Abstract
The small intestine is an important site of infection for many enteric bacterial pathogens, and murine models, including the streptomycin-treated mouse model of infection, are frequently used to study these infections. The environment of the mouse small intestine and the microbiota with which enteric pathogens are likely to interact, however, have not been well described. Therefore, we compared the microbiota and the concentrations of short-chain fatty acids (SCFAs) present in the ileum and cecum of streptomycin-treated mice and untreated controls. We found that the microbiota in the ileum of untreated mice differed greatly from that of the cecum of the same mice, primarily among families of the phylum Firmicutes. Upon treatment with streptomycin, substantial changes in the microbial composition occurred, with a marked loss of population complexity. Characterization of the metabolic products of the microbiota, the SCFAs, showed that formate was present in the ileum but low or not detectable in the cecum while butyrate was present in the cecum but not the ileum. Treatment with streptomycin altered the SCFAs in the cecum, significantly decreasing the concentration of acetate, propionate, and butyrate. In this work, we also characterized the pathology of Salmonella infection in the ileum. Infection of streptomycin-treated mice with Salmonella was characterized by a significant increase in the relative and absolute levels of the pathogen and was associated with more severe ileal inflammation and pathology. Together these results provide a better understanding of the ileal environment in the mouse and the changes that occur upon streptomycin treatment.
The small intestine serves as the site of colonization and attachment and the seat of pathogenesis for a number of important enteric bacterial pathogens of humans and animals. Among the bacteria that cause diarrheal disease in the small bowel are Vibrio cholerae, which colonizes the small intestine and secretes toxins (64), and pathogenic forms of Escherichia coli, including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), and diffusely adherent E. coli (DAEC), all of which either adhere to or affect enterocytes in the small intestine (reviewed in reference 42). Yersinia enterocolitica and Salmonella have also both been shown to preferentially invade the ileum by targeting the Peyer's patches (9, 25). All of these species must thus survive within this region of the intestinal tract in competition with the resident microbiota and must there express determinants necessary for virulence. Additionally, the small intestine is an important site for lesions associated with inflammatory bowel disease (IBD). Although the causes of IBD are complex and multiple, the microbiota of the small intestine is thought to be important to disease development. It has been alternatively theorized that IBD stems from an alteration in the host microbiota present, deficiencies in the host's response to and control of the microbiota, or changes in the function of a particular member of the microbiota (reviewed in references 65 and 72), the last of which is supported by recent findings suggesting a connection between Crohn's disease and strains of Escherichia coli termed adherent and invasive E. coli (5, 19).
The expression of virulence determinants by bacterial pathogens often occurs in response to specific environmental cues. For enteric pathogens including EPEC, ETEC, DAEC, and Y. enterocolitica, a temperature near 37°C is important for virulence (13, 24, 28, 83). For EPEC, physiological osmolarity, near-neutral pH, and quorum sensing have all been implicated in pathogenesis (13, 92). Toxin production and hence virulence in ETEC are regulated by osmolarity and microaerophilic conditions (83), while Dr fimbria production in DAEC is controlled by anaerobic conditions (24). Invasion by Y. enterocolitica is controlled by regulation of the expression of the invasin protein by an acidic pH (28). For Salmonella, low oxygen, log-phase growth, high osmolarity, and slightly alkaline pH have been shown to positively affect expression of genes necessary for Salmonella invasion in vitro (2, 29, 31, 53, 73). Additionally, our previous work has shown that short-chain fatty acid (SCFA) concentrations and pH are important for invasion gene expression. The SCFAs acetate and formate induce invasion gene expression, while butyrate and propionate repress these same genes at pH levels comparable to those of the mammalian intestinal tract (38, 52). Furthermore, previous in vivo studies have shown an association between decreased SCFAs and susceptibility to Salmonella infection in a cecectomized mouse model (87).
Examination of the intestinal environment reveals that enteric pathogens are tuned to respond to cues naturally present in the gut. Host temperature is maintained close to 37°C. There exists an axial oxygen gradient in the gut, with the lumen of the small intestine and colon being anaerobic, whereas the mucosal surface is microaerobic (21). The osmolality of the small intestine in humans is 250 to 425 mosmol/kg depending on the location within the small intestine and the state of digestion (41, 49). In various mammalian species, studies have shown that the SCFAs acetate and propionate are found in both the large and small intestine, while butyrate is present primarily in the cecum and colon and formate is present primarily in the ileum (1, 6, 7, 18, 50, 51, 61, 67). Additionally, it is thought that the resident microbiota present in the intestinal tract produces quorum sensing molecules that may be important for interspecies communication (reviewed in reference 62).
Much of the environment of the intestinal tract is defined by the metabolic processes of the bacterial populations that reside within. There have been several recent comprehensive studies that reveal the large intestine (the cecum, colon, and feces) of mice and humans to be inhabited by a diverse population of bacteria (23, 27, 39, 47, 55, 76, 80, 84). There are 1012 microbes/gram of contents in the distal intestinal tract, and it is estimated that there are 500 to 1,000 different species present (22, 90). Of these species, the majority belong to two phyla, the Firmicutes and Bacteroidetes, this being true for both mice and humans (27, 55). Because the intestinal microbiota plays a critical role in protecting the host against enteric bacterial pathogens or “colonization resistance” (85), increased understanding of the composition of the intestinal microbiota is essential to defining bacterial pathogenesis. In contrast with the wealth of information now available regarding the composition of the large intestine microbiota, relatively little is known about that of the mouse small intestine. A comprehensive survey of the microbiota in the human small intestine has been completed with predominating groups consisting of the Firmicutes (Lachnospiraceae and Bacillus) and the Bacteroidetes (30). However, studies with mice consist of a limited survey of the microbiota of the ileum (69), examination of bacteria present at the phylum level (40), and quantification of particular bacterial groups believed to be common inhabitants of the ileum (3).
The streptomycin-treated mouse model has proven to be an effective method to study enteric disease and has been used to study pathogens such as Vibrio cholerae, enterohemorrhagic E. coli, and Salmonella (4, 60, 64, 88). It is specifically useful for the study of Salmonella, as untreated mice develop systemic salmonellosis without an enteric component, but pretreatment with oral streptomycin produces enterocolitis (4, 70). In this model of infection, the primary pathology occurs in the cecum, and studies have shown that changes in the cecal microbiota occur when mice are pretreated with streptomycin (4, 80). However, the effects of streptomycin treatment on the ileum have not been well characterized.
As the environment of the small intestine is poorly characterized in animals used as models of enteric infection, we have in this study characterized the bacterial populations, fatty acid composition, and histopathological changes of this region in untreated and streptomycin-treated mice, both uninfected and infected with Salmonella enterica serovar Typhimurium. We hypothesized that a unique bacterial population within the ileum, and the environment created by the population, would be important for the protection of the host against invading pathogens. We show here that there exists a stable bacterial community within the ileum that is different from that of the cecum and that the microbial population associated with the ileal mucosa is different from that of the lumenal contents. Additionally, we find that streptomycin treatment alters the microbial populations associated with both the ileum and cecum by decreasing species richness and changing the distribution of phylotypes present. We also find that streptomycin pretreatment allows Salmonella to overcome colonization resistance, increasing by 2 logs the number of Salmonella bacteria present at this intestinal site in treated animals, and thus alters the pathological response of the ileum to Salmonella infection.
MATERIALS AND METHODS
Mouse experiments.
For infection experiments, a spontaneous streptomycin-resistant isolate of Salmonella enterica serovar Typhimurium strain ATCC 14028s was used. Twenty-five milliliters of culture was grown overnight with aeration at 37°C in morpholinopropanesulfonic acid (MOPS) minimal medium with 0.5% glucose (63). The bacteria were then pelleted, washed three times with phosphate-buffered saline (PBS), and resuspended in 1 ml of PBS. Fifty microliters was then used to infect each mouse, equaling a dose of ∼109 Salmonella bacteria per mouse. All mouse experiments were approved by the Institutional Care and Use Committee at Cornell University. Seven-week-old female C57BL/6 mice were obtained from Charles River Laboratories for all experiments and housed in a facility for pathogen containment. Mice were housed three to a cage and fed a standard laboratory diet. Mice were divided into two groups of 12, with one group inoculated orally with 30 μl of sterile water and the other with 30 μl of sterile streptomycin sulfate solution (20 mg per mouse). Twenty-four hours later, six mice in each group of 12 were inoculated orally with 50 μl of sterile PBS, and the remaining six from each group received 50 μl of Salmonella suspension at a total dosage of ∼109 CFU. Forty-eight hours after infection with Salmonella, mice were euthanized and tissues were collected from the ileum and the cecum of each mouse.
