Abstract
Single-chain antibody mutants have been evolved in vitro with antigen-binding equilibrium dissociation constant Kd = 48 fM and slower dissociation kinetics (half-time > 5 days) than those for the streptavidin–biotin complex. These mutants possess the highest monovalent ligand-binding affinity yet reported for an engineered protein by over two orders of magnitude. Optimal kinetic screening of randomly mutagenized libraries of 105–107 yeast surface-displayed antibodies enabled a >1,000-fold decrease in the rate of dissociation after four cycles of affinity mutagenesis and screening. The consensus mutations are generally nonconservative by comparison with naturally occurring mouse Fv sequences and with residues that do not contact the fluorescein antigen in the wild-type complex. The existence of these mutants demonstrates that the antibody Fv architecture is not intrinsically responsible for an antigen-binding affinity ceiling during in vivo affinity maturation.
Over 100 antibody therapeutics are currently in clinical trials for cancer, viral, autoimmune, and other diseases (1). High-affinity antigen recognition is at the heart of all such therapies and is generally attained by affinity maturation in vivo in the mammalian immune system. It has been observed that, during in vivo affinity maturation, the B cell response exhibits an apparent affinity ceiling near Kd > 0.1 nM, because of the inability to discriminate slower dissociation kinetics relative to intrinsic B cell receptor internalization rates (2, 3). Because affinity is a critical variable for therapeutic applications such as antibody tumor targeting (4–7), extensive efforts have been made to generate higher affinity antibodies by directed evolution. The highest antibody affinities reported to date are Kd ≈ 10–20 pM (8–10).
Antibody/hapten recognition in the 4-4-20/fluorescein model system used in this study has been characterized extensively by structural (11, 12), thermodynamic (13), kinetic (14), computational (15), spectroscopic (16), and mutagenic (17) means. The 4-4-20 affinity for the fluorescein–biotin (FL-bio) hapten (Kd = 0.7 ± 0.3 nM in PBS) is near the affinity ceiling of the tertiary immune response. The 4-4-20 scFv antibody fragment can be displayed on the surface of yeast, and mutants with increased affinity can be isolated by flow cytometric cell sorting (18). Herein, we report that optimal kinetic screening of randomly mutagenized libraries of 105–107 yeast surface-displayed 4-4-20 antibodies enabled a >10,000-fold decrease in the rate of dissociation (in PBS) after four cycles of affinity mutagenesis and screening. These in vitro evolved single-chain antibody mutants have an antigen-binding equilibrium dissociation constant Kd = 48 fM and slower dissociation kinetics (half-time > 5 days) than those for the streptavidin–biotin complex, representing the highest reported monovalent ligand-binding affinity for an engineered protein by over two orders of magnitude. The existence of these mutants demonstrates that the antibody Fv architecture does not possess an intrinsic affinity ceiling for antigen binding.
Materials and Methods
Construction of Randomly Mutated scFv Libraries.
A single point mutant of the 4-4-20 scFv (mutant 4 M1.1) with 3-fold higher affinity was isolated previously from a random library generated in a mutator E. coli strain (18). A random scFv library based on this clone was created by adapting the sexual PCR method of Stemmer (19). The expression cassettes of 4 M1.1 and the unselected mutator strain-generated library were amplified by PCR with T3 and T7 promoter standard primers. An equimolar mixture of amplified 4 M1.1 and library PCR products (approximately 30 μg) were fragmented to <200 bp by DNase I, purified by gel-filtration with Centri-Sep columns (Princeton Separations, Princeton), and recombined essentially by following the method of Stemmer (19) replacing Taq polymerase with Pfu polymerase and performing 45 cycles of PCR. Final PCR amplification was performed with Taq polymerase in the presence of 2.25 mM MgCl2 and 0.375 mM MnCl2 to introduce further mutations. Primers for final amplification included sequences of ≈100 bp 5′ and 50 bp 3′ flanking the scFv ORF to allow efficient enzymatic digestion. PCR products were purified by agarose gel electrophoresis and electroelution, digested with NheI and XhoI, gel purified, and ligated into digested pCT302 backbone (18). Ligation reactions were exchanged into distilled water, concentrated with Centricon-30 and Microcon-50 filters (Amicon), and transformed into DH10B Electrocompetent cells (Life Technologies, Grand Island, NY). Transformants were pooled, and aliquots were plated to determine library diversity before amplification and purification of plasmid DNA with the Qiagen (Chatsworth, CA) Maxiprep kit. Library DNA was transformed subsequently into yeast strain EBY100 (11) by using the method of Gietz and coworkers (http://tto.trends.com). Transformants were pooled in SD-CAA medium [6.7% (vol/vol) yeast nitrogen base/2% (vol/vol) glucose/5% (vol/vol) casamino acids/100 mM sodium phosphate, pH 6.0], and aliquots were plated to determine library diversity. Subsequent libraries were constructed with similar methods, but these methods began with plasmid DNA recovered from a mixed yeast population screened for improved binders in the prior round and the unselected library from the prior round. PCR-amplified DNA from these sources was mixed at a 9:1 molar ratio before DNase I fragmentation. Incorporation of 10% unselected library DNA and error-prone final PCR amplification allowed introduction of further mutations as well as recombination of selected mutations in the resulting library. Individual libraries ranged from 3 × 105 to 2 × 107 clones.
