Abstract
Fusobacterium nucleatum is a gram-negative oral anaerobe implicated in periodontal disease and adverse pregnancy outcome. The organism colonizes the mouse placenta, causing localized infection and inflammation. The mechanism of placental colonization has not been elucidated. Previous studies identified a novel adhesin from F. nucleatum, FadA, as being involved in the attachment and invasion of host cells. The fadA deletion mutant F. nucleatum 12230 US1 was defective in host cell attachment and invasion in vitro, but it also exhibited pleiotropic effects with altered cell morphology and growth rate. In this study, a fadA-complementing clone, F. nucleatum 12230 USF81, was constructed. The expression of FadA on USF81 was confirmed by Western blotting and immunofluorescent labeling. USF81 restored host cell attachment and invasion activities. The ability of F. nucleatum 12230, US1, and USF81 to colonize the mouse placenta was examined. US1 was severely defective in placental colonization compared to the wild type and USF81. Thus, FadA plays an important role in F. nucleatum colonization in vivo. These results also represent the first complementation studies for F. nucleatum. FadA may be a therapeutic target for preventing F. nucleatum colonization of the host.
Fusobacterium nucleatum is a gram-negative filamentous anaerobe and a commensal organism of the oral cavity. It is an opportunistic pathogen that has been implicated in various forms of periodontal disease and in infections and abscesses in other parts of the body. F. nucleatum is highly prevalent in intrauterine infections causing adverse pregnancy outcome including spontaneous miscarriage, preterm birth, and stillbirth (1, 2, 6, 9; unpublished results). It was previously suggested that the organism can translocate from the oral cavity to the uterus through hematogenous transmission (5, 8). F. nucleatum adheres to and invades epithelial and endothelial cells (5, 7), which may be a mechanism for transmission. We have shown using mice that once in the blood circulation, F. nucleatum colonizes specifically in the placenta, causing Toll-like receptor 4-mediated localized inflammatory responses leading to preterm or term stillbirth without causing systemic infections (5, 10). The pattern of infection and inflammation mimics that in humans (5). It was unclear, however, which virulence factors allowed the bacteria to colonize the placenta. One possible candidate is the FadA adhesin, the only adhesin identified from F. nucleatum so far to be involved in tissue culture cell attachment and invasion (4).
FadA is a small α-helical peptide consisting of 129 amino acid residues in its nonsecreted form (pre-FadA) and 111 residues in the secreted form (mFadA). It is a unique adhesin in that both pre-FadA and mFadA are required for oligomerization and function (13). The crystal structure of FadA reveals a filamentous structure with the monomers linked together in a head-to-tail pattern via a novel leucine chain motif (11). A fadA deletion mutant, F. nucleatum 12230 US1, was defective in the attachment and invasion of KB oral mucosal carcinoma cells and Chinese hamster ovary (CHO) cells (4). However, the disruption of fadA also exhibited pleiotropic effects, with alterations in bacterial morphology and growth rate (4; unpublished results). Thus, it is necessary to perform genetic complementation studies to confirm the function of FadA. Due to the difficulty in the genetic manipulation of F. nucleatum, no complementing clones in F. nucleatum have been made until now.
In this study, a fadA-complementing clone, F. nucleatum 12230 USF81, was constructed. The ability of F. nucleatum 12230, US1, and USF81 to adhere to and invade epithelial cells was assessed in vitro, and their ability to colonize the mouse placenta was examined in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. F. nucleatum was cultivated using Columbia blood agar supplemented with 5 μg/ml hemin (Sigma), 1 μg/ml menadione (Sigma), and 5% defibrinated sheep blood (Cleveland Scientific, OH) and incubated at 37°C with 5% CO2, 10% H2, and 85% N2. Escherichia coli TOP10 cells were cultivated using LB agar or broth. The fadA gene with a Shine-Dalgarno sequence was amplified using primers fadASDF and fadASDR, creating a 436-bp fragment with a BamHI site at the 5′ end and a SalI site at the 3′ end. The ermF gene was amplified using primers erm1F and erm1R, creating a 622-bp fragment with a BamHI site at the 3′ end. The ermAM gene was amplified using primers erm2F and erm2R, creating a 643-bp fragment with a SalI site at the 5′ end. Following digestion with BamHI and/or SalI, these three fragments were ligated, and the ermF-fadA-ermAM fragment was amplified using primers erm1F and erm2R and cloned into pCR2.1 (Invitrogen). The resulting plasmid, pYH1479, was digested with SalI, and the catP gene from plasmid pJIR418 (12) was inserted into this site to generate pYH1480.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Bacterial strain, plasmid, or primer | Description or sequence (5′-3′) | Reference or source |