DNA isolation.
Samples from the distal ileum and cecum containing both tissue and contents were collected and placed in bead tubes (Mo Bio Laboratories, Inc., Carlsbad, CA), whereas samples of intestinal contents were carefully expressed into microcentrifuge tubes without scraping the mucosa. The mass was determined, and the samples were then flash frozen with liquid nitrogen. Samples were stored at −80°C until they were processed. Total DNA was isolated from samples using a modified version of the Qiagen DNeasy blood and tissue kit (Qiagen). Briefly, 360 μl of buffer ATL was added to samples in their bead tubes and then homogenized for 1 minute using a Mini-Bead-Beater (BioSpec Products, Inc., Bartlesville, OK), a method of mechanical disruption that has been shown to be effective for the isolation of bacterial DNA from fecal samples (56). Then 40 μl of proteinase K was added, and samples were vortexed. Samples were then processed as described in the manufacturer's protocol with one change: prior to the addition of 200 μl of 100% ethanol, the samples were incubated at 70°C for 30 min. Samples of intestinal contents were processed similarly except that bead tubes were not used.
16S rRNA gene clone library construction and analysis.
Clone libraries of 16S rRNA-encoding genes were constructed as previously described (91). Briefly, primers 8F and 1492R were used to amplify the 16S rRNA-encoding genes from the DNA samples (75). For cecal samples, 20 cycles were used for PCR, whereas 24 cycles were used for ileal samples due to the lower concentration of bacterial DNA present in the ileum. The increase in cycles allowed for amplification from all but three ileal samples among the various treatment groups. For the samples that did not amplify, there appeared to be a lower concentration of bacterial DNA leading to mispriming with mouse DNA, which interfered with clone library construction. Purified PCR products were then ligated into a T-tailed cloning vector (pCR 4-TOPO; Invitrogen) and used to transform competent cells. Ninety-four clones per library were sequenced by the Cornell University sequencing facility using the 8F primer. Sequences were then uploaded to the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu), and the pipeline tool available via myRDP was used for quality analysis and alignment (14). The RDP classifier (89), using the default 80% confidence threshold setting, and SeqMatch (14) were then used for taxonomic classification of the aligned sequences. Additionally, distance matrices were generated from the RDP-based sequence alignments and analyzed using the DOTUR program (74). Using a level of 97% sequence identity, DOTUR was used to assign sequences to operational taxonomic units (OTUs), generate rarefaction curves, and calculate diversity indices. To determine the Bray-Curtis measure of β-diversity, the program EstimateS was utilized (15).
Quantitative real-time PCR.
The same DNA used for 16S rRNA gene clone library construction was used for quantitative real-time PCR. The 16S rRNA-encoding gene was used as the target to measure total bacteria and total numbers of Salmonella bacteria present in each sample as previously described (3), except for the bacterial strains used as assay standards. For assays of total bacteria, we used E. coli MG1655 as the standard, and for total Salmonella we used Salmonella enterica serovar Typhimurium ATCC 14028s. Standard curves were constructed using these strains to quantify the total number of 16S rRNA gene copies per sample, with R2 values of ≥0.995 extending to 1,000 copies of 16S rRNA-encoding genes for total bacteria and one copy for Salmonella, defining the lower limits of the test. For each sample, numbers of copies of 16S rRNA-encoding genes per gram of tissue were calculated using the mass of the original sample.
SCFA analysis and pH determination.
Gas chromatography-mass spectrometry (GC-MS) was used to quantify SCFAs in the cecum. Intestinal contents from the cecum were collected in 1% acidified water. The mass of samples was determined, and they were then flash frozen in liquid nitrogen. Samples were processed using a modified version of a previously published protocol (66). Samples were thawed at room temperature, vortexed for 1 minute, and then centrifuged. The supernatant was removed and placed in a 4-ml glass vial (National Scientific, Rockwood, TN) containing 50 μl each of 20 mM stock isotopes, [2H3]acetate and [1-13C]acetate (Cambridge Isotope Laboratories, Inc., Andover, MA), [2H5]propionate, and [13C4]butyrate (Sigma-Aldrich), which were used as internal standards for each sample. Samples were acidified with 10 μl HCl and then extracted with 1 ml of diethyl ether four times. One milliliter of sample was placed in a separate tube containing 2.5 μl of the derivatization reagent 1-tert-butyl-dimethyl-silyl-imidazole (Sigma-Aldrich) and heated at 60°C for 30 min. The derivatization step was performed on duplicate aliquots for each sample. Samples were then transferred to autosampler tubes, and analyses were performed on a Jeol GCMate II MS with an Agilent 6890N GC inlet equipped with a J&W Scientific DB-5MS column (30 m × 0.250 mm, 0.25-μm film thickness). A split injector was used with a 250°C injector temperature and a 50:1 split ratio. The initial oven temperature was 60°C held for 1 min followed by a 5°C/min ramp to 120°C and a 25°C/min ramp to a 270°C final temperature, which was held for 1 min. Total run time was 20 min, including a 3.5-min solvent delay. The derivatized acids were detected using unit-mass resolution selected ion monitoring (SIM) with magnetic field switching at 0.21-s cycle time on “flat-top” mass peaks with a fully open collector slit: formate (retention time [R.T.], 3.98; SIM, 3 to 4.5 min; m/z 75 and 103), acetate (R.T., 4.91/4.95; SIM, 4.5 to 6.4 min; m/z 75, 117, and 121), propionate (R.T., 6.82/6.9; SIM, 6.4 to 7.5; m/z 75, 131, and 136), and butyrate (R.T., 9.07; SIM, 7.5 to 20 min; m/z 75, 145, and 149). The timing and ions for the SIM program were selected based on 35 to 500 m/z full scans on standard samples. The isotopic internal standards were then used for quantification of the SCFAs in each sample. As a result of limitations with the protocol and the small quantity of intestinal contents in the small intestine of mice, we used high-pressure liquid chromatography (HPLC) to quantify all SCFAs in the ileum and formate in the cecum. HPLC and pH determinations were performed as previously described (38) except that a microcombination pH probe (Microelectrodes, Inc., Bedford, NH) was used. For both GC-MS and HPLC analyses, sample preparation included steps to remove bacteria without lysis, and so SCFA concentrations reflect those of the intestinal milieu.
Tissue collection and histology.
The distal ileum (∼4.0 cm) and cecum were harvested, fixed in 10% neutral buffered formalin, and embedded in paraffin, and the entire length was sectioned at 5-μm thickness and stained with hematoxylin and eosin for histopathological assessment. Sections of ileum were scored on a scale of 0 to 4 for the presence and distribution of polymorphonuclear leukocytes in five regions: (i) within the lamina propria of the intestinal mucosa, (ii) inside the intestinal crypts (cryptitis), (iii) at the periphery of Peyer's patches, (iv) within the interfollicular regions of Peyer's patches, and (v) within the subepithelial dome areas of Peyer's patches (0, none; 1, rare; 2, few scattered; 3, many groups; 4, large numbers) by a board-certified blinded veterinary pathologist. Ileitis severity was calculated as a sum of the scores for the five categorical parameters (maximum of 20). The scoring method is a modification of a method used previously (58), and histological designations for each scoring region are widely accepted and consistent with previously published work (10).
Statistical analysis.
Analysis of SCFA concentrations was completed using the Wilcoxon rank-sum test. For quantitative PCR (qPCR) and pH, the Kruskal-Wallis test was used to determine whether there was a significant difference among any of the groups, and then the Wilcoxon rank-sum test using a Bonferroni correction was utilized to determine significance between individual groups. Similarly, for the pathology scores, the Kruskal-Wallis test and then Dunn's posttest were performed. A P value of <0.05 was considered significant. Statistical analysis was performed using Jmp 7.0 software (SAS, Cary, NC) or GraphPad Prism version 5 (GraphPad Software, La Jolla, CA).
Nucleotide sequence accession numbers.
All sequences generated in this study have been submitted to GenBank with accession numbers FJ834458 to FJ838647.
RESULTS
The mouse ileum harbors a defined microbial population.
