi were cloned, and recombinant proteins expressed (data not shown). The abilities of each RevA protein to bind fibronectin were then tested by ELISA. All of the proteins tested bound fibronectin, indicating that this characteristic is likely to be well conserved among RevA alleles (Fig. 2B).
FIG. 2.
(A) Predicted amino acid sequences of RevA proteins of B. burgdorferi strains B31, N40, and 297 and B. garinii strain PBi and of RevB of B. burgdorferi strain B31. Identical amino acid residues found in the majority of proteins are boxed and shaded. Arrows above the sequences indicate the truncation ends of the RevA ΔN, ΔC-1, and ΔC-2 mutants of B31 RevA. 297-4, encoded by cp32-4 prophage of strain 297; 297-12, encoded by cp32-4 prophage of strain 297. (B) RevA proteins from B. burgdorferi strains N40 and 297 and from B. garinii all bind human fibronectin. Binding of human plasma fibronectin to immobilized recombinant RevA proteins was analyzed by ELISA, and bound fibronectin was detected by using specific antibodies. Data represent the means and standard errors of the results from two separate experiments with six replicates per RevA protein. Recombinant ErpA of B. burgdorferi strain B31 was also assayed as a negative control protein that does not detectably bind fibronectin, as assessed by signals compared to background in all types of assays (our unpublished results). The asterisks indicate values significantly different from those for ErpA (P < 0.05, Student's t test assuming unequal variances). (C) RevB binds fibronectin. Binding of human fibronectin to immobilized recombinant RevB, as analyzed by ELISA. Data represent the means and standard errors of the results from two separate experiments with six replicates. The asterisk indicates a value significantly different from that with no fibronectin (P < 0.05, Student's t test assuming unequal variances).
As mentioned above, B. burgdorferi strain B31 naturally contains a cp32 derivative, cp9-1, which contains a gene paralogous to revA that is designated revB (13, 14, 26). Recombinant RevB was produced, and its ability to bind fibronectin was examined by ELISA. Despite having only 28% overall amino acid identity with RevAB31 (Fig. 2A), RevB also bound to fibronectin (Fig. 2C). The large step in binding between 5 and 10 μg/ml of fibronectin was reproducible and may suggest complex interactions between RevB and fibronectin; this possibility is currently under detailed examination. Together, these results demonstrate that both Rev family proteins of B. burgdorferi are able to bind to fibronectin.
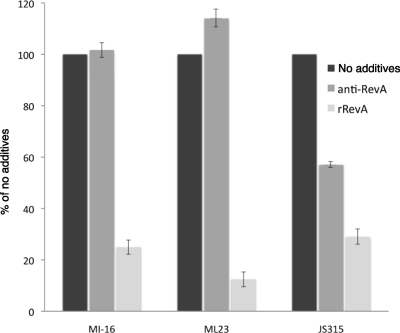
We next examined the biological significance of RevA in the ability of intact, live B. burgdorferi bacteria to adhere to fibronectin. To do so, glass slides were coated with fibronectin or control protein BSA and then incubated with live B. burgdorferi strain B31 MI-16, ML23, or JS315 (which cannot produce the BBK32 fibronectin-binding protein). In some experiments, affinity-purified polyclonal anti-RevA (1:100 dilution) or purified recombinant RevAB31 protein (50 μg/ml) was added prior to the addition of bacteria to test their ability to interfere with spirochetal binding to fibronectin. Recombinant RevAB31 significantly inhibited the adherence of all three strains to fibronectin (Fig. 3). Antibodies specific for RevA significantly interfered with the binding of strain JS315 to fibronectin, reducing the adherence of the BBK32-deficient strain by more than 40%. Anti-RevA antibodies did not detectably affect the adherence of either strain MI-16 or ML23 to fibronectin, consistent with their expression of BBK32 (67). These results demonstrate that RevA plays a role in B. burgdorferi's binding to fibronectin.
FIG. 3.
