Abstract
The aryl hydrocarbon receptor (AHR) repressor (AHRR), an AHR-related basic helix-loop-helix/Per-AHR nuclear translocator-Sim protein, is regulated by an AHR-dependent mechanism and acts as a transcriptional repressor of AHR function. Resulting from a teleost-specific genome duplication, zebrafish have two AHRR genes (AHRRa and AHRRb), but their functions in vivo are not well understood. We used antisense morpholino oligonucleotides (MOs) in zebrafish embryos and a zebrafish liver cell line (ZF-L) to characterize the interaction of AHRRs and AHRs in normal embryonic development, AHR signaling, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) toxicity. Zebrafish embryos exposed to TCDD (2 and 8nM) during early development showed strong induction of CYP1A, AHRRa, and AHRRb at 48 and 72 hours post-fertilization (hpf). An MO targeting AHR2 inhibited TCDD-induced expression of CYP1A, AHRRa, and AHRRb by 84–95% in 48 hpf embryos, demonstrating a primary role for AHR2 in mediating AHRR induction. Dual MO knockdown of both AHRRs in ZF-L cells enhanced TCDD induction of CYP1A, but not other CYP1 genes. In embryos, dual knockdown of AHRRs, or knockdown of AHRRb alone, enhanced the induction of CYP1A, CYP1B1, and CYP1C1 by TCDD and decreased the constitutive expression of Sox9b. In contrast, knockdown of AHRRa did not affect Sox9b expression or CYP1 inducibility. Embryos microinjected with each of two different MOs targeting AHRRa and exposed to dimethyl sulfoxide (DMSO) displayed developmental phenotypes resembling those typical of TCDD-exposed embryos (pericardial edema and lower jaw malformations). In contrast, no developmental phenotypes were observed in DMSO-exposed AHRRb morphants. These data demonstrate distinct roles of AHRRa and AHRRb in regulating AHR signaling in vivo and suggest that they have undergone subfunction partitioning since the teleost-specific genome duplication.
Keywords: dioxin, AHR, AHR repressor, zebrafish, evolution
The aryl hydrocarbon receptor (AHR), a member of the basic helix-loop-helix/Per-AHR nuclear translocator (ARNT)-Sim (bHLH-PAS) protein family, is a ligand-activated transcription factor that plays important roles in gene regulation, toxicology, and development (Okey, 2007). The AHR regulates a battery of genes, including several cytochrome P450 (CYP) genes such as CYP1A1, the most highly induced and well-studied target gene. Upon activation by a ligand, the AHR translocates to the nucleus and forms a heterodimer with the ARNT transcription factor. The AHR/ARNT heterodimer recognizes and binds to AHR-responsive enhancer elements (AHREs) in the promoter regions of a variety of target genes. In addition to the induction of several CYP genes, AHR agonists also induce the expression of the AHR repressor (AHRR), which has been shown to have functional AHREs in its proximal promoter in a variety of vertebrate species (Baba et al., 2001; Haarmann-Stemmann et al., 2007; Karchner et al., 2002; Mimura et al., 1999). The AHRR is a member of the bHLH-PAS protein family and is closely related to the AHR. However, unlike the AHR, which has two PAS domains, the AHRR only has a single PAS domain and lacks a ligand-binding region (Mimura et al., 1999). The AHRR, which is capable of dimerizing with ARNT, forms a negative feedback loop with AHR, repressing AHR signaling by competition for binding to AHREs (Evans et al., 2008; Mimura et al., 1999), as well as by novel mechanisms that are independent of AHRE binding by AHRR (Evans et al., 2008).