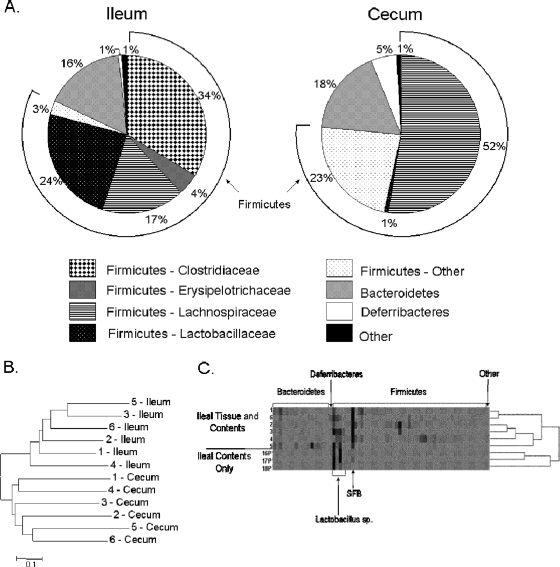
To increase our understanding of the mouse model of enteric infection and the potential interaction of pathogens with both the host and the resident microbiota in the small intestine, we first characterized the bacteria of the most distal segment, the ileum, using conventional mice and a culture-independent method. Ileal and cecal samples, including both tissue and contents, were taken from C57BL/6 mice, and 16S rRNA gene clone libraries were created from these samples. We analyzed a total of 536 partial rRNA-encoding gene sequences from the ileum and 533 from the cecum. The clone libraries of the cecum were examined to provide a point of reference for our characterization of the ileum, since much work has previously been done to characterize the microbiota of the cecum (47, 55, 80, 84). From these sequences, we determined that there exists a bacterial community within the ileum that consists primarily of the phyla Firmicutes (82%) and Bacteroidetes (16%). At the taxonomic level of family, the most predominant Firmicutes were of the families Clostridiaceae (34%), Lactobacillaceae (24%), and Lachnospiraceae (17%) (Fig. 1A). Of the Clostridiaceae, the most predominant clones (32% of the total bacterial population) had high sequence identity with a bacterial group termed the segmented filamentous bacteria (SFB). The SFB have previously been described as unculturable bacteria that are closely associated with the ileal epithelium (20). Previous phylogenetic analysis has also shown the SFB to be most closely related to the genus Clostridium, a member of the family Clostridiaceae (78). Thus, SFB constituted a substantial portion of the ileal microbiota. In contrast to the bacterial composition of the ileum, the predominant family of the Firmicutes in the cecum was the Lachnospiraceae (52%), a member of the order Clostridiales, while merely 1% of the cecal microbiota consisted of the Lactobacillaceae and less than 1% consisted of the Clostridiaceae (Fig. 1A). These differences between the bacterial composition of the ileum and that of the cecum at the family level suggest that there is a defined microbial community in the ileum that differs greatly from that found in the cecum.
FIG. 1.
The microbial composition of the ileum. 16S rRNA gene clone libraries were created from the ileum and cecum of C57BL/6 mice. (A) Taxonomic analysis of the clone library sequences was performed using RDP Classifier and SeqMatch. Assignment to the phylum level for the sequences from the ileum and cecum of untreated mice is displayed with further distinction among the phylum Firmicutes to the family level. The total of the Firmicutes is shown by the arc extending around the pie chart. (B) Cluster analysis of the Bray-Curtis distance measure of diversity between the microbial communities for the ileum and cecum from untreated mice. For the Bray-Curtis distance measure, an OTU was defined as 97% sequence similarity. (C) 16S rRNA gene clone libraries were created from samples containing ileal tissue and contents or contents alone from untreated mice. Sequences were assigned to OTUs using a definition of 97% sequence similarity. A heat map is used to show the relative abundance of OTUs, with specific OTUs detected in the sample oriented along the horizontal axis, and the dendrogram showing the distribution of OTUs. Darker coloring within the heat map indicates greater representation of specific OTUs. Phyla are shown above the figure, and highly represented OTUs are shown below. Numbers 1 to 6 and 16P to 18P represent individual mice.
Rarefaction analysis was next used both to identify differences in richness among the microbial populations of the cecum and ileum and to assess the efficacy of our sampling technique in obtaining representative samples from both of these sites (34, 36). This analysis revealed that the ileum had lower overall species richness than did the cecum, with observed OTUs of 67 and 156, respectively (Table 1 and Fig. 2A). The analysis, however, also suggested an underrepresentation of the number of OTUs present at both sites, as shown by the positive slope of the curves in Fig. 2A and the Chao1 analysis of estimated richness (11, 12) (Table 1). This was nevertheless not surprising, as much is still unknown about the complete microbial richness of the intestinal tract. In addition to taxonomic analysis and determination of species richness, we next examined the diversity of the ileal and cecal samples using the Bray-Curtis distance measure of diversity (57). Cluster analysis of the Bray-Curtis distance using an OTU definition of 97% sequence similarity showed that the ileal samples clustered with each other but separately from samples derived from the cecum (Fig. 1B). This analysis demonstrated that there were differences in the distribution of microbial OTUs in the ileal and cecal samples and that the ileal sample taken from each mouse was more similar to other ileal samples than it was to the cecal sample taken from the same mouse. Together these analyses show that there is a unique microbial population associated with the ileum that is different from that of the cecum.
TABLE 1.
Observed and estimated OTUs
| Sample | No. of OTUs | Chao1 richness estimator value (95% confidence interval) |
|---|---|---|
| Ileum | 67 | 152 (101-282) |
| Ileal contents | 9 | 12 (9-32) |
| Ileum, streptomycin treated | 49 | 62 (53-93) |
| Cecum | 156 | 227 (194-288) |
| Cecum, streptomycin treated | 97 | 138 (115-189) |
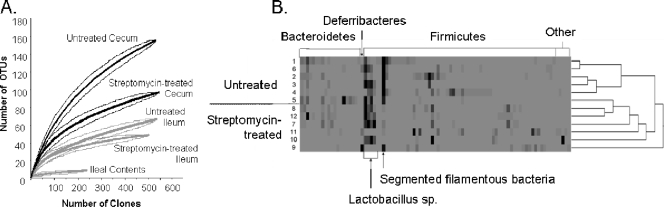
FIG. 2.
Streptomycin treatment alters the microbial composition of both the ileum and the cecum. (A) Rarefaction analysis and 95% confidence intervals are shown for the 16S rRNA-encoding sequences obtained from each site in the untreated and streptomycin-treated mice. Central lines represent rarefaction analysis of each site with the surrounding lines representing the upper and lower 95% confidence intervals. For rarefaction analysis, an OTU was defined as 97% sequence similarity. (B) 16S rRNA gene clone libraries were created from samples containing ileal tissue and contents from untreated mice and those pretreated with streptomycin. Sequences were assigned to OTUs using a definition of 97% sequence similarity. A heat map is used to show the relative abundance of OTUs, with specific OTUs detected in the sample oriented along the horizontal axis, and the dendrogram showing the distribution of OTUs. Darker coloring within the heat map indicates greater representation of specific OTUs. Phyla are shown above the figure, and highly represented OTUs are shown below. Untreated mice shown in this figure are the same as those shown in Fig. 1C.
Previous work with humans has shown that the microbial population associated with the feces is different from that of the colonic tissue (27). Thus, we further characterized the ileum to determine whether differences existed between the population of bacteria present within the intestinal contents and that associated with the ileal tissue. Samples containing ileal contents but not the tissue of the ileum itself were taken from three C57BL/6 mice, and 238 16S rRNA gene clones were compared to those derived from ileal samples containing both tissue and contents. We found that there was decreased species richness in the samples that contained only contents (Fig. 1C and 2A). The total number of OTUs observed was nine in the samples containing only contents, compared to 67 in the samples that included both tissue and contents. Additionally, analysis utilizing the Chao1 richness estimator further supported this difference in the richness of OTUs, with no overlap in the 95% confidence intervals (Table 1). Furthermore, the Shannon-Weiner diversity index, an indicator of diversity within a population (57), was 2.87 in samples that included both tissue and contents compared to 0.98 in samples that contained only contents, indicating lower diversity within the bacterial community derived from the contents of the ileum. Analysis of the composition of OTUs present in the ileum (Fig. 1C) also showed that there was a larger number of OTUs present in the samples containing both tissue and contents and that there was a difference in the distribution of these OTUs. Specifically, we found upon comparison that the relative abundance of the SFB was much lower in the samples containing only contents, 3% ± 2.6% compared to 31% ± 13% in samples with both tissue and contents. These results are consistent with previous reports of the SFB being closely associated with the ileal epithelium (20). Thus, these results show that there is greater bacterial diversity in the samples that contain tissue and contents than in those that contain contents alone and suggest the presence of a diverse bacterial population in close association with the intestinal mucosa.