B. burgdorferi binding to fibronectin is inhibited by soluble recombinant RevA (rRevA) and by antibodies specific for RevA. Glass microscope slides were coated by overnight incubation with 10 μg/ml human plasma fibronectin or control protein BSA in PBS. After being blocked, slides were covered with suspended B. burgdorferi strain B31 MI-16, ML23, or JS315 (BBK32 mutant), incubated, and washed extensively, and then bacteria were visualized by dark-field microscopy. Numbers of adherent bacteria observed at ×200 magnification in 10 fields per slide were counted. Data represent the means and standard errors of the results from three separate experiments.
Mapping the fibronectin-binding region of RevA.
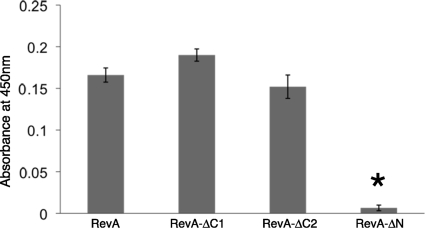
Truncated variants of RevAB31 were produced to identify the region(s) of RevA involved with binding to fibronectin. The last 20 amino acid residues of RevA carboxy termini are almost identical among known alleles (Fig. 2A), but their deletion had no appreciable effect on fibronectin binding, as determined by ELISA (Fig. 4). The truncation of 30 additional amino acids from the carboxy terminus also had no significant effect on fibronectin binding. However, removal of the first 60 amino acids beyond the lipidation site on the amino terminus of RevA significantly inhibited fibronectin binding. Modeling analyses predicted that this amino-terminal region is largely alpha helical and highly ordered, with a strong probability (40 to 90%) of forming coiled-coils.
FIG. 4.
The amino terminus of RevA is essential for ligand binding. Binding of human fibronectin to immobilized wild-type and truncated RevAB31 recombinant proteins was analyzed by ELISA. Sequences of mutant proteins are indicated in Fig. 2. Data represent the means and standard errors of the results from two separate experiments with six replicates per RevA protein. The asterisk indicates a value significantly different from that for the full-length protein (P = < 0.001, Student's t test assuming unequal variances).
Fibronectin-RevA interactions.
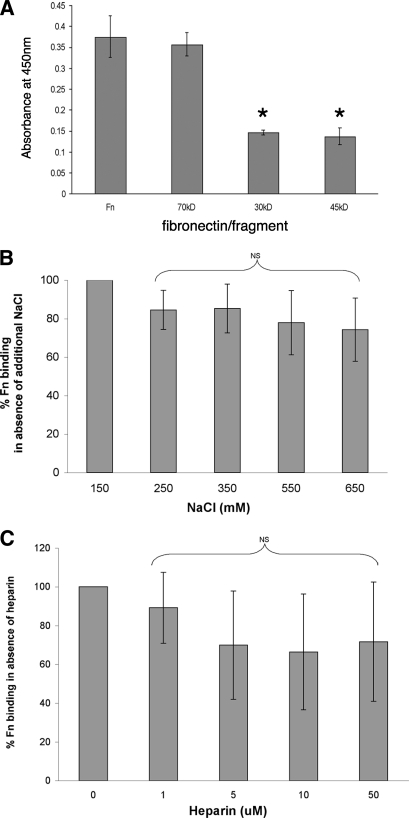
Fibronectin is a multidomain protein, with various regions of the protein exhibiting different biophysical properties. Many known fibronectin-binding proteins adhere to specific ligand domains and may be identified through the use of specific proteolytic fragments of fibronectin. The amino-terminal 70-kDa fragment of fibronectin can be subdivided into two functional domains, the 30-kDa amino-terminal domain and the adjacent gelatin-binding domain (45-kDa fragment). The ability of RevA to bind to the 70-kDa, 45-kDa, and 30-kDa fibronectin fragments was tested by ELISA. RevA bound the 70-kDa fragment equally as well as it did full-length fibronectin (Fig. 5A). RevA bound only about half as well to the 30-kDa or 45-kDa fragments.
FIG. 5.