Most of the current knowledge of AHRR function comes from transient transfection assays and other in vitro methods used to characterize AHRR expression or the role of this protein in transcriptional regulation of AHR signaling. However, very little is known regarding the role of AHRR in development or AHR signaling in vivo. A recent study (Hosoya et al., 2008) showed that AHRR-deficient mice exhibit enhanced CYP1A1 induction in a tissue-specific manner after treatment with 3-methylcholanthrene. AHRR deficiency in mice does not appear to have any gross effects on development, growth, or reproduction (Hosoya et al., 2008), although a systematic histological assessment of AHRR-deficient mice has not yet been reported. Many other proteins in the AHR signaling pathway are vital for normal development, independent of their roles in regulating xenobiotic-metabolizing enzymes. Loss of AHR signaling causes a variety of developmental phenotypes, such as abnormal liver development caused by persistence of the ductus venosus, as well as vascular abnormalities in the eye and kidney (Bunger et al., 2003, 2008; Lahvis et al., 2000). ARNT-deficient mice die in utero due to major failure in placental vascularization (Kozak et al., 1997), and AHR-associated protein 9 (ARA9)-deficient mice die in utero due to cardiac malformation (Lin et al., 2007). ARA9 is an immunophilin-like protein that participates as one of the chaperone proteins that stabilizes the cytosolic AHR complex (LaPres et al., 2000). In addition, partial inhibition of ARNT or ARA9 function by the production of hypomorphs recapitulates the ductus venosus phenotype found in AHR-deficient mice (Lin et al., 2008; Walisser et al., 2004). Given these observations and recent findings suggesting that AHRR is involved in human reproductive physiology and in the regulation of cell growth (reviewed in Haarmann-Stemmann and Abel, 2006; Hahn et al., 2009), we hypothesized that AHRR may play a role in basic biological processes, in addition to its role in toxicology.
The zebrafish (Danio rerio) has emerged recently as an excellent model for investigations into both development and developmental toxicology (reviewed in Carney et al., 2006; Hill et al., 2005). Recent studies have revealed that fish—including zebrafish—often possess two or more homologs (co-orthologs) of human transcription factors, the result of a genome duplication that occurred at the base of the ray-finned fish lineage, after its divergence for the lineage leading to mammals (Meyer and Van de Peer, 2005). Many of the duplicated (paralogous) genes in fish have undergone subfunctionalization, resulting in paralogs with complementary functions or expression patterns (Postlethwait et al., 2004). In such cases, the multiple functions of a single mammalian gene can be elucidated by investigating the duplicated fish genes between which the subfunctions have been partitioned (Amores et al., 1998). As a result of the teleost-specific genome duplication (and an earlier vertebrate-specific genome duplication), zebrafish have multiple AHR genes (AHR1a, AHR1b, and AHR2) (Andreasen, Hahn, et al., 2002; Hahn et al., 2006; Karchner et al., 2005; Tanguay et al., 1999), ARNT genes (ARNT1 and ARNT2) (Prasch et al., 2006; Tanguay et al., 2000), and AHRR genes (AHRRa and AHRRb) (These two zebrafish AHRR genes correspond to AHRR1 (AHRRa) and AHRR2 (AHRRb) from our earlier report in Evans et al., 2005)). The presence of multiple isoforms for these components of the AHR signaling pathway make zebrafish an ideal model organism for studying the diversification and subfunction partitioning of this superfamily of transcription factors (Hahn et al., 2006; Postlethwait et al., 2004). Investigation of the subfunctionalization of these duplicated transcription factors may offer further insight into the differing roles of the AHR signaling pathway in biological processes, such as development or cell growth, and toxicology. Here, we report new findings concerning the regulation and function of both isoforms of AHRR (AHRRa and AHRRb) in zebrafish. Using antisense morpholino oligonucleotides (MOs), we demonstrate the role of AHR2 in regulating the 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced expression of both AHRRa and AHRRb. We also demonstrate separate roles for AHRRa and AHRRb in development, AHR signaling in response to TCDD exposure, and mediation of TCDD-induced toxicity.
MATERIALS AND METHODS
Fish Husbandry
The Tupfel/Long fin mutation wild-type strain of zebrafish were used for all experiments and were maintained as previously described (Jönsson, Orrego, et al., 2007). Fertilized eggs were obtained from multiple group breedings from tanks of 30 female and 15 male fish. Embryos were reared as described previously (Jönsson, Jenny, et al., 2007). Procedures used in these experiments were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution.