Streptomycin alters the microbiota of the ileum.
To better understand the streptomycin-treated mouse model of infection and specifically the potential changes that occur in the small intestine, we next characterized the microbiota of the ileum in streptomycin-treated mice using 16S rRNA gene clone libraries. Mice were treated orally with streptomycin, and samples from the ileum and cecum were obtained 72 h after that treatment. A total of 503 and 545 sequences were analyzed from the ileum and cecum of these mice, respectively. To allow comparisons with the untreated mice described above, we performed the two experiments simultaneously, using animals obtained as a single lot. We found that streptomycin treatment altered the composition of microbial communities in both the ileum and the cecum. Rarefaction analysis demonstrated that streptomycin treatment decreased species richness in both the ileum and cecum (Fig. 2A). In this case, the Chao1 estimator of richness supported this conclusion as well, as there was no overlap in the 95% confidence intervals of the Chao1 values of the treated sites compared to the respective untreated sites (Table 1). We also examined the total number of 16S rRNA-encoding genes/gram in the ileum and cecum by using qPCR. Interestingly, we found that there was a 10-fold decrease in bacterial numbers, from 109 to 108 16S rRNA gene copies/gram, in the ileum but that there was no significant change in the cecum. Additionally, the Bray-Curtis distance measure of diversity between the microbial communities showed that in both the ileum and the cecum the majority of samples from untreated mice clustered together but separately from the respective samples taken from streptomycin-treated mice (data not shown). Collectively, these results show that streptomycin treatment caused a decrease in the richness of the OTUs and altered the distribution of OTUs in the majority of mice. Some variation did occur within the groups. Specifically, for one mouse in the group that had not been treated with streptomycin, cluster analysis of both the ileum and cecum placed the composition of the microbiota between those of the other untreated mice and those of the streptomycin-treated group (not shown). These results thus show variation among individual animals but demonstrate differences between the treated and untreated groups. Among ileal samples, we found that changes due to streptomycin treatment were not uniform among individual mice, with differing OTUs predominating after treatment with this antimicrobial. In particular, two mice had a single family of organisms predominate after treatment that was different from the other, in one case the family Lachnospiraceae and in the other the family Deferribacteraceae, whereas the remaining mice did not have a particular predominating group. Therefore, to further analyze these population changes, we did not combine these libraries of sequences but instead characterized the changes in the ileum caused by streptomycin by examining the microbiota of individual mice for the presence and prevalence of specific OTUs (Fig. 2B; untreated mice shown in this figure are the same as those shown in Fig. 1C). Overall, there were changes in the distribution of OTUs as well as in the relative abundance of particular OTUs. More specifically, there was a decrease in the relative abundance of the SFB in the streptomycin-treated mice, 3% ± 4% compared to 31% ± 13% in the ileum of untreated mice, consistent with previous reports that this group of bacteria is sensitive to streptomycin (43).
Streptomycin pretreatment enhances Salmonella infection of the ileum.
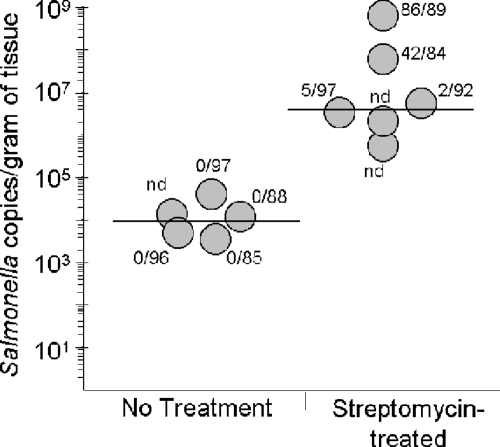
In the murine model of Salmonella infection, streptomycin treatment has been shown to enhance infection of the cecum (4, 80), but little is known about effects on the small intestine. We therefore next characterized the microbiota in the ileum of mice infected with Salmonella to determine whether infection was also altered in this region of the intestinal tract. C57BL/6 mice were pretreated orally with either sterile water or streptomycin, and 24 h later both groups were orally infected with a streptomycin-resistant strain of Salmonella. To examine the microbiota present in each of these groups, 16S rRNA gene clone libraries were created. A sufficient bacterial DNA concentration for clone library construction was obtained from five of six untreated mice and four of six streptomycin-treated mice, for a total of 452 and 362 sequences from the respective groups. Taxonomic analysis of these clone libraries showed that only one animal in the untreated group had detectable Salmonella in the ileum, with one Salmonella 16S rRNA-containing clone identified among the 86 clones sequenced (not shown). In contrast, for mice pretreated with streptomycin prior to infection, 2 to 97% of the total bacterial population observed in the ileum consisted of Salmonella. To more precisely determine the extent of Salmonella infection of the ileum, we used qPCR to determine the number of Salmonella-specific 16S rRNA gene sequences present. We were able to detect Salmonella in all of the animals, whether or not they were pretreated with streptomycin, but found that streptomycin treatment significantly increased the number of Salmonella bacteria at this site (P < 0.05). The number of copies of the Salmonella 16S rRNA gene per gram of tissue was increased in streptomycin-treated animals, with a median in pretreated mice of 4 × 106, more than 2 logs greater than that of the untreated mice, which had a median of 1 × 104. There was, however, much variation among individual animals, with the number of Salmonella gene copies obtained from mice treated with streptomycin prior to infection ranging from 5 × 105 to 6 × 108/gram of tissue (Fig. 3). One possible explanation for this variation could be various levels of Salmonella infection in these mice. However, histopathological examination confirmed the presence of severe diffuse colitis similar to that previously reported for mice orally administered streptomycin prior to infection (4). Mucosal damage and inflammation ranged from moderate and multifocal in one mouse to severe and focally extensive or diffuse in the other mice (not shown). Therefore, taken together these results show that, although variation exists in the number of Salmonella bacteria able to colonize the ileum, pretreatment with streptomycin significantly alters the composition of the ileal microbiota and leads to enhanced survival of and colonization by Salmonella in this important region of the intestine.
FIG. 3.
Streptomycin pretreatment increases Salmonella infection of the ileum. Real-time PCR was used to compare the numbers of Salmonella bacteria in the ilea of individual mice without additional treatment to those in mice pretreated with oral streptomycin. The number of copies of the Salmonella 16S rRNA gene amplified using Salmonella-specific primers was determined and is shown relative to the mass of the ileal sample. The limit of detection for Salmonella was one copy/100 ng total DNA. Represented adjacent to the circles are the numbers of Salmonella 16S rRNA gene sequences identified over the total size of the clone library analyzed from this same animal. Circles with “nd” (not determined) show animals in which clone libraries could not be produced, primarily due to low bacterial DNA yields. Horizontal lines represent the medians for each group.
Streptomycin treatment alters the fatty acid composition of the intestine.
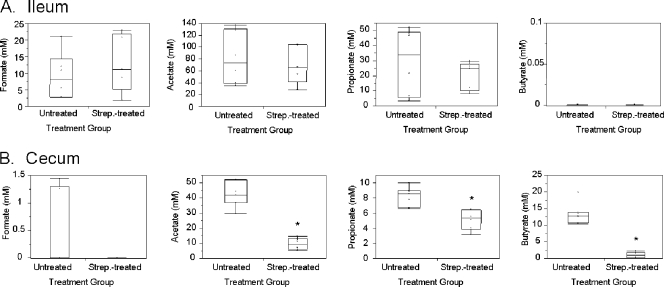
Since there are differences in the microbiota found in the ileum and cecum, we next considered whether these differences might manifest themselves as variations in the presence and concentrations of SCFAs in each intestinal location, suggesting a possible means by which Salmonella virulence might be modulated in vivo, as SCFAs have been shown to have differing effects on Salmonella invasion depending upon their composition and concentration (38, 52). We therefore characterized the SCFAs in these regions of the intestine using GC-MS and HPLC. As anticipated, there were significant differences in the SCFAs present in the ileum and cecum. We found that formate, previously identified as a positive signal for Salmonella invasion in vitro, was present in the ileum at a median concentration of 8.1 mM but was low or below the level of detection (median concentration of 0 mM) in the cecum (Fig. 4). In contrast, butyrate, previously shown to be a negative signal for invasion, was not detectable in the ileum but was present in the cecum at a median concentration of 12.8 mM (Fig. 4). We also determined the pH of the intestinal lumen, as acidity affects the penetration of SCFAs into bacteria and thus their effects on gene expression. The median pH of the ileum was 6.68 and that of the cecum was 6.39, both at levels below neutrality, thus promoting increased bacterial uptake of SCFAs.