RevA-fibronectin interactions. (A) Binding of RevAB31 to immobilized fibronectin fragments, as analyzed by ELISA. Bound RevA was detected by using purified specific antibody. Data represent the means and standard errors of the results from two separate experiments with six replicates. The asterisks indicate values significantly different from those with binding of RevA to whole fibronectin (P < 0.05, Student's t test assuming unequal variances). Fn, full-length fibronectin; 70kDa, amino-terminal 70-kDa fragment of fibronectin; 30kDa, 30-kDa amino-terminal domain of the 70-kDa fragment; 45kDa, 45-kDa gelatin-binding domain of the 70-kDa fragment. (B) Role of ionic interactions in RevAB31 binding of fibronectin. Binding of fibronectin to immobilized RevA in the presence of increasing concentrations of NaCl was analyzed by ELISA. Data represent the means and standard errors of the results from three experiments with six replicates per concentration of NaCl. Fn, fibronectin; NS, not statistically significant. (C) Effects of heparin on RevAB31-fibronectin interactions. Data represent the means and standard errors of the results from three experiments with six replicates per concentration of heparin. Fn, fibronectin; NS, not statistically significant.
Some adhesin-ligand interactions, such as those between the Leptospira interrogans LenA protein and laminin, are adversely affected by ionic strength (73). Therefore, RevA binding to fibronectin was assessed by the addition of increasing concentrations of NaCl to ELISA reaction mixtures. The inclusion of NaCl at a concentration more than fourfold greater than the physiological concentration did not have a significant impact upon fibronectin binding by RevA, suggesting that nonionic interactions are chiefly responsible for fibronectin binding by RevA (Fig. 5B).
Fibronectin contains heparin-binding domains, and the presence of heparin can inhibit the binding of some fibronectin adhesins (58). Therefore, fibronectin binding by RevA was examined in the presence of increasing concentrations of heparin. Heparin did not have any significant effect on RevA-fibronectin binding (Fig. 5C). These results indicate that RevA binds fibronectin through the non-heparin-binding domains of the 70-kDa amino terminus of fibronectin (Fig. 5C).
Regulation of RevA expression during the mammalian tick infection cycle.
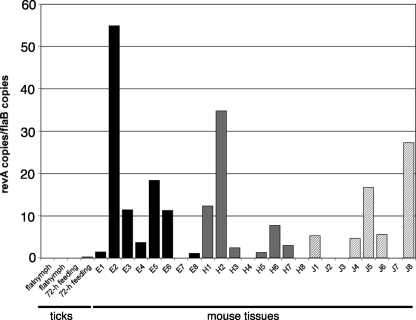
Serological studies indicated that humans and laboratory animals are frequently exposed to RevA during B. burgdorferi infection (30, 56). A previous study found that B. burgdorferi ceased transcription of revA shortly after ticks acquired spirochetes from feeding on infected mice (30). To further investigate the expression of this protein, we used qRT-PCR to examine revA transcript levels during mammalian infection. Two weeks after they had been infected via feeding by nymphal ticks, mice were killed and tissues were collected from all animals. In addition, we examined two cohorts of unfed (“flat”), infected tick nymphs and two cohorts of infected tick nymphs that had fed for 72 h on previously naïve mice. B. burgdorferi transcribed abundant levels of revA in infected mouse hearts, tibiotarsal joints, and ears (Fig. 6). Variations in revA mRNA expression levels were observed from tissue to tissue and mouse to mouse, which may be associated with variations in bacterial loads, host responses, or other vagaries of mammalian infection (28, 52). Importantly, note that all experimental animals yielded at least one revA-positive tissue sample. In contrast, revA transcripts were largely undetectable in both unfed and feeding nymphal ticks.
FIG. 6.
Temporal analysis of revA expression during mammalian and tick infections. Illustrated are qRT-PCR results from two independently collected and processed pools of 20 to 30 infected, unfed nymphs; three independent pools of infected nymphs that had fed on naïve mice for 72 h; and the ear pinnae (E), hearts (H), and tibiotarsal joints (J) of eight mice that had been infected for 2 weeks following tick bite transmission of B. burgdorferi strain B31 MI-16. Gene expression levels were calculated as nanograms of target gene per nanogram of the constitutively expressed B. burgdorferi flaB gene. Note that at least one tissue from each of the eight mice contained detectable levels of revA mRNA.