Morpholino Oligonucleotides
Morpholinos designed to block initiation of translation of zebrafish AHR2, AHRRa, and AHRRb were obtained from Gene Tools, LLC (Philomath, OR). The previously described MO (AHR2 MO: 5′-TGTACCGATACCCGCCGACATGGTT-3′) (Carney et al., 2004; Prasch et al., 2003) was used for AHR2. Two MOs that overlap by 5 bp were designed for AHRRa (AHRRa MO1: 5′-ATCCTCAAGATGCCTGCTGTGTGTG-3′; AHRRa MO2: 5′-GACAGTCTCCAGGCGGAATCATCCT-3′). AHRRa MO1 (25 bp) complements 23 bp of the 5′ untranslated region (UTR) and the first two residues of the start codon. AHRRa MO2 (25 bp) complements the first three residues upstream of the start codon and the first 22 residues of the coding region. One morpholino was designed for AHRRb (AHRRb MO1: 5′-CATCTTCTCCAAATATCTTACAACT-3′). AHRRb MO1 complements the start codon and 22 additional residues of the 5′ UTR. Gene Tools’ standard control morpholino (Ctrl MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used as an injection control. All morpholinos were fluorescein tagged for visualization by fluorescence microscopy. Early attempts at designing splice-blocking morpholinos were made; however, we were limited in our targeting with respect to the splice-blocking morpholinos. To avoid the production of an intact DNA-binding fragment that could potentially serve as a dominant negative or still have repressive capabilities (Evans et al., 2008), we limited the splice-blocking morpholino targeting to the second and third exons of the AHRR genes. Multiple attempts were made to design splice-blocking morpholinos, particularly for the AHRRa gene, but we were unable to demonstrate any effective or significant knockdown of “wild-type” transcript using RT-PCR techniques. In addition, a polyclonal antibody to AHRRa was used to attempt to confirm knockdowns using Western blot techniques but was not sufficiently sensitive to detect AHRRa in zebrafish embryos. Based on this outcome, we chose to use multiple translational blocking morpholinos to confirm phenotypes.
Confirmation of MO Translation Inhibition by In Vitro Protein Synthesis
The TNT T7 Quick Coupled Reticulocyte Lysate System (Promega, Madison, WI) was used to synthesize [35S]methionine-labeled zebrafish AHRRb protein, and the TNT T3 Coupled Reticulocyte Lysate System (Promega) was used to synthesize [35S]methionine-labeled zebrafish AHR2 and AHRRa proteins as per manufacturer’s protocols. Briefly, TNT reagents were combined with 1 μl of [35S]methionine (> 1000 Ci/mmol at 10 mCi/ml), 2 μl AHRRb in pcDNA 3.1/Zeo, or 2 μl AHR2 or AHRRa in pBK-CMV (0.5 μg/μl) and adjusted to a final volume of 25 μl with H2O. To test the efficacy of the target MOs, 0.5 μl of a 25μM stock of the standard Ctrl MO, gene-specific MO, or an MO specific for a paralogous gene was added to the reaction for a final concentration of 500nM. Mixtures were incubated at 30°C for 90 min to allow for sufficient in vitro transcription and translation of the target proteins. Fifteen microliters of the labeled protein were resolved by SDS-polyacrylamide gel electrophoresis. Fluorography was used to amplify the signal and visualize proteins on film. Densitometric analysis was performed with the ImageJ software program from the National Institute of Health (http://rsb.info.nih.gov/ij/). The relative densitometric units were determined by normalizing the target MO treatments to the Ctrl MO treatments after all densitometric values were adjusted for local background and band size.
Microinjection of Zebrafish Embryos with Morpholino Antisense Oligonucleotides
All morpholinos were fluorescein tagged for screening purposes to guarantee that only successfully injected embryos were used for the subsequent experiments. All morpholinos were diluted to 0.18mM in deionized water. A Narishige IM-300 microinjector was used to inject 2.1 nl of morpholino into the yolk of two- to four-cell stage embryos, resulting in approximately 3.3–3.4 ng of morpholino per embryo. Injection volumes were calibrated by injecting solutions into mineral oil and measuring the diameter of the sphere with a stage micrometer (volume = 4/3πr3; 160 μm diameter is equivalent to 2.1 nl). At 3 hours post-fertilization (hpf), embryos were sorted to remove damaged or unfertilized eggs, and the remaining embryos were screened by fluorescence for successful MO incorporation (> 95%).
Exposure of Zebrafish Embryos to TCDD and Experimental Designs
Groups of noninjected (No MO) or MO-injected embryos were placed in glass petri dishes with no more than three embryos per milliliter of 0.3× Danieau's and then exposed to carrier (0.1% dimethyl sulfoxide [DMSO]) or TCDD (dissolved in DMSO) for 1 h, starting at 6 hpf. After TCDD exposure, the embryos were washed three times in fresh 0.3× Danieau's, then placed in petri dishes with 25 ml fresh 0.3× Danieau's, and held in an incubator at 28.5°C with a 1400 h light/1000 h dark cycle. Danieau's solution was renewed at 48 and 96 hpf as needed. Total RNA was prepared using the RNA STAT60 protocol.
Developmental expression of AHRRa and AHRRb.
Embryos were exposed to 0.1% DMSO, 2nM TCDD, or 8nM TCDD for 1 h at 6 hpf. Three biological replicates of 20 pooled embryos were collected for each treatment at 24, 48, and 72 hpf. Embryos were flash frozen in liquid nitrogen and stored at −80°C until total RNA isolation used for quantitative RT-PCR.