Screening of Yeast-Displayed scFv Libraries.
Yeast cells [108 in 2.5 ml of TBS (20 mM Tris base/137 mM sodium chloride, pH 7.6)] displaying mutagenized 4-4-20 scFvs were first incubated with 1 μM FL-bio to saturate surface binding, washed, and then incubated in 1 μM 5-aminofluorescein competitor for a fixed period at 25°C. Competition times were calculated from a mathematical model (20). Labeling with streptavidin-R-phycoerythrin after competition allowed isolation by flow cytometric sorting of those cells retaining the highest levels of bound FL-bio. Cells were costained with 12CA5 mAb (Roche Molecular Biochemicals) to normalize for cell-to-cell variation in expression level as described (18). Cells were sorted with gate settings determined by using a mathematical analysis (20); four rounds of sorting and regrowth were performed to isolate highly enriched (≈80%) populations of improved mutants.
Kinetic and Sequence Analysis of Isolated Clones.
Individual clones chosen at random were assayed for dissociation rate constants (kdiss). Cells were saturated with FL-bio (1.0 μM; room temperature; 30 min), washed, labeled with streptavidin-R-phycoerythrin [1:100 streptavidin-phycoerythrin (PharMingen); 30 min on ice], and incubated at 25°C with at least a 100-fold molar excess of 5-aminofluorescein (10 μM vs. ≈1 nM surface-displayed antibody in the reaction). Aliquots were removed and analyzed by flow cytometry at various times; dissociation rate constants were determined by fitting fluorescence data to a first-order kinetic model. Plasmid DNA was recovered from yeast cells by glass bead lysis and phenol:chloroform extraction (21), followed by purification with the Wizard DNA Cleanup kit (Promega) and transformation into competent DH5α, DH0B, or XL1-Blue cells. scFv ORF sequences were determined by the University of Illinois Biotechnology Center by dideoxy terminator sequencing with an ABI Prism 377. Sequenced clones were chosen based on differing values of kdiss to avoid sequencing multiple isogenic clones.
Fluorescence Quenching of Antigen by Soluble scFv Protein.
scFvs were subcloned and expressed solubly in Saccharomyces cerevisiae under conditions as described (22). Protein was recovered from raw culture supernatants by affinity chromatography with FITC-coupled BSA. scFv-containing fractions were purified further by FPLC on a Hi-TrapQ anion exchange column (Amersham Pharmacia). Protein concentration, binding constants, and maximal fluorescence quenching constants were determined by direct titration with FL-bio in an SLM Aminco SPF500 spectrofluorometer. Excitation wavelength was 492 nm, and emission was collected at 510 nm; all experiments were conducted in quartz cuvettes thermostated to 25°C. Dissociation rate constants were measured by addition of a 500-fold molar excess of 5-aminofluorescein (nonfluorescent competitor) at time 0 to an equilibrated mixture of scFv and FL-bio in PBS (150 mM NaCl/10 mM sodium phosphate buffer, pH 8) or low salt buffer (LSB; 1 mM sodium phosphate, pH 8). Association rate constants were determined by injection of FL-bio into equilibrated scFv samples and fitting quenching data to a model with kdiss values determined from competitive dissociation experiments. Control experiments lacking 4 M5.3 scFv, FL-bio, or 5-aminofluorescein were performed in parallel to ensure the absence of artifacts from adsorption, evaporation, photobleaching, or other processes over the course of the experiment.