FIG. 4.
SCFAs of the untreated and streptomycin-treated ileum and cecum. Intestinal contents were collected from the ileum and cecum of both untreated and streptomycin-treated mice 72 h after treatment. SCFAs were quantified using HPLC analysis for the ileum (A) or GC-MS for the cecum (B). The box plots show the medians with 25% and 75% quartiles. The bars in each box plot extend to the outermost points located within the quartile ± 1.5× the interquartile range. An asterisk indicates a significant difference at a P value of <0.01 between the untreated mice and streptomycin-treated mice for SCFA concentrations.
Since alterations in the microbiota occur with streptomycin treatment, and the microbiota produces SCFAs, we next determined whether changes occurred to the SCFA concentrations as a result of this treatment. We found that streptomycin treatment altered the SCFAs of the cecum, with a significant decrease in acetate, propionate, and butyrate, but did not significantly alter the SCFAs of the ileum or the pH of either site (Fig. 4 and data not shown). The median acetate concentration was substantially reduced, from 42.0 mM to 9.4 mM, and the median butyrate concentration was reduced from 12.8 mM to 1.0 mM in the cecum, while the median propionate concentration was more modestly reduced from 8.5 mM to 5.4 mM. These results, taken with previous findings, show that SCFAs exist in concentrations capable of signaling the modulation of Salmonella virulence. Additionally, the results suggest that the ileal environment in normal mice is conducive to tissue invasion, while that of the cecum represses invasion, but that treatment with streptomycin alters the environment of the cecum to reduce repressive signals, perhaps explaining the cecal pathology that is commonly observed during Salmonella infection of streptomycin-treated mice.
Streptomycin treatment prior to infection with Salmonella enhances ileal inflammation.
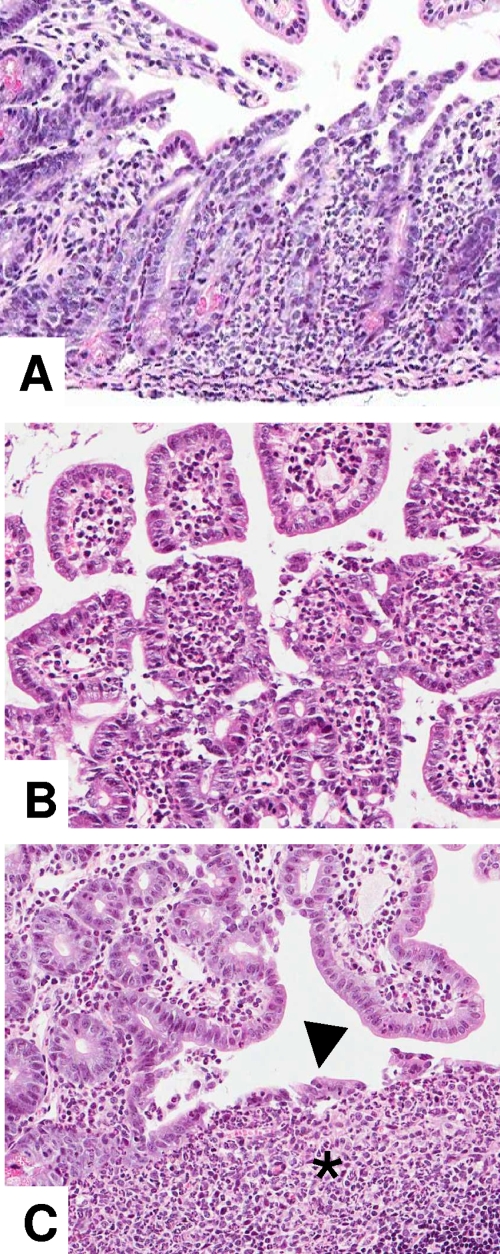
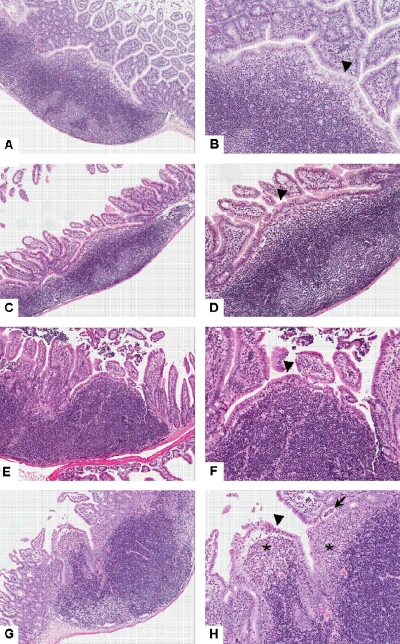
Previous work on the streptomycin-treated mouse model of Salmonella infection primarily focused on development of severe colitis without detailed examination of the ileum (4). Therefore, we characterized the histopathological changes present in the ileum of control mice and mice infected with Salmonella with and without oral administration of streptomycin. Compared to mice infected with Salmonella without antibiotic treatment, streptomycin administration prior to infection with Salmonella enhanced the extent and degree of inflammation present in the distal ileum (Fig. 5 and 6). Five of the six mice administered streptomycin prior to Salmonella showed multifocal to segmental infiltration of the lamina propria at the base of intestinal crypts by clusters of polymorphonuclear neutrophils accompanied by epithelial cell damage and cryptitis (Fig. 5). Similarly, large numbers of neutrophils were present within interfollicular regions and at the periphery of Peyer's patches extending into the lamina propria of adjacent mucosa. In contrast, mice infected with Salmonella without streptomycin pretreatment had only a few neutrophils admixed with a few eosinophils at the periphery of Peyer's patches (six of six mice) and interfollicular regions (two of six mice). The subepithelial dome areas of Peyer's patches in three of six mice inoculated with Salmonella after oral streptomycin administration also were markedly expanded by large numbers of neutrophils, a change that was absent in mice from the other groups (Fig. 5 and 6). Moreover, the follicle-associated epithelium overlying the dome areas of ileal Peyer's patches of mice receiving streptomycin with or without Salmonella was diffusely low cuboidal, compared to the tall columnar epithelium in untreated control mice (Fig. 6). To better quantify histopathological changes in the ileum of mice in each of the four treatment groups, lesions were scored on a 0 to 4 scale, and ileitis severity was calculated as the sum of the scores for the five categorical parameters described in Materials and Methods, with 20 representing maximum severity. Using this scoring system, the median score for untreated mice was 1, while those mice that had received both streptomycin and Salmonella and displayed ileitis had a median score of 15 (P < 0.05). Slight increases in ileal pathology were present in groups that received Salmonella or streptomycin alone (medians of 4.5 and 7, respectively), but these values were not significantly different from those for untreated mice. In addition to the ileal mucosal changes, histopathological examination of the ileum revealed a complete absence of long filamentous bacteria in the lumen of the ileum of mice treated with streptomycin with or without Salmonella infection (not shown). These results are thus consistent with the microbial ecology data demonstrating a decrease in the numbers of SFB in mice receiving streptomycin and show that antibiotic administration exacerbates the pathology of the ileum caused by Salmonella.
FIG. 5.
Streptomycin administration prior to infection with Salmonella enhances the extent and degree of ileal inflammation in mice. Hematoxylin- and eosin-stained ileum was obtained from mice pretreated with streptomycin and infected with Salmonella. (A) Necrosis of ileal villi accompanied by focally extensive infiltration of lamina propria by large numbers of polymorphonuclear neutrophils extending along the base of intestinal crypts. Magnification, ×12. (B) Infiltration of villous lamina propria by large numbers of polymorphonuclear neutrophils with segmental sloughing of intestinal epithelial cells. Magnification, ×20. (C) Infiltration of ileal Peyer's patch subepithelial dome area by large numbers of polymorphonuclear neutrophils (asterisk) together with focal disruption of the follicle-associated epithelium (arrowhead) and transepithelial migration of neutrophils. Magnification, ×20.