AHR2 morpholino knockdowns for gene expression assessment.
Embryos were microinjected (No MO) with Ctrl MO or AHR2 MO as described above. Embryos were exposed to either 0.1% DMSO or 2nM TCDD for 1 h at 6 hpf. Three biological replicates of 20 pooled embryos were collected for each treatment at 48 hpf. Embryos were flash frozen in liquid nitrogen and stored at −80°C until total RNA isolation used for quantitative RT-PCR.
AHRR morpholino knockdowns for gene expression assessment.
Embryos were microinjected with Ctrl MO, AHRRa MO1 or MO2, or AHRRb MO as described above. Embryos were exposed to 0.1% DMSO, 2nM TCDD or 8nM TCDD for 1 h at 6 hpf. Three biological replicates of 20 pooled embryos were collected for each treatment at 48 and 72 hpf. Embryos were flash frozen in liquid nitrogen and stored at −80°C until total RNA isolation used for quantitative RT-PCR.
AHRR morpholino knockdowns for phenotypic assessment.
Embryos were microinjected with Ctrl MO, AHRRa MO1 or MO2, or AHRRb MO as described above. Embryos were exposed to 0.1% DMSO, 0.5nM TCDD, 1nM TCDD, or 2nM TCDD for 1 h at 6 hpf. Ten embryos per treatment were sampled at 72 hpf. The incidence and severity of pericardial edema in embryos from each treatment group were determined by quantification of the pericardial sac area. Embryos were anesthetized by addition of MS-222 (0.015% final concentration) to the 0.3× Danieau's and photographed in a lateral view. The area of the pericardial sac was measured using the ImageJ software program. A relative value for the severity of pericardial edema was determined by dividing the size of the pericardial sac for each individual embryo by the mean size of the pericardial sac from No MO DMSO Ctrl (No MO Ctrl).
Gene Knockdown with Morpholino Antisense Oligonucleotides in ZF-L Cells
Using a novel transfection peptide developed by Gene Tools, LLC, a recent study (Tyson-Capper and Europe-Finner, 2006) demonstrated the successful transfection of MO into human cell lines and corresponding knockdown of the target gene. This transfection reagent was used to assess the effect of AHRRa and AHRRb knockdown on Cyp1 expression in a zebrafish liver cell line (ZF-L). The ZF-L cell line was maintained in LDF medium (50% Leibovitz's L-15 medium, 35% Dulbecco's modification of eagle's medium, and 15% Ham's F12) supplemented with 5% heat-inactivated fetal bovine serum (FBS) at 28°C. To determine the effect of AHRRa and AHRRb knockdown on Cyp1 expression, ZF-L cells were plated in 48-well plates at a density of 55,000 cells per well in 200 μl of LDF medium. Cells were allowed to adhere to the plate for 30 min, and an additional 100 μl of LDF medium containing 15% heat-inactivated FBS was added to each well, and the cells were incubated overnight at 28°C. The next day, fresh LDF medium containing the Endo-Porter (Gene Tools, LLC) transfection peptide (6 μl/ml LDF medium) and 3μM of Ctrl MO or a 6μM combination of AHRRa MO1 and AHRRb MO was placed in each well. The ZF-L cells were incubated in the MO-containing medium for 48 h to allow for full incorporation of the MOs. After 48 h, the LDF medium was exchanged for new medium containing 0.1% DMSO or TCDD dissolved in 0.1% DMSO (final concentration of TCDD—0.5, 2, and 10nM), and cells were incubated for 24 h. Each biological replicate was collected by pooling all the cells from six wells. Three biological replicates were collected per treatment. Cells were lysed by adding 100 μl of RNA STAT60 (Tel-Test B) to each well (600 μl pool per replicate). Total RNA was isolated using the standard protocol for RNA STAT60.