Results and Discussion
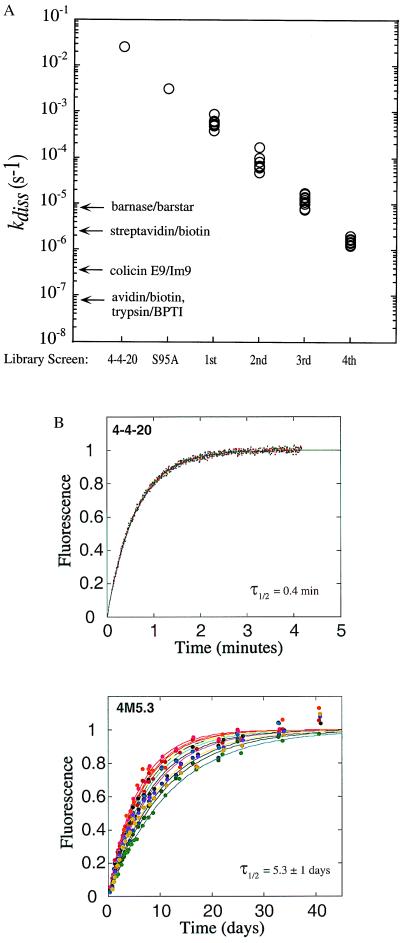
The complete 4-4-20 scFv ORF was mutagenized by error-prone DNA shuffling (19). Fluorescently labeled clones exhibiting slowed antibody-hapten dissociation kinetic constants were identified and isolated by flow cytometry with optimal screening and sorting conditions calculated from a mathematical model (20). Up to 20 improved clones were selected randomly for individual measurement of the dissociation rate constant kdiss, and 10 improved clones exhibiting the widest range of kdiss values were selected for further analysis and are represented in Fig. 1A. The complete collection of isolated mutants was then recombined by modified DNA shuffling, together with further error-prone PCR mutagenesis. This cycle of mutagenesis and screening was repeated three times, resulting in mutant scFvs with dissociation rate constants over four orders of magnitude slower than the kdiss of the 4-4-20 scFv (Fig. 1). As a basis for comparison, the dissociation rates for other high-affinity protein–ligand complexes are also shown in Fig. 1. The half-time for dissociation of the 4 M5.3 mutant is over 5 days, such that optimal screening of a fifth library would require kinetic competition times longer than 3 weeks (20) for each of four subsequent enrichment sorting steps.
Figure 1.
Dissociation kinetics of mutant scFv at 25°C. (A) Yeast displaying mutant scFv isolated from a random library was assayed for antigen dissociation rate. The 4-4-20 and VHS95A values were calculated from fluorescence quenching data with soluble scFvs. Dissociation rates for barnase/barstar (45), streptavidin/biotin (46), avidin/biotin (46), and trypsin/bovine pancreatic trypsin inhibitor (BPTI) (47) are indicated for comparison. (B) Dissociation kinetics of purified, soluble 4 M5.3 and 4-4-20 scFvs were assayed by fluorescence quenching. Triplicate traces for 4-4-20 scFv in PBS (150 mM NaCl/10 mM sodium phosphate buffer, pH 8) are shown, and 12 independent replicates are shown for 4 M5.3 in PBS or LSB. τ1/2, half-time for dissociation.
A clone from the fourth library screen (4 M5.3) was solubly expressed, and the association and dissociation constants were measured in solution (Table 1; Fig. 1B). These kinetic values are in quantitative agreement with those measured on the cell surface (Fig. 1A). Electrostatic steering of fluorescein into the binding pocket (23, 24) is evidenced by the salt dependence of kass. The association rate constant of 4 M5.3 is ≈14-fold lower than that of 4-4-20 scFv in both PBS and LSB. This decrease somewhat offsets the 16,000-fold improvement in kdiss (3,000-fold in LSB), resulting in Kd = 270 fM in PBS and Kd = 48 fM in LSB. To our knowledge, the 4 M5.3/FL-bio interaction is the highest affinity noncovalent protein–ligand complex yet engineered.
Table 1.
Binding and dissociation rate constants for soluble scFvs
| scFv | Buffer | kass (× 10−7 M−1·s−1) | kdiss (× 106 s−1) |
Kd, fM
|
|
|---|---|---|---|---|---|
| kdiss/kass | Direct titration | ||||
| 4-4-20 | PBS | 8.0 ± 3.5 | 25,100 ± 500 | 310,000 | 700,000 ± 300,000 |
| LSB | 41.0 ± 8.0 | 4,270 ± 100 | 10,000 | ND | |
| 4 M5.3 | PBS | 0.6 ± 0.03 | 1.6 ± 0.3 | 270 | ND |
| LSB | 2.9 ± 0.9 | 1.4 ± 0.4 | 48 | ND | |
ND, not determined.