|---|---|---|
| Bacterial strains | ||
| F. nucleatum | ||
| 12230 | Wild type | 7 |
| US1 | fadA deletion mutant (12230 ΔfadA::ermF-ermAM) | 4 |
| USF81 | fadA-complementing strain (12230 US1 ermF-ermAM::fadA-catP) | This study |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR2.1 | Cloning vector for PCR fragments | Invitrogen |
| pJIR418 | Carrying catP, conferring thiamphenicol resistance | 12 |
| pYH1479 | Carrying the ermF-fadA-ermAM fragment with the SalI site between fadA and ermAM | This study |
| pYH1480 | catP from pJIR418 cloned into the SalI site in pYH1479, used for sonoporation to generate a fadA-complementing clone | This study |
| Primers | ||
| fadASDF | TTTGGATCCTAAATAAATAATTTGGGAGGTAACAAAAATG | This study |
| fadASDR | TTTGTCGACTTAGTTACCAGCTCTTAAAGC | This study |
| erm1F | TGCATACCTTTGTTCCTCGG | This study |
| erm1R | TTTTTTGGATCCGAAGGACAATGGAACCTCCC | This study |
| erm2F | TTTTTTGTCGACAAATTGGAACAGGTAAAGGGC | This study |
| erm2R | CTCATAGAATTATTTCCTCCCG | This study |
Construction of the fadA-complementing clone.
Log-phase F. nucleatum US1 cells were washed and resuspended in phosphate-buffered saline (PBS) supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2. The bacterial suspension (approximately 1 × 109 cells) was mixed with 50 μg plasmid pYH1480 and 50 μl Definity:Perflutren lipid microsphere (Bristol-Myers Squibb) in 0.4-ml wells of an eight-well flat-bottom immunomodule with a frame (Nalge Nunc International). The mixture was treated by sonoporation as previously described (3). The sonoporated suspension was plated onto Columbia blood agar plates and incubated without selection for 24 h at 37°C under anaerobic conditions, followed by replication on Columbia blood agar plates containing 5 μg/ml thiamphenicol. The plates were incubated for approximately 3 to 5 days at 37°C. The thiamphenicol-resistant colonies were isolated and inoculated in Columbia broth containing 5 μg/ml thiamphenicol.
Western blotting analysis.
Western blotting analysis was performed as previously described (13), with slight modifications. Whole-cell F. nucleatum cultures, at approximately 108 CFU, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by transfer to polyvinylidene difluoride membranes (Millipore). Anti-FadA monoclonal antibody (MAb) 5G11-3G8 (13) was used as the primary antibody at a 1:5,000 dilution, and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Pierce) was used as a secondary antibody at a 1:2,000 dilution, followed by chemiluminescence development.
Immunofluorescent staining of FadA.
Immunofluorescent staining of FadA was performed as previously described, with modifications (11). F. nucleatum cells were harvested by centrifugation and resuspended in PBS. The suspension was spotted onto a glass slide and allowed to air dry. After methanol fixation, the bacteria were first incubated with 5% skim milk in PBS for 1 h at room temperature, followed by incubation with anti-FadA MAb 5G11-3G8 (13) at a 1:50 dilution or its mouse IgG1 isotype for 30 min at 37°C. Following washes with PBS, the bacteria were incubated for 1 h at 37°C with Alexa Fluor 594 chicken anti-mouse IgG (Invitrogen) at a 1:100 dilution. For controls, conjugated secondary antibodies were incubated with the bacteria without prior incubation with anti-FadA MAb.
Tissue culture attachment and invasion assays.
Tissue culture attachment and invasion by F. nucleatum were determined as previously described (7). Briefly, CHO (ATCC CRL10154) and immortalized human oral keratinocyte OKF6/Tert (Tert) cells (13) were grown to near confluence in 24-well trays, followed by the addition of bacteria at a multiplicity of infection of approximately 50:1 to 100:1 (bacteria:cells). For attachment assays, the infected monolayers were incubated at 37°C in 5% CO2 for 1 h and then washed four times with PBS. The cells were lysed by water, and serial dilutions were plated onto blood agar plates to enumerate cell-associated bacteria. For invasion assays, the infected monolayers were incubated at 37°C for 3 to 4 h, followed by washes with PBS. Fresh media containing 300 μg/ml gentamicin and 200 μg/ml metronidazole were added to the monolayer and incubated for an additional hour to eliminate extracellular bacteria. The cells were washed again and lysed with water. The serial dilutions were plated to enumerate the invaded bacteria. The level of attachment (or invasion) was expressed as the percentage of cell-associated (or invaded) bacteria over the total number of bacteria initially added to the monolayers.