Gene Expression Assessment by Real-Time RT-PCR
cDNA was synthesized from 2 μg total RNA using random hexamers and the Omniscript cDNA Synthesis Kit (Qiagen, Valencia, CA). Quantitative PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). At the end of each PCR run, a melt curve analysis was performed to ensure that only a single product was amplified. Three technical replicates were used for each sample. A standard curve for each gene (with the exception of Sox9b) was generated by serially diluting plasmids containing a full-length copy of each gene. Total molecule numbers were calculated for each sample and normalized by a β-actin correction factor. Changes in expression are reported as changes in fold induction by normalizing molecule numbers to the appropriate control. No plasmid template was available for Sox9b, so relative expression was calculated as the fold change compared with the Ctrl MO DMSO treatment according to the following equation (Livak and Schmittgen, 2001): Relative mRNA expression = 2−ΔΔCt; where ΔΔCt = [Ct(Sox9b) − Ct(actin)]treatment − [Ct(Sox9b) − Ct(actin)]control. Real-time PCR primers and the thermocycler protocols for AHRRb, CYP1A, and β-actin are provided in Evans et al., (2005). Real-time PCR primers and the thermocycler protocols for CYP1B1, CYP1C1, and CYP1C2 are provided in Jönsson, Jenny, et al., (2007). Real-time PCR primers for Sox9b are provided in Xiong et al. (2008). The PCR conditions used here for Sox9b were 95°C for 3 min and 95°C for 15 s/62°C for 1 min (40 cycles). Real-time PCR primers for AHRRa are provided in Andreasen et al. (2007). The PCR conditions used here for AHRRa were 95°C for 3 min and 95°C for 15 s/64°C for 1 min (40 cycles).
Statistical Analysis
All statistical analyses were performed with the Prism 5 software package (GraphPad Software Inc., San Diego, CA). Data were logarithmically transformed as needed to improve equality of variances. The differences among transcript levels determined by real-time RT-PCR or quantitative changes in phenotype (pericardial edema) in response to TCDD were evaluated using one-way analysis of variance (ANOVA, p value < 0.05), followed by Dunnett's Multiple Comparison test. Multifactor analyses for determining combined effects of gene knockdown and TCDD exposure were evaluated using two-way ANOVA (p value < 0.05), followed by Bonferroni post-tests. Specific analyses are described in the Results section and figure legends. All experiments used for the manuscript were repeated at least twice to confirm the observations. Each experiment in which real-time RT-PCR was performed consisted of 3 replicates of 20 pooled embryos. The statistical analysis was consistent between replicated experiments in all cases; however, data from only one representative experiment are presented.
RESULTS
Expression of Zebrafish AHRRa and AHRRb during Development
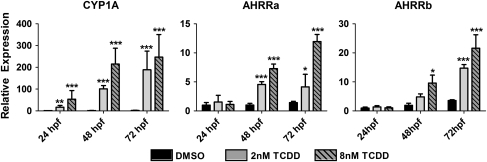
To assess the constitutive expression of AHRRa and AHRRb during development and determine their inducibility by TCDD, embryos were exposed to 0.1% DMSO, 2nM TCDD, or 8nM TCDD at 6 hpf and sampled at 24, 48, and 72 hpf for measurement of AHRR expression by real-time RT-PCR (Fig. 1). To assess the efficacy of the TCDD exposures, CYP1A expression was also determined. All three transcripts displayed modest increases in constitutive expression (approximately two to threefold) prior to hatching (72 hpf) (Fig. 1). Although CYP1A was induced by 2 and 8nM TCDD at all three time points, TCDD exposure did not affect the expression of AHRRa or AHRRb at 24 hpf. However, both AHRRa and AHRRb were responsive to TCDD by 48 and 72 hpf (Fig. 1). At 48 hpf, AHRRa was induced 4.6-fold and 7.3-fold by 2 and 8nM TCDD, respectively (Supplementary Table 1 provides fold induction data, i.e., values for TCDD-treated fish normalized to DMSO controls at each time point). At 72 hpf, AHRRa was induced 2.9-fold and 8.3-fold by 2 and 8nM TCDD, respectively. AHRRb was only induced by 8nM TCDD (5.1-fold) at 48 hpf, whereas at 72 hpf, both 2 and 8nM TCDD significantly induced AHRRb expression by 4.1- and 6.0-fold, respectively. In comparison, the induction of CYP1A by TCDD ranged between 16.8-fold and 180-fold during this developmental period (Supplementary Table 1).
FIG. 1.
Developmental time course of expression of AHR signaling pathway components in response to TCDD exposure. Real-time RT-PCR was used to quantify transcript expression in whole zebrafish embryos during development after exposure to moderate and high concentrations of TCDD. Standard curves derived from plasmid dilutions were used to calculate transcript abundance. To highlight changes in relative gene expression as a result of either changes in developmental time point or TCDD exposure, transcript abundance for each sample (time and exposure) was normalized to the 24 hpf DMSO control. Error bars represent one standard deviation; n = 3 replicates of 20 pooled embryos. Statistically significant difference between DMSO and TCDD at each time point is represented by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001).