Sequences were obtained of each of the mutants represented in Fig. 1,
and mutated residues are shown in Table 2. With
the exception of 4 M4.15, 16, and 19 (which are identical isolates),
all mutant clones are distinct. In each round, at least one mutation
was found in a majority of the clones (henceforth termed
“consensus” mutations). No consensus mutations were identified in
the linker or epitope tag regions; thus, these are not represented in
Table 2 for conciseness.

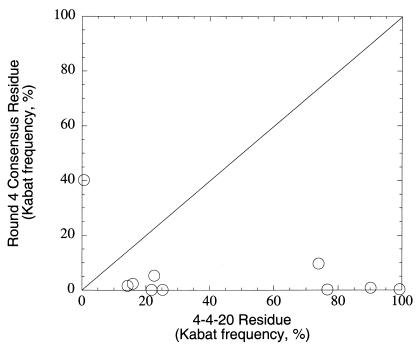
Consensus mutations accumulate in each round; once fixed in an early round, they generally are present in subsequent rounds. It is noteworthy that particular consensus mutations dominate the screened population at each round. It is not possible to ascertain from the available information whether mutations added in subsequent rounds were introduced by recombination of preexisting mutations or de novo mutagenesis during error-prone PCR and DNA shuffling. Of 10 consensus mutations after the fourth round of screening, 9 of 10 are in the VH domain, and 6 of 10 are in CDR loops. The relative paucity of mutations in the VL domain is striking, with 6-fold, 7-fold, 7-fold, and 4-fold excess mutations in VH relative to VL in each of the four rounds, respectively. Comparison of frequency in the Kabat database (25) for consensus mutations vs. 4-4-20 wild-type residues indicates that 9 of 10 consensus changes are to rare amino acids at those positions, as shown in Fig. 2. Most striking in this regard is the VH103 mutation (fixed in the mutant population in round 1), which alters the nearly invariant VH103 tryptophan residue (99.3% of mouse VH sequences in the Kabat database). Of the 10 consensus mutant substitutions, 9 occur in fewer than 10% of known mouse antibody sequences, by comparison to only one such nonconserved residue at these sites in the wild-type 4-4-20 sequence. Thus, this in vitro directed evolution approach generally sampled areas of antibody sequence space infrequently accessed by the in vivo process, perhaps because of constraints arising from the mechanism of somatic hypermutation.
Figure 2.
Natural conservation of mutated 4-4-20 residues. Comparison of occurrence frequencies in mouse VH and VL κ genes of wild-type (abscissa) and mutation (ordinate) amino acid identities are shown. Frequencies were calculated at mutated sequence positions from statistics in the Kabat database (25).
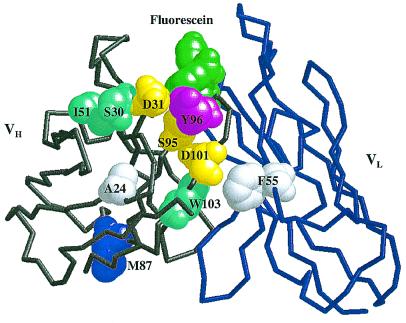
The locations of consensus mutations are identified on the 4-4-20 Fab crystal structure shown in Fig. 3. The consensus mutations cluster strikingly around VH CDR3; it has been argued previously that the VH CDR3 loop exerts the greatest influence on antigen-binding specificity (26–28). The 4-4-20 CDR loops can be grafted onto a different scFv framework without loss of affinity (29), indicating that fluorescein recognition is dominated by the CDR loops. In fact, 9 of the 10 consensus mutations identified in the present work are located at 4-4-20 residues that were present in the loop-grafted 4D5Flu hybrid protein, indicating that they lie within the portion of the scFv largely responsible for binding specificity. Of particular interest is the VLF55V mutation enriched in the fourth round of this study, which was also identified independently as one of two mutations that together improved the stability of 4D5Flu by 4 kcal/mol, with an unexpected 20-fold increase in affinity (30).
Figure 3.