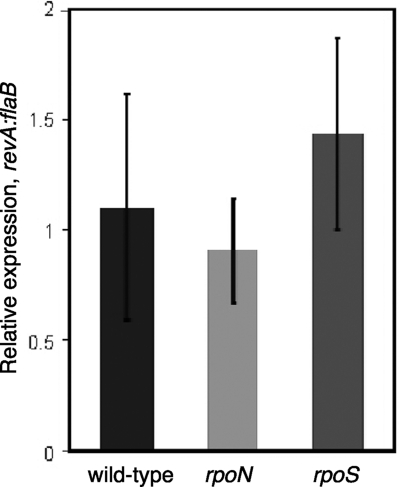
B. burgdorferi encodes only three sigma subunits of RNA polymerase, the “housekeeping” protein RpoD (σ70) and the alternative sigma factors RpoN (σ54) and RpoS (σS) (26). The two borrelial alternative sigma factors have been implicated in the transcription of some B. burgdorferi genes that are differentially regulated between the mammalian and tick environments (9-11, 19, 20, 25, 41, 77). Notably, production of the previously identified fibronectin-binding protein BBK32 is controlled through RpoN and RpoS (39). Therefore, the expression levels of revA in the wild type and isogenic mutants with either rpoN or rpoS deleted were analyzed by real-time qRT-PCR. The level of transcription of revA was not statistically different from the level in the wild type in either the rpoN or the rpoS mutant (Fig. 7). These data demonstrate that the expression of revA is not directly dependent upon either of the alternative sigma factors found in B. burgdorferi and indicate that revA transcription must be RpoD dependent.
FIG. 7.
Expression of revA is dependent upon RpoD (σ70)-containing RNA polymerase. Gene expression levels were calculated as nanograms of target gene per nanogram of the constitutively expressed B. burgdorferi flaB gene. Each experiment was repeated three times, and the error bars represent standard errors.
DISCUSSION
B. burgdorferi can adhere to many components of its vertebrate hosts’ ECM, suggesting that B. burgdorferi-ECM interactions are likely to be important for the persistence of this pathogen (8). Several ECM-binding proteins have been identified in Lyme disease spirochetes, including the glycosaminoglycan (GAG) and decorin-binding proteins Bgp and DbpA/DbpB and the fibronectin- and GAG-binding protein BBK32 (33, 59, 63). None of these proteins have been demonstrated to be absolutely required for infectivity, suggesting a redundancy among B. burgdorferi ECM adhesins (2, 34, 46, 67, 76). The present studies identified RevA, a 17-kDa outer-surface lipoprotein, as an additional fibronectin-binding protein of B. burgdorferi. Previous serological studies and our gene-specific qRT-PCR demonstrated that revA mRNA and protein are produced during mammalian infection (29, 30, 69; this study). RevA is thus appropriately placed on the outer surface of B. burgdorferi to interact with host fibronectin.
In addition to RevA, B. burgdorferi has another, well-characterized fibronectin-binding protein, BBK32. BBK32, like RevA, is produced during mammalian infection, and its binding to fibronectin is unaffected by heparin (22, 23, 45, 47, 66). However, unlike that of RevA, BBK32's expression is controlled by the RpoN-RpoS sigma factor cascade (39). B. burgdorferi bacteria lacking BBK32 still bind fibronectin and retain infectivity (46, 67), leading us to hypothesize that the fibronectin-binding activity of BBK32 is redundant to that of the RevA/RevB proteins. A recent study suggested that B. burgdorferi requires BBK32 and host fibronectin for initial interactions with the host microvasculature; however, additional spirochetal proteins appear to be necessary for stationary adhesion (55). Rev proteins are reasonable candidates for being among these additional adhesins and may be part of the borrelial repertoire needed for hematogenous dissemination and/or adhesion in the host. Indeed, the observations showing that BBK32 and RevA expression are regulated through different mechanisms suggest that these proteins could be expressed in different host niches or at different stages of infection. The apparently different mechanisms by which RevA and BBK32 interact with fibronectin also suggest different roles for each borrelial surface protein.