Confirmation of Morpholino Target Specificity by Inhibition of In Vitro Translation
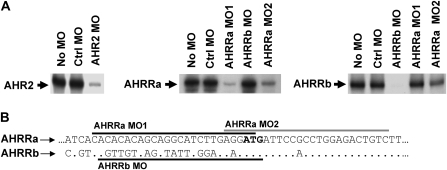
Prior to conducting experiments using MOs to investigate the regulation of AHRRa or AHRRb, we determined the efficacy and specificity of the MOs by assessing the inhibitory effects of the MO on the in vitro translation of the target proteins (Fig. 2A). The AHR2 MO significantly reduced the translation of AHR2 in the in vitro reaction by approximately 98%, similar to previous reports (Prasch et al., 2003). AHRRa MO1 and AHRRa MO2 inhibited AHRRa translation by approximately 97 and 90%, respectively. AHRRa and AHRRb are highly conserved at the N-terminal end of the protein. Thus, the AHRRa MO2, which targets the start codon and significantly extends into the coding sequence of AHRRa, shares 92% identity with the same region of AHRRb (Fig. 2B). Despite the high identity, the two mismatches in AHRRa MO2 are sufficient to significantly reduce the effective inhibition of AHRRb translation (50% inhibition) compared to the efficacy of the other target MOs. In contrast, the AHRRb MO caused greater than 99% inhibition of AHRRb translation in the in vitro reaction.
FIG. 2.
Morpholino specificity and inhibition efficacy confirmed by in vitro translation. (A) In vitro transcription/translation of zebrafish AHR2, AHRRa, and AHRRb in the presence or absence of Ctrl MO, target-specific MO, or nonspecific MO. (B) Schematic of the proximal 5′ UTR of AHRRa and AHRRb highlighting the MO target sequences.
Effect of AHR2 Knockdown on AHRRa and AHRRb Expression and Induction
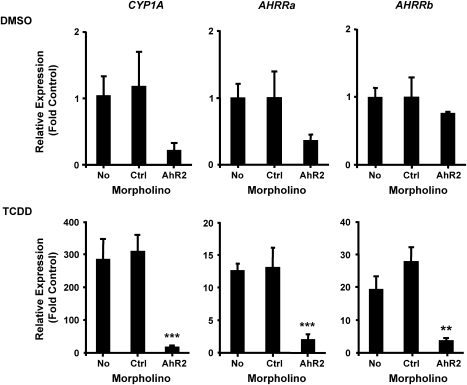
To determine whether the induction of AHRRa and AHRRb by TCDD is AHR2 dependent, embryos were injected with a standard Ctrl MO or the AHR2 MO, exposed to DMSO or 2nM TCDD at 6 hpf, and sampled at 48 hpf for measurement of AHRRa and AHRRb expression by real-time RT-PCR. To confirm effective knockdown of AHR2 signaling, CYP1A expression was also assessed. Knockdown of AHR2 had no significant effect on the basal expression of the three genes. Both the No MO and the Ctrl MO–injected embryos exposed to 2nM of TCDD responded with strong induction of CYP1A, AHRRa, and AHRRb. The AHR2 MO–injected embryos exhibited significantly reduced induction of all three genes: CYP1A (94% reduction), AHRRa (85%), and AHRRb (87%) (Fig. 3). Thus, AHR2 appears to be a major regulator of AHRRa and AHRRb induction by TCDD in zebrafish embryos.
FIG. 3.
Effect of AHR2 knockdown on CYP1A, AHRRa, and AHRRb expression in response to TCDD exposure. AHR2 translation was blocked by an AHR2 MO injected at the two- to four-cell stage in zebrafish embryos. Real-time RT-PCR was used to quantify transcript abundance in whole zebrafish embryos at 48 hpf after exposure to 0.1% DMSO (carrier) or 2nM TCDD. Standard curves derived from plasmid dilutions were used to calculate transcript abundance. Relative fold induction was determined by comparing transcript abundance for each sample to the noninjected (No MO) DMSO control. Error bars represent one standard deviation; n = 3 replicates of 20 pooled embryos. Statistically significant difference in transcript expression compared to Ctrl MO is represented by asterisks (**p < 0.01, ***p < 0.001). Results presented are from one experiment, but data are representative of two separate experiments.