F. nucleatum colonization in the mouse placenta.
F. nucleatum colonization in the mouse placenta was determined as previously described (5). Briefly, approximately 2 × 107 CFU of F. nucleatum were injected into each pregnant outbred CF-1 mouse via the tail vein on day 16 of gestation. The placentas were harvested at 6 or 24 h postinjection, followed by homogenization. Serial dilutions were plated onto blood agar plates to enumerate the live bacteria. Live bacterial titers in the placentas were expressed as log(CFU/gram tissue).
Statistical analysis.
The Student t test was used for data analysis. A P value of <0.05 was considered to be significant.
RESULTS
Construction of the fadA-complementing strain.
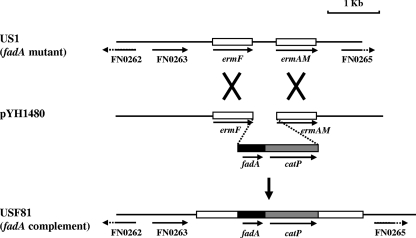
fadA deletion mutant strain US1 was constructed via sonoporation by replacing fadA on the chromosome of F. nucleatum 12230 with the ermF-ermAM cassette, conferring clindamycin resistance (4). To complement the fadA deletion, an exogenous wild-type copy of fadA needs to be introduced into US1. Complementation by shuttle plasmids was not a viable option because to date, no shuttle plasmid has been successfully introduced into F. nucleatum 12230. Taking advantage of the important characteristic of sonoporation, i.e., one-step double-crossover recombination, and the fact that fadA is monocistronic and the replacement of fadA with the ermF-ermAM cassette did not affect the transcription of the downstream genes (3, 4), we constructed the complementing clone by inserting wild-type fadA back onto the chromosome in the ermF-ermAM cassette. Suicide plasmid pYH1480 was generated, carrying the ermF-fadA-catP-ermAM fragment, with wild-type fadA and catP flanked by a 622-bp fragment of ermF upstream and a 643-bp ermAM fragment downstream, respectively (Fig. 1). All four genes or gene fragments were aligned such that they transcribed in the same direction as the ermF-ermAM cassette on the chromosome of US1. The >600-bp flanking fragments of ermF and ermAM provide enough homology for the double crossover to take place. Plasmid pYH1480 was delivered into US1 by sonoporation using established protocols (3). Thiamphenicol-resistant transformants, conferred by catP, were selected. As reported previously, the transformation efficiency was low, at approximately 0.02 transformants/μg DNA (4). Nevertheless, PCR examination of the transformants showed that, as expected, they were double-crossover constructs with wild-type fadA and catP inserted between ermF and ermAM on the chromosome of US1. One such clone was designated USF81.
FIG. 1.
Construction of fadA-complementing clone USF81. Suicide plasmid pYH1480 carrying ermF-fadA-catP-ermAM was sonoporated into fadA deletion mutant US1 (ΔfadA::ermF-ermAM), in which the fadA gene was almost completely replaced by the ermF-ermAM cassette. Through double-crossover homologous recombination in ermF and ermAM, the wild-type fadA-catP fragment was inserted into the chromosome of US1, generating thiamphenicol-resistant, fadA-complementing clone USF81.
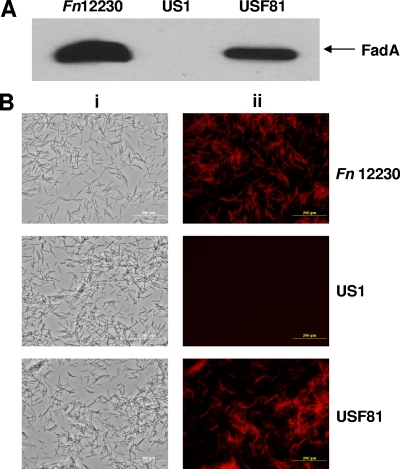
Expression of FadA in the fadA-complementing strain.
The expression of FadA in USF81 was examined by Western blotting analysis and immunofluorescent labeling using anti-FadA MAb 5G11-3G8 (13) (Fig. 2). F. nucleatum 12230 and US1 were included as positive and negative controls, respectively. While the results from the Western blotting analysis indicated that FadA was expressed at a lower level in USF81 than in the wild type, the results from immunofluorescent labeling were comparable between the two strains. Furthermore, since the immunofluorescent staining was performed using non-detergent-treated cells, the positive staining of USF81 indicates that, like in F. nucleatum 12230, FadA is exposed on the bacterial cell surface.
FIG. 2.