Sites of consensus mutations in the 4-4-20 Fv. Backbone structure of the first 118 heavy-chain residues (gray) and the first 112 light-chain residues (blue) are shown. Fluorescein ligand (green) and mutated residues are depicted by space-filling models. Mutation sites are color-coded by distance from the binding site: first-shell residues are magenta; second-shell residues are yellow; third-shell residues are cyan; and fourth-shell residues are white. Residues in the first shell were defined as those with one or more atoms directly contacting ligand; second-shell residues were defined as those with one or more atoms directly contacting any residue in the first shell; third- and fourth-shell residues contact second- and third-shell residues, respectively. Definitions of contact were interatomic distances (in Å) equal to or less than 4.1 C-C, 3.3 O-O, 3.4 N-N and N-O, 3.8 C-N, and 3.7 C-O (48). Atomic coordinates were from the high-resolution crystal structure of Whitlow, et al. (ref. 11; PDB ID code 1FLR).
Only 1 of 10 4 M5.3 consensus mutations is in a fluorescein-contact residue; 3 are in the second shell; 3 are in the third shell; and 2 are in the fourth shell (Fig. 3). VH87 is on the scFv face opposite the fluorescein-binding pocket. This spatial distribution generally supports the observation that further affinity maturation of antibodies with affinity in the low nanomolar range occurs most effectively via changes in “vernier” or second-sphere residues (31, 32) rather than contact residues (33). In addition, 4 of 10 consensus mutations occur at VH–VL interfacial sites, suggesting that improved stability and/or orientation of the VH–VL domain pairing may be important in affinity improvement for the scFv.
A molecular dynamics comparison of liganded and unliganded 4-4-20 demonstrates a significant increase in interresidue correlated motions on fluorescein binding, particularly in the VH domain (15). The 4-4-20 VH domain possesses generally larger temperature factors than the VL domain (11), indicating greater flexibility. Of the 10 consensus mutations, 9 lie within VH, which could be consistent with the binding site preorganization mechanism for affinity improvement proposed previously for antibody affinity maturation from the germ-line sequence (34). Dissection of the thermodynamic, kinetic, and structural mechanisms by which the mutations in 4 M5.3 increase binding free energy by 3–4 kcal/mol should contribute to an improved understanding of protein recognition. Information gleaned from these gain-of-function mutations would complement loss-of-function mutational studies of strong protein–ligand interactions such as streptavidin–biotin (35).
The majority of current antibody engineering strategies focus mutagenesis on the antibody CDR loops (e.g., refs. 8, 10, 36, and 37), an approach that would not identify 4 of the 10 consensus mutations from this study. In fact, the first consensus mutation to become fixed (VHW103L) occurred at a highly conserved framework position removed from the binding site; the possibility that 1 or more subsequently selected mutations depended on the context of the VH103 mutation must be considered. The experimentally simpler strategy of error-prone PCR of the whole scFv gene was therefore arguably more effective given these results. Concerns regarding potential immunogenicity of framework mutations in a therapeutic antibody might be addressed by judicious choice of buried residues from among the selected mutations.
The relative ease with which extremely high affinity has been attained in this study might be attributed to (i) quantitatively optimized screening methodology and (ii) minimization of expression bias by use of a eukaryotic expression host. In the first instance, labeling for optimal discrimination of improved clones (20) and reduced stochastic variation in dissociation kinetics because of assay of 104–105 scFv molecules per yeast cell enable fine discrimination of affinity (reproducibility of ±10% in kdiss assayed by flow cytometry for yeast-displayed scFvs and ±30% for Kd; ref. 38). In the second instance, yeast's secretory biosynthetic apparatus effectively folds and displays or secretes (22) the 4-4-20 scFv, a molecule that forms inclusion bodies and is 98% insoluble when expressed in the periplasm of wild-type E. coli (39). Thus, a greater proportion of protein shape space will be sampled by yeast display because of elimination of prokaryotic expression biases against scFvs such as 4-4-20.
Generation of antibodies that bind essentially irreversibly relative to the relevant physiological time scale could improve efficacy for cancer immunotherapy with noninternalizing tumor-associated antigens (4–7) and passive immunization against viral and microbial pathogens (40–43). The mutagenesis and screening methodology described herein has also been applied successfully to an antibody against hen egg lysozyme (data not shown) and an antibody against the T cell receptor (44). Beyond the implications of the general capability to engineer femtomolar affinity antibodies, more specifically, a molecule such as 4 M5.3 that binds and quenches fluorescein with an affinity similar to that of streptavidin–biotin may enable interesting approaches in analytical biochemistry when combined with the array of available fluorescence-detection methodologies.