Differential Roles for AHRRa and AHRRb in AHR Signaling
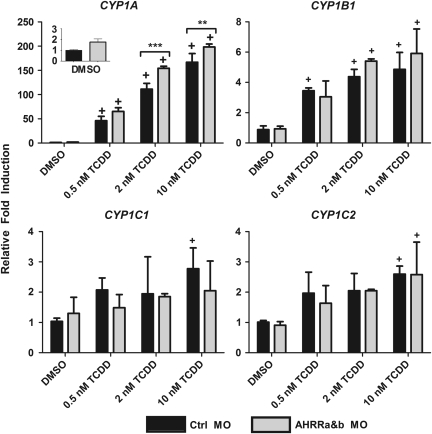
To begin to determine the role of AHRR in regulating AHR signaling, experiments utilizing the ZF-L cell line were used to assess changes in CYP1 expression and inducibility after dual AHRRa and AHRRb knockdown. In addition to measuring expression of CYP1A, we also measured that of other members of the zebrafish CYP1 family (1B1, 1C1, and 1C2). Previous studies have shown that these genes are responsive to AHR agonists and are regulated in an AHR2-dependent manner in vivo (Jönsson, Jenny, et al., 2007; Jönsson, Orrego, et al., 2007), but their expression in ZF-L cells has not been determined. ZF-L cells were transfected with either Ctrl MO or an equal mix of AHRRa MO1 and AHRRb MO and exposed to carrier (0.1% DMSO) or one of three concentrations of TCDD (0.5, 2, or 10nM), chosen to bracket the EC50 value of 1.27nM TCDD for CYP1A induction in ZF-L cells (Evans et al., 2005). In cells treated with Ctrl MO, CYP1A expression was significantly induced at all three concentrations of TCDD (Fig. 4). CYP1B1, CYP1C1, and CYP1C2 also were induced by TCDD, although the degree of induction was moderate (approximately two to fivefold) as compared to the strong induction of CYP1A (∼167-fold). CYP1B1 was significantly induced at all three TCDD concentrations, whereas for CYP1C1 and 1C2, significant induction occurred only at 10nM TCDD. Knockdown of both AHRRs resulted in enhanced CYP1A induction, with a statistically significant, 19–39% increase in induction at 2 and 10nM TCDD (Fig. 4). In contrast to the results for CYP1A, the induction of CYP1B1, CYP1C1, and CYP1C2 was not enhanced by the dual AHRR MO treatment (Fig. 4).
FIG. 4.
Enhancement of TCDD-induced expression of endogenous CYP1A by dual AHRRa/AHRRb MO knockdown in ZF-L cells. ZF-L cells were cotreated with Endo-Porter (MO transfection agent) and MOs targeting AHRRa and AHRRb for 48 h and then exposed to 0.1% DMSO or various concentrations of TCDD for 24 h. Real-time RT-PCR was used to quantify CYP1 transcript expression. Standard curves derived from plasmid dilutions were used to calculate transcript abundance. Relative fold induction was determined by comparing transcript abundance for each sample to the Ctrl MO DMSO. Error bars represent one standard deviation; n = 3 replicates of 6 pooled wells of cells from a 48-well plate. Statistically significant difference in CYP1 induction compared to DMSO for each individual MO treatment is represented by (+) sign (p < 0.05). Statistically significant difference in CYP1 induction between Ctrl MO and AHRRa MO1/AHRRb MO is represented by asterisks (**p < 0.01, ***p < 0.001).