Expression of FadA in F. nucleatum 12230, US1, and USF81. (A) Western blotting analysis using MAb 5G11-3G8. The arrow points to FadA detected in F. nucleatum 12230 (Fn12230) and USF81. (B) Immunofluorescent labeling of FadA on non-detergent-treated bacteria using MAb 5G11-3G8. The same fields were captured by both light (i) and fluorescent (ii) microscopes using a 40× objective.
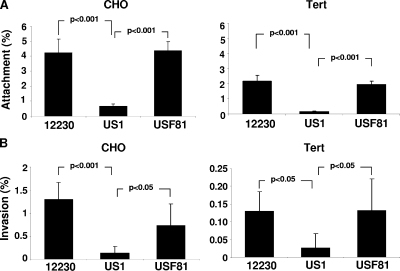
FadA complementation restores wild-type levels of attachment and invasion of host cells.
We have previously shown that US1 was defective in attachment to host cells. However, since morphological and growth rate changes were observed in US1, the question remained as to whether or not the decreased adherence was due to the absence of FadA. Such a question can now be addressed using USF81. USF81 adhered to both CHO and Tert cells to levels comparable to those of F. nucleatum 12230, confirming that FadA was responsible for restoring the attachment activity (Fig. 3A). Furthermore, previous studies showed that the purified FadA protein could be internalized into host cells (13), but it was unclear if FadA was a true invasin responsible for the internalization of the bacteria into the host cells. Thus, F. nucleatum 12230, US1, and USF81 were tested for invasion of CHO and Tert cells (Fig. 3B). While US1 was severely defective in the invasion of both cell types, USF81 restored the invasion levels to wild-type levels. The results shown here represent the first demonstration of the direct involvement of FadA in F. nucleatum invasion.
FIG. 3.
Attachment (A) and invasion (B) of CHO and immortalized human oral keratinocyte OKF6/Tert (Tert) cells by wild-type F. nucleatum 12230, fadA deletion mutant US1, and the fadA-complementing clone USF81. The results shown are averages of data from at least four independent experiments, with each experiment performed in triplicate. The standard deviations are expressed as lines above the bars. In all experiments, no significant difference was found between F. nucleatum 12230 and USF81.
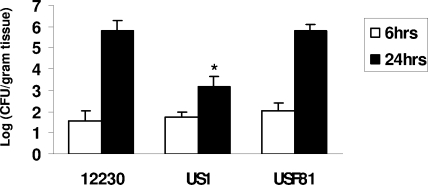
FadA is involved in bacterial colonization in mouse placenta.
To determine if FadA is involved in bacterial colonization in vivo, we took advantage of the previously established pregnant murine model (5) by injecting F. nucleatum 12230, US1, and USF81 into the mouse tail veins on day 16 of gestation. Bacterial titers in the mouse placenta were determined at 6 and 24 h postinjection according to our established protocol (5). At 6 h, the bacterial titers for all three strains were similar to each other (Fig. 4). This result confirms that the doses of the bacterial strains were comparable and that all three strains survived in mice equally well during that period. At 24 h, the titer of US1 was >1,000-fold lower than those of F. nucleatum 12230 and USF81 (Fig. 4), indicating that FadA may be responsible for F. nucleatum colonization in vivo. FadA is the first F. nucleatum component identified to be involved in intrauterine infection in the mouse model.
FIG. 4.
Colonization of F. nucleatum 12230, US1, and USF81 in the mouse placenta. Approximately 2 × 107 CFU of different F. nucleatum strains were injected into each pregnant CF-1 mouse via tail vein on day 16 of gestation. The placentas were harvested 6 or 24 h later. Live bacterial titers in the placentas were determined and expressed as log(CFU/gram tissue). The results shown are averages of data from three to seven mice per strain. The standard deviations are expressed as lines above the bars. *, P < 0.05 compared to F. nucleatum 12230.
DISCUSSION
F. nucleatum is refractory to genetic manipulation, possibly due to the numerous restriction and modification systems which the organism encodes (4). Using sonoporation, we constructed the first allelic exchange mutant, US1, in F. nucleatum (4). In this study, a fadA-complementing clone, USF81, was constructed by inserting a wild-type copy of fadA onto the chromosome of US1. The original deletion was not restored. Thus, our approach fulfills the cis-trans testing criteria of complementation. An advantage of inserting the complementing gene onto the chromosome is that only one copy of the gene is provided, as in the wild type, compared to multiple copies provided by plasmids. To the best of our knowledge, this is the first genetic complementation reported for F. nucleatum. The study reported here confirms that FadA is expressed on the bacterial cell surface. The loss of FadA detection by fluorescent labeling in US1 was not due to changed cell morphology. Furthermore, complementation of FadA restored the ability of the bacteria to adhere to and invade CHO and oral keratinocyte OKF6/Tert cells to levels similar to those of wild-type F. nucleatum. Attachment precedes invasion; thus, the reduced invasion activity observed in US1 may be attributable to its reduced ability to adhere. However, since previous studies have demonstrated both binding and internalization of purified recombinant FadA (13), it is plausible that FadA is required for both attachment and invasion.