FIG. 6.
Streptomycin administration prior to infection with Salmonella is associated with ileal Peyer's patch inflammation in mice. Hematoxylin- and eosin-stained ileal Peyer's patches of uninfected mice and mice infected with Salmonella with and without streptomycin pretreatment. (A and B) Untreated and uninfected; (C and D) streptomycin treated; (E and F) Salmonella infected; (G and H) streptomycin treated and Salmonella infected. The subepithelial dome area is diffusely infiltrated by large numbers of polymorphonuclear neutrophils in the streptomycin-treated and Salmonella-infected mouse (G and H) (asterisks), while the follicle-associated epithelium is markedly attenuated in mice infected with Salmonella with (H) (arrowhead) or without (F) (arrowhead) streptomycin compared with tall columnar epithelium in uninfected mice (B and D) (arrowheads). The streptomycin-treated and Salmonella-infected mouse also displays focal disruption of the follicle-associated epithelium along with multifocal transepithelial migration of polymorphonuclear neutrophils and focal crypt abscess (H) (arrow). Magnifications, ×6 (A, C, E, and G) and ×12 (B, D, F, and H).
DISCUSSION
There has been much recent work describing the bacterial composition and chemical environment of the large intestine, including the cecum, colon, and feces, but relatively little is known about the comparable conditions of the small intestine. In this work we have characterized the ileum of the mouse, the species used frequently as a model of infection for many of the enteric bacterial pathogens that cause disease in the small intestine, and have examined the effects on this organ of both antibiotic administration and infection with Salmonella. As an effective rodent model of human enteric disease requires an understanding of the types of bacteria that normally reside within the intestine, we first identified the constituents of the ileal microbiota. We found the microbiota of the mouse ileum to be quite different from that of the cecum, with the predominant Firmicutes present being the Clostridiaceae, Lactobacillaceae, and Lachnospiraceae, while in the cecum the predominant family was the Lachnospiraceae, with much lower relative abundance of the Lactobacillaceae and Clostridiaceae. These findings are consistent with previous results for humans which showed a similar higher relative abundance of the Lachnospiraceae in the large intestine and Lactobacillaceae in the small intestine (30). In contrast to those results, however, our findings also show a large relative abundance of the Clostridiaceae in the mouse ileum that was not observed in the human small intestine. In mice, 94% of the bacteria classified as Clostridiaceae belonged to a group of bacteria called the SFB. The SFB are a group of unculturable, gram-variable, long, segmented, and filamentous bacteria that have been shown to be closely associated with the ileal epithelium in mice and other vertebrates (20, 44). They have previously been observed in one human ileal sample (44); however, less is known about the prevalence in the normal human ileum. It is possible that they are not as abundant in the human ileum as in the mouse ileum or that more studies are necessary to look specifically for this group of bacteria in human samples.
Our characterization of the ileal microbiota also showed that there were differences in the microbial population associated with samples containing tissue and lumenal contents compared to those that contained only contents. In the samples containing only contents, there was lower species richness, with the majority of bacteria classified within the family Lactobacillaceae (Fig. 1C). This suggests that the great majority of the microbial diversity observed in the ileum arises from a population of bacteria intimately associated with the intestinal mucosa rather than within the ileal lumen. The increased relative abundance of the Lactobacillaceae in lumenal samples could be attributed to several factors. Previous work using qPCR has shown that Lactobacillus bacteria are present in quantities between 107 and 109 in the distal small intestine (3), indicating that they are a predominant group in this location. Current data give conflicting reports on the ability of Lactobacillus species to adhere to the intestinal mucosa (35); therefore, one explanation for the relative abundance of Lactobacillaceae in intestinal contents is that the Lactobacillaceae may not be as closely associated with the ileal epithelium as are members of the Lachnospiraceae, Clostridiaceae, and Bacteroidaceae. Supporting this hypothesis is previous work showing that the SFB, members of the Clostridiaceae, are very closely associated with the ileal epithelium (20), which is consistent with our results showing the relative abundance of the SFB in samples containing tissue to be 10-fold greater than that in samples with intestinal contents alone. Additionally, Lactobacillus species have previously been shown to be abundant in the upper intestinal tract, including the stomach and upper small intestine (71, 82). Thus, the bacterial population in the ileal contents may be a transient population consisting primarily of bacteria shed from the upper intestinal tract. The disparity between the microbial composition of the mucosa and that of the contents is important, because these results suggest that sampling the intestinal contents alone severely underrepresents the diversity present in the ileum. Thus, to fully understand the interaction of invading pathogens with the host, it is important to characterize the bacteria associated with the mucosa as well as within the lumenal contents.
Upon treatment of mice with streptomycin, we observed changes in the composition of the microbiota in both the ileum and cecum. Specifically, in the ileum there was a 10-fold decrease in the SFB. Previous work has suggested a role for the SFB in host-pathogen interactions; specifically, they were shown to be important in providing protection against Salmonella and EPEC colonization of ileal surfaces (33, 37) and for stimulating the mucosal immune response in mice (46, 81). Correlated with this loss of microbiota was the enhanced infection of the ileum by Salmonella after treatment with streptomycin. We observed a significant increase in the number of Salmonella bacteria residing in the ileum in streptomycin-treated mice, suggesting that the administration of this antimicrobial alters the ileal environment sufficiently to allow improved colonization by the invading pathogen. In addition to the changes in the SFB observed with streptomycin treatment, we also observed inflammation in the ileum of mice treated with streptomycin prior to infection with Salmonella that was not seen in animals infected with Salmonella without streptomycin treatment. In this study, mice were infected with Salmonella 24 h after streptomycin treatment, while the characterization of the ileal environment and microbiota was conducted in streptomycin-treated mice 72 h after administration of the antibiotic. A previous study has shown that recovery of the normal microbiota, as measured at the phylum level, occurred 5 days after streptomycin treatment (80). Therefore, it is possible that antibiotic treatment elicited changes in the numbers of bacterial species not observed in this study that contributed to Salmonella colonization. However, even so, these results, along with those of previous studies, suggest a possible role for the SFB in preventing colonization and infection of the ileal surface of normal mice, either by physically blocking colonization or by creating an immune response and thus limiting Salmonella entry to the Peyer's patches and systemic disease. Although mice that are monoassociated with SFB exist (45), these mice would not allow a full understanding of the interaction of the SFB and surrounding commensals with the invading pathogen. Thus, further work examining the relationship between the SFB and Salmonella infection in the mouse is hampered by the intractable nature of this intestinal resident. In addition to the effects of the microbiota on Salmonella infection, the genetic background of the mouse might also play an important role. The strain used in this work, C57BL/6, is Nramp1−/− and thus highly susceptible to Salmonella infection. A previous study noted greater ileal inflammation after Salmonella infection in untreated Nramp1+/+ strains than in C57BL/6 mice (79), suggesting innate differences in their responses to infection. That work, however, did not examine the effects of streptomycin pretreatment on this pathology, and so the relative utility of mouse strains for this infection model remains to be investigated.
SCFAs are produced by the intestinal microbiota and thus are largely responsible for the composition of the intestinal chemical environment. Furthermore, previous work has shown that SCFAs affect both viability and virulence gene expression in enteric pathogens (8, 32, 38, 48, 52, 54, 59, 77; reviewed in reference 86). Therefore, in addition to characterizing the microbiota of the ileum and cecum, we also determined the SCFA concentrations in these regions to better understand the chemical cues likely to be sensed by pathogenic bacteria. We found that formate was present in the ileum but was at a low concentration or was undetectable in the cecum and, conversely, that butyrate was present in the cecum but was not detectable in the ileum. These differences in SCFAs may be a result of the variation in the microbiota between these two locations. For example, the family Lachnospiraceae contains genera that are classified as butyrate producers (16), and we found that there was a higher relative abundance of the Lachnospiraceae in the cecum (52%) than in the ileum (17%). Additionally, there is a higher relative abundance of the Lactobacillaceae in the ileum (24%) than in the cecum (1%), and under certain conditions species of Lactobacillus have been shown to be heterofermentative, producing formate and acetate in addition to lactic acid (17, 68). Upon treatment with streptomycin, we observed significant changes in the SCFA concentrations in the cecum, as was previously observed (67); however, we observed more substantial decreases in both the acetate and butyrate concentrations than those that were previously reported. It is likely that these changes in SCFAs occurred as a result of the changes in the microbiota in this region. Interestingly, at the family level, there did not appear to be a change in the relative abundance of the Lachnospiraceae, which contain several butyrate producers. However, we did observe decreased species richness and a change in the distribution of phylotypes at the level of 97% sequence similarity in the cecum of streptomycin-treated mice. These results suggest changes at the species level; thus, it is possible that there were changes in the relative abundance of the butyrate-producing species present within the family Lachnospiraceae leading to the changes in the butyrate concentration in the cecum. Another possibility is that streptomycin eliminates a particular group of bacteria important for acetate production. Previous work has shown that some butyrate producers use exogenous acetate for butyrate production (26); therefore, a decrease in the population of acetate-producing bacteria could potentially lead to a decrease in both acetate and butyrate. Thus, it is possible that the changes in microbiota as a result of streptomycin treatment produced these changes in SCFA concentrations in the cecum.