Acknowledgments
The 4-4-20 scFv was obtained from D. M. Kranz, who also provided helpful comments on the manuscript. We thank G. Durack for assistance optimizing cell-sorting instrumentation. Funding was provided by the Whitaker Foundation and National Institutes of Health Grant GM55767.
Abbreviations
- FL-bio
fluorescein–biotin
- LSB
low salt buffer
Footnotes
See commentary on page 10679.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170297297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170297297
References
- 1.Halim N S. The Scientist. 2000;14:16–17. [Google Scholar]
- 2.Batista F D, Neuberger M S. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 3.Foote J, Eisen H N. Proc Natl Acad Sci USA. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams G P, Schier R, Marshall K, Wolf E J, McCall A M, Marks J D, Weiner L M. Cancer Res. 1998;58:485–490. [PubMed] [Google Scholar]
- 5.Colcher D, Pavlinkova G, Beresford G, Booth B J, Choudhury A, Batra S K. Q J Nucl Med. 1998;42:225–241. [PubMed] [Google Scholar]
- 6.Roovers R C, Henderikx P, Helfrich W, van der Linden E, Reurs A, de Bruine A P, Arends J W, de Leij L, Hoogenboom H R. Br J Cancer. 1998;78:1407–1416. doi: 10.1038/bjc.1998.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson H, Bacon L, Pedley R B, Derbyshire E, Field A, Osbourn J, Allen D. Br J Cancer. 1998;78:181–188. doi: 10.1038/bjc.1998.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schier R, Bye J, Apell G, McCall A, Adams G P, Malmqvist M, Weiner L M, Marks J D. J Mol Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Wiesmann C, Fuh G, Li B, Christinger H W, McKay P, de Vos A M, Lowman H B. J Mol Biol. 1999;293:865–881. doi: 10.1006/jmbi.1999.3192. [DOI] [PubMed] [Google Scholar]
- 10.Yang W-P, Green K, Pinz-Sweeney S, Briones A T, Burton D R, Barbas C F I. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 11.Whitlow M, Howard A J, Wood J F, Voss E W, Jr, Hardman K D. Protein Eng. 1995;8:749–761. doi: 10.1093/protein/8.8.749. [DOI] [PubMed] [Google Scholar]
- 12.Herron J N, Terry A H, Johnston S, He X M, Guddat L W, Voss E W, Jr, Edmundson A B. Biophys J. 1994;67:2167–2183. doi: 10.1016/S0006-3495(94)80738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herron J N, Kranz D M, Jameson D M, Voss E W., Jr Biochemistry. 1986;25:4602–4609. doi: 10.1021/bi00364a022. [DOI] [PubMed] [Google Scholar]
- 14.Kranz D M, Herron J N, Voss E W., Jr J Biol Chem. 1982;257:6987–6995. [PubMed] [Google Scholar]
- 15.Lim K, Herron J N. Biochemistry. 1995;34:6962–6974. doi: 10.1021/bi00021a008. [DOI] [PubMed] [Google Scholar]
- 16.Lim K, Jameson D M, Gentry C A, Herron J N. Biochemistry. 1995;34:6975–6984. doi: 10.1021/bi00021a009. [DOI] [PubMed] [Google Scholar]
- 17.Denzin L K, Gulliver G A, Voss E W., Jr Mol Immunol. 1993;30:1331–1345. doi: 10.1016/0161-5890(93)90094-r. [DOI] [PubMed] [Google Scholar]
- 18.Boder E T, Wittrup K D. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 19.Stemmer W P. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 20.Boder E T, Wittrup K D. Biotechnol Progr. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 21.Ward A C. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shusta E V, Raines R T, Pluckthun A, Wittrup K D. Nat Biotechnol. 1998;16:773–777. doi: 10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- 23.Omelyanenko V G, Jiskoot W, Herron J N. Biochemistry. 1993;32:10423–10429. doi: 10.1021/bi00090a018. [DOI] [PubMed] [Google Scholar]