We have shown that F. nucleatum colonizes and proliferates in the mouse placenta following hematogenous infection (5). Specific colonization in the placenta was observed at 24 h postinfection, when the bacteria were completely eliminated from the liver and spleen (5). It was unclear which virulence factor(s) allowed the bacteria to colonize the placenta. In this study, we have demonstrated that FadA plays a key role in this process. When injected into the tail veins of pregnant mice, F. nucleatum 12230, US1, and USF81 were isolated from the placenta in similar quantities at 6 h, confirming that equivalent doses of these strains were administered. In addition, the results at 6 h indicate that all three strains survived equally well in mice during that period. At 24 h postinjection, the amount of US1 recovered was severely reduced compared to amounts of F. nucleatum 12230 and USF81, indicating that FadA is involved in placental colonization and proliferation. Previously reported observations revealed that F. nucleatum invaded endothelial cells in infected mouse placentas in vivo (5). It is possible that the lack of FadA prevented the bacteria from crossing the endothelial lining in the placenta, rendering them unable to colonize. The results reported here represent the first demonstration of FadA function in vivo. It is also the first virulence factor identified in F. nucleatum to be involved in the pathogenesis of intrauterine infection. FadA may be a potential therapeutic target for the prevention and treatment of adverse pregnancy outcome caused by F. nucleatum infection.
Acknowledgments
We thank Yann Fardini for assistance in manuscript preparation and Howard Kuramitsu for reading the manuscript.
This work was supported in part by NIH grants RO1 DE 14924, KO2 DE 16102, and R21 DE17165 to Y.W.H.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 27 April 2009.
REFERENCES
- 1.Altshuler, G., and S. Hyde. 1985. Fusobacteria. An important cause of chorioamnionitis. Arch. Pathol. Lab. Med. 109739-743. [PubMed] [Google Scholar]
- 2.Chaim, W., M. Mazor, and J. R. Leiberman. 1997. The relationship between bacterial vaginosis and preterm birth. A review. Arch. Gynecol. Obstet. 25951-58. [DOI] [PubMed] [Google Scholar]
- 3.Han, Y. W., A. Ikegami, P. Chung, L. Zhang, and C. X. Deng. 2007. Sonoporation is an efficient tool for intracellular fluorescent dextran delivery and one-step double-crossover mutant construction in Fusobacterium nucleatum. Appl. Environ. Microbiol. 733677-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han, Y. W., A. Ikegami, C. Rajanna, H. I. Kawsar, Y. Zhou, M. Li, H. T. Sojar, R. J. Genco, H. K. Kuramitsu, and C. X. Deng. 2005. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 1875330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han, Y. W., R. W. Redline, M. Li, L. Yin, G. B. Hill, and T. S. McCormick. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 722272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, Y. W., T. Shen, P. Chung, I. A. Buhimschi, and C. S. Buhimschi. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J. Clin. Microbiol. 4738-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han, Y. W., W. Shi, G. T. Huang, S. K. Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 683140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill, G. B. 1993. Investigating the source of amniotic fluid isolates of fusobacteria. Clin. Infect. Dis. 16(Suppl. 4)S423-S424. [DOI] [PubMed] [Google Scholar]
- 9.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3222-232. [DOI] [PubMed] [Google Scholar]
- 10.Liu, H., R. W. Redline, and Y. W. Han. 2007. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J. Immunol. 1792501-2508. [DOI] [PubMed] [Google Scholar]
- 11.Nithianantham, S., M. Xu, M. Yamada, A. Ikegami, M. Shoham, and Y. W. Han. 2009. Crystal structure of FadA adhesin from Fusobacterium nucleatum reveals a novel oligomerization motif, the leucine chain. J. Biol. Chem. 2843865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sloan, J., T. A. Warner, P. T. Scott, T. L. Bannam, D. I. Berryman, and J. I. Rood. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27207-219. [DOI] [PubMed] [Google Scholar]
- 13.Xu, M., M. Yamada, M. Li, H. Liu, S. G. Chen, and Y. W. Han. 2007. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 28225000-25009. [DOI] [PubMed] [Google Scholar]