On the basis of previous in vitro observations indicating modulation of Salmonella invasion gene expression by certain SCFAs, we hypothesized that alterations in the microbiota associated with streptomycin administration and the resulting changes in relative concentrations of SCFAs in the intestinal tract may affect the pathogenesis of Salmonella infection in the mouse model. Previous in vitro work from our laboratory and those of others has shown that specific SCFAs can positively or negatively affect the expression of Salmonella invasion genes. In particular, formate has been characterized as a positive signal and butyrate as a negative signal (8, 32, 38, 52). Since in the mouse model of infection Salmonella invades primarily in the ileum, in particular the Peyer's patches, causing septicemia (9, 70), the observation that formate is detectable in the ileum and not the cecum suggests that it may be an important positive signal in vivo for Salmonella invasion gene expression. In contrast, the presence of butyrate in the cecum and not the ileum suggests that it may be an important negative signal in vivo. Therefore, these results, along with previously published results for cecectomized mice (87), indicate that the distribution and abundance of the SCFAs in the intestinal tract may play an important role in Salmonella virulence. Upon treatment with streptomycin prior to Salmonella infection, there are changes in the intestinal pathology, with the appearance of pronounced cecal inflammation (4) and, as shown in this work, exacerbated pathology of the ileum as well. Changes in the ileum cannot be attributed to alterations in the concentrations of SCFAs present at this site, as untreated mice have an SCFA composition that is likely already conducive to bacterial invasion, and streptomycin treatment did not elicit a change in that composition. In contrast, the cecal contents of treated mice showed a significant reduction in SCFA concentrations, with butyrate being substantially reduced. These results thus suggest that streptomycin treatment prior to infection decreases the negative signal provided by butyrate, allowing Salmonella invasion in the cecum and the cecal pathology observed in this infection model.
Acknowledgments
We thank Kevin Cummings for invaluable assistance with statistical analysis and Yanyan Huang and Kellie Cicconi for assistance with the mouse experiments.
This project was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. N01-AI-30054.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Argenzio, R. A., and M. Southworth. 1975. Sites of organic acid production and absorption in gastrointestinal tract of the pig. Am. J. Physiol. 228454-460. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 3.Barman, M., D. Unold, K. Shifley, E. Amir, K. Hung, N. Bos, and N. Salzman. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgart, M., B. Dogan, M. Rishniw, G. Weitzman, B. Bosworth, R. Yantiss, R. H. Orsi, M. Wiedmann, P. McDonough, S. G. Kim, D. Berg, Y. Schukken, E. Scherl, and K. W. Simpson. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1403-418. [DOI] [PubMed] [Google Scholar]
- 6.Bohnhoff, M., C. P. Miller, and W. R. Martin. 1964. Resistance of the mouse's intestinal tract to experimental salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med. 120805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohnhoff, M., C. P. Miller, and W. R. Martin. 1964. Resistance of the mouse's intestinal tract to experimental salmonella infection. II. Factors responsible for its loss following streptomycin treatment. J. Exp. Med. 120817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyen, F., F. Haesebrouck, A. Vanparys, J. Volf, M. Mahu, F. Van Immerseel, I. Rychlik, J. Dewulf, R. Ducatelle, and F. Pasmans. 2008. Coated fatty acids alter virulence properties of Salmonella Typhimurium and decrease intestinal colonization of pigs. Vet. Microbiol. 132319-327. [DOI] [PubMed] [Google Scholar]
- 9.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 1391189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesta, M. F. 2006. Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 34599-608. [DOI] [PubMed] [Google Scholar]
- 11.Chao, A. 1984. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 11265-270. [Google Scholar]
- 12.Chao, A., M. C. Ma, and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80193-201. [Google Scholar]
- 13.Clarke, S. C., R. D. Haigh, P. P. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colwell, R. K. 2006. EstimateS: statistical estimation of species richness and shared species from samples, 8th ed. http://purl.oclc.org/estimates.
- 16.Cotta, M., and R. Forster. 2006. The family Lachnospiraceae, including the genera Butyrivibrio, Lachnospira and Roseburia, p. 1002-1021. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
- 17.Cselovszky, J., G. Wolf, and W. P. Hammes. 1992. Production of formate, acetate, and succinate by anaerobic fermentation of Lactobacillus pentosus in the presence of citrate. Appl. Microbiol. Biotechnol. 3794-97. [Google Scholar]
- 18.Cummings, J. H. 1981. Short chain fatty acids in the human colon. Gut 22763-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darfeuille-Michaud, A., J. Boudeau, P. Bulois, C. Neut, A. L. Glasser, N. Barnich, M. A. Bringer, A. Swidsinski, L. Beaugerie, and J. F. Colombel. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127412-421. [DOI] [PubMed] [Google Scholar]
- 20.Davis, C. P., and D. C. Savage. 1974. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 10948-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson, A. M., D. Trenchard, and A. Guz. 1965. Small bowel tonometry: assessment of small gut mucosal oxygen tension in dog and man. Nature 206943-944. [DOI] [PubMed] [Google Scholar]
- 22.Dethlefsen, L., P. B. Eckburg, E. M. Bik, and D. A. Relman. 2006. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21517-523. [DOI] [PubMed] [Google Scholar]
- 23.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diard, S., A. L. Toribio, Y. Boum, F. Vigier, I. Kansau, O. Bouvet, and A. Servin. 2006. Environmental signals implicated in Dr fimbriae release by pathogenic Escherichia coli. Microbes Infect. 81851-1858. [DOI] [PubMed] [Google Scholar]
- 25.Doyle, M. P. 1990. Pathogenic Escherichia coli, Yersinia enterocolitica, and Vibrio parahaemolyticus. Lancet 3361111-1115. [DOI] [PubMed] [Google Scholar]
- 26.Duncan, S. H., G. Holtrop, G. E. Lobley, A. G. Calder, C. S. Stewart, and H. J. Flint. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91915-923. [DOI] [PubMed] [Google Scholar]
- 27.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 3081635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellison, D. W., M. B. Lawrenz, and V. L. Miller. 2004. Invasin and beyond: regulation of Yersinia virulence by RovA. Trends Microbiol. 12296-300. [DOI] [PubMed] [Google Scholar]
- 29.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 582014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 10413780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 581879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, I. Hautefort, A. Thompson, J. C. Hinton, and F. Van Immerseel. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72946-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garland, C. D., A. Lee, and M. R. Dickson. 1982. Segmented filamentous bacteria in the rodent small-intestine—their colonization of growing animals and possible role in host-resistance to salmonella. Microb. Ecol. 8181-190. [DOI] [PubMed] [Google Scholar]
- 34.Gotelli, N. J., and R. K. Colwell. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4379-391. [Google Scholar]
- 35.Hammes, W. P., and C. Hertel. 2006. The genera Lactobacillus and Carnobacterium, p. 320-403. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
- 36.Heck, K., and G. Belle. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 561459-1461. [Google Scholar]
- 37.Heczko, U., A. Abe, and B. B. Finlay. 2000. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J. Infect. Dis. 1811027-1033. [DOI] [PubMed] [Google Scholar]
- 38.Huang, Y., M. Suyemoto, C. D. Garner, K. M. Cicconi, and C. Altier. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J. Bacteriol. 1904233-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huse, S. M., L. Dethlefsen, J. A. Huber, D. M. Welch, D. A. Relman, and M. L. Sogin. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov, I. I., R. Frutos, N. Manel, K. Yoshinaga, D. B. Rifkin, R. B. Sartor, B. B. Finlay, and D. R. Littman. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4337-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jantratid, E., N. Janssen, C. Reppas, and J. B. Dressman. 2008. Dissolution media simulating conditions in the proximal human gastrointestinal tract: an update. Pharm. Res. 251663-1676. [DOI] [PubMed] [Google Scholar]