- 24.Leckband D E, Kuhl T, Wang H K, Herron J, Muller W, Ringsdorf H. Biochemistry. 1995;34:11467–11478. doi: 10.1021/bi00036a020. [DOI] [PubMed] [Google Scholar]
- 25.Johnson G, Wu T T, Kabat E A. In: Immunochemistry and Molecular Immunology, Weir's Handbook of Experimental Immunology. Herzenberg L A, Weir W M, Herzenberg L A, Blackwell C, editors. I. Cambridge, MA: Blackwell Science; 1996. pp. 6.1–6.21. [Google Scholar]
- 26.Davis M M, Lyons D S, Altman J D, McHeyzer-Williams M, Hampl J, Boniface J J, Chien Y. Ciba Found Symp. 1997;204:94–100. doi: 10.1002/9780470515280.ch7. [DOI] [PubMed] [Google Scholar]
- 27.VanDyk L, Meek K. Int Rev Immunol. 1992;8:123–133. doi: 10.3109/08830189209055568. [DOI] [PubMed] [Google Scholar]
- 28.Kabat E A, Wu T T. J Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 29.Jung S, Pluckthun A. Protein Eng. 1997;10:959–966. doi: 10.1093/protein/10.8.959. [DOI] [PubMed] [Google Scholar]
- 30.Jung S, Honegger A, Pluckthun A. J Mol Biol. 1999;294:163–180. doi: 10.1006/jmbi.1999.3196. [DOI] [PubMed] [Google Scholar]
- 31.Arkin M R, Wells J A. J Mol Biol. 1998;284:1083–1094. doi: 10.1006/jmbi.1998.2234. [DOI] [PubMed] [Google Scholar]
- 32.Foote J, Winter G. J Mol Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- 33.Burks E A, Chen G, Georgiou G, Iverson B L. Proc Natl Acad Sci USA. 1997;94:412–417. doi: 10.1073/pnas.94.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wedemayer G J, Patten P A, Wang L H, Schultz P G, Stevens R C. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 35.Freitag S, Chu V, Penzotti J E, Klumb L A, To R, Hyre D, Le Trong I, Lybrand T P, Stenkamp R E, Stayton P S. Proc Natl Acad Sci USA. 1999;96:8384–8389. doi: 10.1073/pnas.96.15.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Beuerlein G, Nie Y, Smith H, Lee B A, Hensler M, Huse W D, Watkins J D. Proc Natl Acad Sci USA. 1998;95:6037–6042. doi: 10.1073/pnas.95.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daugherty P S, Chen G, Olsen M J, Iverson B L, Georgiou G. Protein Eng. 1998;11:825–832. doi: 10.1093/protein/11.9.825. [DOI] [PubMed] [Google Scholar]
- 38.VanAntwerp J J, Wittrup K D. Biotechnol Progr. 2000;16:31–37. doi: 10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- 39.Nieba L, Honegger A, Krebber C, Pluckthun A. Protein Eng. 1997;10:435–444. doi: 10.1093/protein/10.4.435. [DOI] [PubMed] [Google Scholar]
- 40.Bachmann M F, Kalinke U, Althage A, Freer G, Burkhart C, Roost H, Aguet M, Hengartner H, Zinkernagel R M. Science. 1997;276:2024–2027. doi: 10.1126/science.276.5321.2024. [DOI] [PubMed] [Google Scholar]
- 41.Lamarre A, Talbot P J. J Immunol. 1995;154:3975–3984. [PubMed] [Google Scholar]
- 42.Gauduin M C, Parren P W, Weir R, Barbas C F, Burton D R, Koup R A. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 43.Ma J K, Hikmat B Y, Wycoff K, Vine N D, Chargelegue D, Yu L, Hein M B, Lehner T. Nat Med. 1998;4:601–606. doi: 10.1038/nm0598-601. [DOI] [PubMed] [Google Scholar]
- 44.Kieke M C, Cho B K, Boder E T, Kranz D M, Wittrup K D. Protein Eng. 1997;10:1303–1310. doi: 10.1093/protein/10.11.1303. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber G, Fersht A R. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 46.Piran U, Riordan W J. J Immunol Methods. 1990;133:141–143. doi: 10.1016/0022-1759(90)90328-s. [DOI] [PubMed] [Google Scholar]
- 47.Fritz H, Wunderer G. Arzneimittelforschung. 1983;33:479–494. [PubMed] [Google Scholar]
- 48.Braden B C, Goldman E R, Mariuzza R A, Poljak R J. Immunol Rev. 1998;163:45–57. doi: 10.1111/j.1600-065x.1998.tb01187.x. [DOI] [PubMed] [Google Scholar]