The deletion of 60 amino acids from the amino terminus eliminated fibronectin binding, implicating that region in ligand binding. The amino-terminal region of RevA possesses no similarities to any other known bacterial fibronectin-binding proteins, including BBK32 of B. burgdorferi, or to those of other spirochetes, such as L. interrogans, Treponema pallidum, or Treponema denticola (3, 12, 18, 32, 36, 48, 50, 63, 64, 73). Thus, RevA may represent a novel type of bacterial adhesin. The amino-terminal domain is predicted to be comprised of structured alpha helices, likely arranged into coiled-coil tertiary structures. In contrast, the carboxy-terminal 50 residues of RevA are not essential for fibronectin binding and are predicted to form a largely disordered domain.
We also noted that RevA appears to also bind mammalian laminin, although to a significantly lesser extent than fibronectin. Additional studies are under way to assess the ability of RevA to adhere to other mammalian ECM components. Indeed, an ability of RevA to bind additional host ligands might be expected, as the ability of a bacterial adhesin to bind multiple host ligands is more often the rule than the exception (60). As examples, B. burgdorferi BBK32 binds host GAGs in addition to fibronectin (23), the B. burgdorferi ErpACP/OspE/CRASP3-5 proteins can simultaneously bind both human factor H and plasminogen (4, 5, 38), and the streptococcal M protein binds a diverse variety of host tissue components (24). Moreover, there are numerous examples of mammalian proteins which adhere to multiple, distinct self-ligands (27, 75).
In conclusion, we identified RevA and RevB as being novel fibronectin-binding proteins of B. burgdorferi. The adherence of B. burgdorferi to fibronectin via RevA/RevB proteins, in conjunction with the previously identified BBK32 protein, is hypothesized to facilitate colonization of mammalian tissues. In addition, fibronectin can interact with host cells through integrins and thereby influence cellular activities (35, 58). It is therefore possible that the RevA/RevB proteins might affect mammalian host responses to infection. The identification of these new, distinctive fibronectin adhesins, in conjunction with previous knowledge of other borrelial host-interactive proteins, provides insight into mechanisms employed by the Lyme disease spirochete to colonize its mammalian hosts. ECM adhesins like RevA are also attractive targets for the development of preventative and curative therapies for Lyme disease.
Acknowledgments
This work was supported by exploratory funds provided by the University of Kentucky College of Medicine to B. Stevenson and by NIH Ruth L. Kirschstein Individual National Research Service Award F32-AI081480 to C. A. Brissette. The University of Kentucky Center for Structural Biology Protein Core Facility is supported in part by funds from NIH National Center for Research Resources (NCRR) grant P20-RR020171.
We thank Carol Beach of the University of Kentucky Center for Structural Biology Protein Core Facility for mass spectrometric analyses. We thank Peter Kraiczy, Patricia Rosa, and Jonathan Skare for sharing bacterial strains and Logan Burns, Sherwood Casjens, Henry Choy, Ashutosh Verma, and Michael Woodman for helpful comments and technical advice.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 27 April 2009.