- 42.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 43.Klaasen, H. L., J. P. Koopman, F. G. Poelma, and A. C. Beynen. 1992. Intestinal, segmented, filamentous bacteria. FEMS Microbiol. Rev. 8165-180. [DOI] [PubMed] [Google Scholar]
- 44.Klaasen, H. L., J. P. Koopman, M. E. Van den Brink, M. H. Bakker, F. G. Poelma, and A. C. Beynen. 1993. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27141-150. [DOI] [PubMed] [Google Scholar]
- 45.Klaasen, H. L., J. P. Koopman, M. E. Van den Brink, H. P. Van Wezel, and A. C. Beynen. 1991. Mono-association of mice with non-cultivable, intestinal, segmented, filamentous bacteria. Arch. Microbiol. 156148-151. [DOI] [PubMed] [Google Scholar]
- 46.Klaasen, H. L., P. J. Van der Heijden, W. Stok, F. G. Poelma, J. P. Koopman, M. E. Van den Brink, M. H. Bakker, W. M. Eling, and A. C. Beynen. 1993. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 61303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuehl, C. J., H. D. Wood, T. L. Marsh, T. M. Schmidt, and V. B. Young. 2005. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect. Immun. 736952-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon, Y. M., and S. C. Ricke. 1998. Induction of acid resistance of Salmonella typhimurium by exposure to short-chain fatty acids. Appl. Environ. Microbiol. 643458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladas, S. D., P. E. Isaacs, G. M. Murphy, and G. E. Sladen. 1986. Fasting and postprandial ileal function in adapted ileostomates and normal subjects. Gut 27906-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laerke, H. N., and B. B. Jensen. 1999. D-tagatose has low small intestinal digestibility but high large intestinal fermentability in pigs. J. Nutr. 1291002-1009. [DOI] [PubMed] [Google Scholar]
- 51.Laerke, H. N., B. B. Jensen, and S. Hojsgaard. 2000. In vitro fermentation pattern of D-tagatose is affected by adaptation of the microbiota from the gastrointestinal tract of pigs. J. Nutr. 1301772-1779. [DOI] [PubMed] [Google Scholar]
- 52.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 461451-1464. [DOI] [PubMed] [Google Scholar]
- 53.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 874304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levison, M. E. 1973. Effect of colon flora and short-chain fatty acids on growth in vitro of Pseudomonas aeruginosa and Enterobacteriaceae. Infect. Immun. 830-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 10211070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li, F., M. A. Hullar, and J. W. Lampe. 2007. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J. Microbiol. Methods 68303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magurran, A. 2004. Measuring biological diversity. Blackwell Scientific Ltd., Oxford, United Kingdom.
- 58.McBee, M. E., P. Z. Zheng, A. B. Rogers, J. G. Fox, and D. B. Schauer. 2008. Modulation of acute diarrheal illness by persistent bacterial infection. Infect. Immun. 764851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McHan, F., and E. B. Shotts. 1993. Effect of short-chain fatty acids on the growth of Salmonella typhimurium in an in vitro system. Avian Dis. 37396-398. [PubMed] [Google Scholar]
- 60.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34836-849. [DOI] [PubMed] [Google Scholar]
- 61.Meynell, G. G. 1963. Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br. J. Exp. Pathol. 44209-219. [PMC free article] [PubMed] [Google Scholar]
- 62.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55165-199. [DOI] [PubMed] [Google Scholar]
- 63.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olivier, V., N. H. Salzman, and K. J. Satchell. 2007. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect. Immun. 755043-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Packey, C. D., and R. B. Sartor. 2008. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J. Intern. Med. 263597-606. [DOI] [PubMed] [Google Scholar]
- 66.Pouteau, E., F. Rochat, A. Jann, I. Meirim, J. L. Sanchez-Garcia, K. Ornstein, B. German, and O. Ballevre. 2008. Chicory increases acetate turnover, but not propionate and butyrate peripheral turnovers in rats. Br. J. Nutr. 99287-296. [DOI] [PubMed] [Google Scholar]
- 67.Que, J. U., S. W. Casey, and D. J. Hentges. 1986. Factors responsible for increased susceptibility of mice to intestinal colonization after treatment with streptomycin. Infect. Immun. 53116-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rhee, S. K., and M. Y. Pack. 1980. Effect of environmental pH on fermentation balance of Lactobacillus bulgaricus. J. Bacteriol. 144217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 1483651-3660. [DOI] [PubMed] [Google Scholar]
- 70.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 31335-1344. [DOI] [PubMed] [Google Scholar]
- 71.Sarma-Rupavtarm, R. B., Z. Ge, D. B. Schauer, J. G. Fox, and M. F. Polz. 2004. Spatial distribution and stability of the eight microbial species of the altered Schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 702791-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sartor, R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134577-594. [DOI] [PubMed] [Google Scholar]
- 73.Schiemann, D. A., and S. R. Shope. 1991. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect. Immun. 59437-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 711501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmidt, T. M., and D. A. Relman. 1994. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 235205-222. [DOI] [PubMed] [Google Scholar]
- 76.Sekirov, I., N. M. Tam, M. Jogova, M. L. Robertson, Y. Li, C. Lupp, and B. B. Finlay. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 764726-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shin, R., M. Suzuki, and Y. Morishita. 2002. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J. Med. Microbiol. 51201-206. [DOI] [PubMed] [Google Scholar]
- 78.Snel, J., H. J. Blok, H. M. P. Kengen, W. Ludwig, F. G. J. Poelma, J. P. Koopman, and A. D. L. Akkermans. 1994. Phylogenetic characterization of clostridium related segmented filamentous bacteria in mice based on 16S ribosomal-RNA analysis. Syst. Appl. Microbiol. 17172-179. [Google Scholar]
- 79.Stecher, B., G. Paesold, M. Barthel, M. Kremer, J. Jantsch, T. Stallmach, M. Heikenwalder, and W. D. Hardt. 2006. Chronic Salmonella enterica serovar Typhimurium-induced colitis and cholangitis in streptomycin-pretreated Nramp1+/+ mice. Infect. Immun. 745047-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stecher, B., R. Robbiani, A. W. Walker, A. M. Westendorf, M. Barthel, M. Kremer, S. Chaffron, A. J. Macpherson, J. Buer, J. Parkhill, G. Dougan, C. von Mering, and W. D. Hardt. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talham, G. L., H. Q. Jiang, N. A. Bos, and J. J. Cebra. 1999. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 671992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tannock, G. W., and D. C. Savage. 1974. Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect. Immun. 9591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trachman, J. D., and M. Yasmin. 2004. Thermo-osmoregulation of heat-labile enterotoxin expression by Escherichia coli. Curr. Microbiol. 49353-360. [DOI] [PubMed] [Google Scholar]
- 84.Turnbaugh, P. J., F. Backhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Waaij, D., J. M. Berghuis-de Vries, and L.-V. Lekkerkerk. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (London) 69405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Immerseel, F., J. B. Russell, M. D. Flythe, I. Gantois, L. Timbermont, F. Pasmans, F. Haesebrouck, and R. Ducatelle. 2006. The use of organic acids to combat Salmonella in poultry: a mechanistic explanation of the efficacy. Avian Pathol. 35182-188. [DOI] [PubMed] [Google Scholar]
- 87.Voravuthikunchai, S. P., and A. Lee. 1987. Cecectomy causes long-term reduction of colonization resistance in the mouse gastrointestinal tract. Infect. Immun. 55995-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 582438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 735261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu, J., and J. I. Gordon. 2003. Inaugural article: honor thy symbionts. Proc. Natl. Acad. Sci. USA 10010452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Young, V. B., and T. M. Schmidt. 2004. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 421203-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu, C., S. Feng, V. Sperandio, Z. Yang, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2007. The possible influence of LuxS in the in vivo virulence of rabbit enteropathogenic Escherichia coli. Vet. Microbiol. 125313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]