REFERENCES
- 1.Barthold, S. W., K. D. Moody, G. A. Terwilliger, P. H. Duray, R. O. Jacoby, and A. C. Steere. 1988. Experimental Lyme arthritis in rats infected with Borrelia burgdorferi. J. Infect. Dis. 157842-846. [DOI] [PubMed] [Google Scholar]
- 2.Blevins, J. S., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkman, M. B., M. A. McGill, J. Pettersson, A. Rogers, P. Matejkova, D. Smajs, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2008. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect. Immun. 761848-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brissette, C. A., A. E. Cooley, L. H. Burns, S. P. Riley, A. Verma, M. E. Woodman, T. Bykowski, and B. Stevenson. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 298(S1)257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brissette, C. A., K. Haupt, D. Barthel, A. E. Cooley, A. Bowman, C. Skerka, R. Wallich, P. F. Zipfel, P. Kraiczy, and B. Stevenson. 2009. The Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77300-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brissette, C. A., A. Verma, A. Bowman, A. E. Cooley, and B. Stevenson. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bykowski, T., M. E. Woodman, A. E. Cooley, C. A. Brissette, V. Brade, R. Wallich, P. Kraiczy, and B. Stevenson. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 754227-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabello, F. C., H. P. Godfrey, and S. A. Newman. 2007. Hidden in plain sight: Borrelia burgdorferi and the extracellular matrix. Trends Microbiol. 15350-354. [DOI] [PubMed] [Google Scholar]
- 9.Caimano, M. J., C. H. Eggers, C. A. Gonzalez, and J. D. Radolf. 2005. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp45-borne ospA and lp6.6 genes. J. Bacteriol. 1877845-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caimano, M. J., C. H. Eggers, K. R. O. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 726433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron, C. E., E. L. Brown, J. M. Kuroiwa, L. M. Schnapp, and N. L. Brouwer. 2004. Treponema pallidum fibronectin-binding proteins. J. Bacteriol. 1867019-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll, J. A., N. El-Hage, J. C. Miller, K. Babb, and B. Stevenson. 2001. Borrelia burgdorferi RevA antigen is a surface-exposed outer membrane protein whose expression is regulated in response to environmental temperature and pH. Infect. Immun. 695286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35490-516. [DOI] [PubMed] [Google Scholar]
- 15.Coburn, J. 2001. Adhesion mechanisms of the Lyme disease spirochete, Borrelia burgdorferi. Curr. Drug Targets Infect. Disord. 1171-179. [DOI] [PubMed] [Google Scholar]
- 16.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 571182-1195. [DOI] [PubMed] [Google Scholar]
- 17.de Souza, M. S., A. L. Smith, D. S. Beck, G. A. Terwilliger, E. Fikrig, and S. W. Barthold. 1993. Long-term study of cell-mediated responses to Borrelia burgdorferi in the laboratory mouse. Infect. Immun. 611814-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards, A. M., H. F. Jenkinson, M. J. Woodward, and D. Dymock. 2005. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect. Immun. 732891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2006. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 591859-1875. [DOI] [PubMed] [Google Scholar]
- 20.Elias, A. F., J. L. Bono, J. A. Carroll, P. Stewart, K. Tilly, and P. Rosa. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 1822909-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 702139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fikrig, E., W. Feng, S. W. Barthold, S. R. Telford III, and R. A. Flavell. 2000. Arthropod- and host-specific Borrelia burgdorferi bbk32 expression and the inhibition of spirochete transmission. J. Immunol. 1645344-5351. [DOI] [PubMed] [Google Scholar]
- 23.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 1025162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 27.Furlan, M. 2002. Sticky and promiscuous plasma proteins maintain the equilibrium between bleeding and thrombosis. Swiss Med. Wkly. 132181-189. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore, R. D., Jr., R. R. Howison, V. L. Schmit, and J. A. Carroll. 2008. Borrelia burgdorferi expression of the bba64, bba65, bba66, and bba73 genes in tissues during persistent infection in mice. Microb. Pathog. 45355-360. [DOI] [PubMed] [Google Scholar]
- 29.Gilmore, R. D., Jr., and M. L. Mbow. 1998. A monoclonal antibody generated by antigen inoculation via tick bite is reactive to the Borrelia burgdorferi Rev. protein, a member of the 2.9 gene family locus. Infect. Immun. 66980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3799-808. [DOI] [PubMed] [Google Scholar]
- 31.Grab, D. J., C. Givens, and R. Kennedy. 1998. Fibronectin-binding activity in Borrelia burgdorferi. Biochim. Biophys. Acta 1407135-145. [DOI] [PubMed] [Google Scholar]
- 32.Haapasalo, M., K. H. Muller, V. J. Uitto, W. K. Leung, and B. C. McBride. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect. Immun. 602058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 662674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 684759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauck, C. R., and K. Ohlsen. 2006. Sticky connections: extracellular matrix protein recognition and integrin-mediated cellular invasion by Staphylococcus aureus. Curr. Opin. Microbiol. 95-11. [DOI] [PubMed] [Google Scholar]
- 36.Hauk, P., F. Macedo, E. C. Romero, S. A. Vasconcellos, Z. M. de Morais, A. S. Barbosa, and P. L. Ho. 2008. In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect. Immun. 762642-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Häupl, T., G. Hahn, M. Rittig, A. Krause, C. Schoerner, U. Schonherr, J. R. Kalden, and G. R. Burmester. 1993. Persistence of Borrelia burgdorferi in ligamentous tissue from a patient with chronic Lyme borreliosis. Arthritis Rheum. 361621-1626. [DOI] [PubMed] [Google Scholar]
- 38.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 2768427-8435. [DOI] [PubMed] [Google Scholar]
- 39.Herzberger, P., C. Siegel, C. Skerka, V. Fingerle, U. Schulte-Spechtel, A. van Dam, B. Wilske, V. Brade, P. F. Zipfel, R. Wallich, and P. Kraiczy. 2007. Human pathogenic Borrelia spielmanii sp. nov. resists complement-mediated killing by direct binding of immune regulators factor H and factor H-like protein 1. Infect. Immun. 754817-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 41.Hübner, A., X. Yang, D. M. Nolen, T. G. Popova, P. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 9812724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, J. H., J. Singvall, U. Schwartz-Linek, B. J. B. Johnson, J. R. Potts, and M. Höök. 2004. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J. Biol. Chem. 27941706-41714. [DOI] [PubMed] [Google Scholar]
- 43.Kopp, P. A., M. Schmitt, H. J. Wellensiek, and H. Blobel. 1995. Isolation and characterization of fibronectin-binding sites of Borrelia garinii N34. Infect. Immun. 633804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornblatt, A. N., A. C. Steere, and D. G. Brownstein. 1984. Experimental Lyme disease in rabbits: spirochetes found in erythema migrans and blood. Infect. Immun. 46220-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lahdenne, P., J. Panelius, H. Saxen, T. Heikkila, H. Sillanpaa, M. Peltomaa, M. Arnez, H. I. Huppertz, and I. J. Seppala. 2003. Improved serodiagnosis of erythema migrans using novel recombinant borrelial BBK32 antigens. J. Med. Microbiol. 52563-567. [DOI] [PubMed] [Google Scholar]
- 46.Li, X., X. Liu, D. S. Beck, F. S. Kantor, and E. Fikrig. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 743305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. DNA microarray assessment of putative Borrelia burgdorferi lipoprotein genes. Infect. Immun. 703300-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, Y. P., and Y. F. Chang. 2008. The C-terminal variable domain of LigB from Leptospira mediates binding to fibronectin. J. Vet. Sci. 9133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lünemann, J. D., S. Zarmas, S. Priem, J. Franz, R. Zschenderlein, E. Aberer, R. Klein, L. Schouls, G. R. Burmester, and A. Krause. 2001. Rapid typing of Borrelia burgdorferi sensu lato in specimens from patients with different manifestations of Lyme borreliosis. J. Clin. Microbiol. 391130-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merien, F., J. Truccolo, G. Baranton, and P. Perolat. 2000. Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 18517-22. [DOI] [PubMed] [Google Scholar]
- 51.Miller, J. C. 2005. Example of real-time quantitative reverse transcription-PCR (Q.-RT-PCR) analysis of bacterial gene expression during mammalian infection: Borrelia burgdorferi in mouse tissues, p. 1D. 3. In R. T. Coico, T. F. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 52.Miller, J. C., K. Narayan, B. Stevenson, and A. R. Pachner. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 3927-33. [DOI] [PubMed] [Google Scholar]
- 53.Miller, J. C., K. von Lackum, K. Babb, J. D. McAlister, and B. Stevenson. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 716943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moody, K. D., S. W. Barthold, G. A. Terwilliger, D. S. Beck, G. M. Hansen, and R. O. Jacoby. 1990. Experimental chronic Lyme borreliosis in Lewis rats. Am. J. Trop. Med. Hyg. 42165-174. [DOI] [PubMed] [Google Scholar]
- 55.Norman, M. U., T. J. Moriarty, A. R. Dresser, B. Millen, P. Kubes, and G. Chaconas. 2008. Molecular mechanisms involved in vascular interactions of the Lyme disease pathogen in a living host. PLoS Pathog. 4e1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowalk, A. J., R. D. Gilmore, Jr., and J. A. Carroll. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect. Immun. 743864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pachner, A. R., J. Basta, E. Delaney, and D. Hulinska. 1995. Localization of Borrelia burgdorferi in murine Lyme borreliosis by electron microscopy. Am. J. Trop. Med. Hyg. 52128-133. [DOI] [PubMed] [Google Scholar]
- 58.Pankov, R., and K. M. Yamada. 2002. Fibronectin at a glance. J. Cell Sci. 1153861-3863. [DOI] [PubMed] [Google Scholar]
- 59.Parveen, N., M. Caimano, J. D. Radolf, and J. M. Leong. 2003. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Mol. Microbiol. 471433-1444. [DOI] [PubMed] [Google Scholar]
- 60.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48585-617. [DOI] [PubMed] [Google Scholar]
- 61.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. D. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 1783293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preac-Mursic, V., B. Wilske, and G. Schierz. 1986. European Borrelia burgdorferi isolated from humans and ticks: culture conditions and antibiotic susceptibility. Zentralbl. Bakteriol. Hyg. A 263112-118. [DOI] [PubMed] [Google Scholar]
- 63.Probert, W. S., and B. J. B. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 301003-1015. [DOI] [PubMed] [Google Scholar]
- 64.Probert, W. S., J. H. Kim, M. Hook, and B. J. Johnson. 2001. Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 694129-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raibaud, S., U. Schwarz-Linek, J. H. Kim, H. T. Jenkins, E. R. Baines, S. Gurusiddappa, M. Hook, and J. R. Potts. 2005. Borrelia burgdorferi binds fibronectin through a tandem beta-zipper, a common mechanism of fibronectin binding in staphylococci, streptococci, and spirochetes. J. Biol. Chem. 28018803-18809. [DOI] [PubMed] [Google Scholar]
- 66.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seshu, J., M. D. Esteve-Gassent, M. Labandeira-Rey, J. H. Kim, J. P. Trzeciakowski, M. Hook, and J. T. Skare. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 591591-1601. [DOI] [PubMed] [Google Scholar]
- 68.Simpson, W. J., W. Burgdorfer, M. E. Schrumpf, R. H. Karstens, and T. G. Schwan. 1991. Antibody to a 39-kilodalton Borrelia burgdorferi antigen (P39) as a marker for infection in experimentally and naturally inoculated animals. J. Clin. Microbiol. 29236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skare, J. T., D. M. Foley, S. R. Hernandez, D. C. Moore, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1999. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect. Immun. 674407-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stanek, G., and F. Strle. 2003. Lyme borreliosis. Lancet 3621639-1647. [DOI] [PubMed] [Google Scholar]
- 71.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308733-740. [DOI] [PubMed] [Google Scholar]
- 72.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 662648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. DeMoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szczepanski, A., M. B. Furie, J. L. Benach, B. P. Lane, and H. B. Fleit. 1990. Interaction between Borrelia burgdorferi and endothelium in vitro. J. Clin. Investig. 851637-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Timpl, R., T. Sasaki, G. Kostka, and M.-L. Chu. 2003. Fibulins: a versatile family of extracellular matrix proteins. Nat. Rev. Mol. Cell Biol. 4479-489. [DOI] [PubMed] [Google Scholar]
- 76.Weening, E. H., N. Parveen, J. P. Trzeciakowski, J. M. Leong, M. Hook, and J. T. Skare. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect. Immun. 765694-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, X. F., M. C. Lybecker, U. Pal, S. M. Alani, J. Blevins, A. T. Revel, D. S. Samuels, and M. V. Norgard. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 1874822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zückert, W. R. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr. Protoc. Microbiol. Chapter 12Unit 12C.1. [DOI] [PubMed]