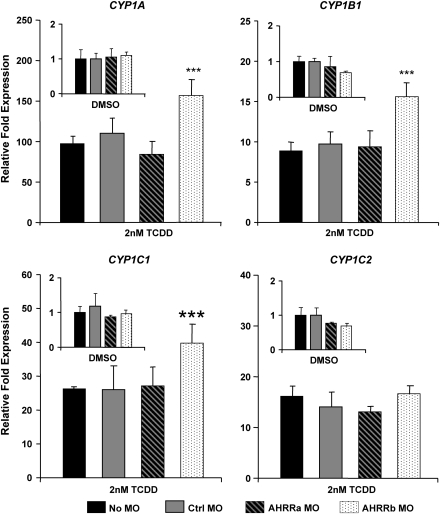
Following the initial results in ZF-L cells, we performed a series of experiments with zebrafish embryos to determine if dual AHRR knockdown would enhance CYP1 expression in vivo. Zebrafish embryos were microinjected with either Ctrl MO or an equal mixture of AHRRa MO1 and AHRRb MO and exposed to 0.1% DMSO or 2nM TCDD for 1 h at 6 hpf. In initial experiments in which CYP1A expression was measured at 48 hpf by real-time RT-PCR, there was no significant enhancement of induction (Supplementary Fig. 1A). Two additional experiments were performed with both the Ctrl MO and an equal mix of AHRRa MO1 and AHRRb MO, with sampling at 72 hpf after exposure to 0.1% DMSO, 2, or 8nM TCDD. CYP1A induction was significantly enhanced (48–51%) by the dual AHRR knockdown at this time point at both concentrations of TCDD (Supplementary Fig. 1B). To determine if individual AHRR knockdowns had differential effects on CYP1 expression, embryos were injected with the Ctrl MO, AHRRa MO1, or AHRRb MO and exposed to 0.1% DMSO or 2nM TCDD, and CYP1 expression was assessed by real-time RT-PCR at 72 hpf. All four CYP1 transcripts (1A, 1B1, 1C1, and 1C2) were strongly induced by 2nM TCDD compared to DMSO treatments (Fig. 5). AHRRb knockdown caused an approximately 50% enhancement of CYP1A, 1B1, and 1C1 induction, whereas AHRRa knockdown had no effect on expression or induction of any CYP1 genes (Fig. 5). CYP1C2 induction was not affected by either AHRR MO. When compared in the same experiment, coinjection of AHRRa MO1 and AHRRb MO enhanced CYP1A induction to the same extent as the individual AHRRb knockdown (Supplementary Fig. 1C), suggesting that enhanced CYP1 induction was primarily caused by knockdown of AHRRb.
FIG. 5.
Enhancement of TCDD-induced expression of endogenous CYP1 transcripts by individual AHRRa and AHRRb knockdowns in embryos. AHRRa or AHRRb translation was blocked by MO injection at the two- to four-cell stage in zebrafish embryos. Real-time RT-PCR was used to quantify CYP1 transcript abundance in whole zebrafish embryos at 72 hpf after exposure to 0.1% DMSO (carrier) or 2nM TCDD. Standard curves derived from plasmid dilutions were used to calculate transcript abundance. Relative fold expression was determined by comparing transcript abundance for each sample to the noninjected (No MO) DMSO control. Error bars represent one standard deviation; n = 3 replicates of 20 pooled embryos. Statistically significant difference in transcript expression compared to Ctrl MO is represented by asterisks (***p < 0.001). Results presented are from one experiment, but data are representative of two separate experiments.
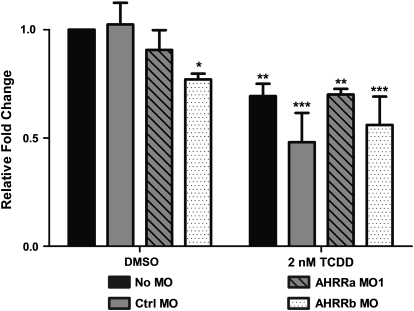
Previous studies have suggested a role for Sox9b in mediating the TCDD-induced developmental phenotypes, including craniofacial malformations (Andreasen et al., 2006; Xiong et al., 2008). In those studies, TCDD caused decreased expression of Sox9b in developing jaw and regenerating fins. To determine if AHRR knockdown would affect Sox9b expression or its repression by TCDD at the level of the whole embryo, embryos were microinjected with a Ctrl MO or individual AHRR MOs, exposed to 0.1% DMSO or 2nM TCDD, and sampled at 72 hpf for assessment of Sox9b expression by real-time RT-PCR. In both No MO and Ctrl MO–injected embryos, TCDD exposure caused a significant reduction in Sox9b expression (Fig. 6). While AHRRa MO1 had no statistically significant effect on Sox9b expression in DMSO-treated embryos, AHRRb knockdown caused a significant decrease in Sox9b mRNA in the absence of TCDD (Fig. 6). However, neither AHRRa knockdown nor AHRRb knockdown had any effect on the TCDD-induced repression of Sox9b expression in whole embryos (Fig. 6).
FIG. 6.
Sox9b expression after AHRR knockdown or TCDD exposure. AHRRa or AHRRb translation was blocked by MO injection at the two- to four-cell stage in zebrafish embryos. Real-time RT-PCR was used to quantify Sox9b expression in whole zebrafish embryos at 72 hpf after exposure to 0.1% DMSO (carrier) or 2nM TCDD. Relative fold change was calculated by the 2−ΔΔCt method by comparison to the Ctrl MO DMSO treatment. Error bars represent one standard deviation; n = 3 replicates of 20 pooled embryos. Statistically significant difference in transcript expression compared to Ctrl MO DMSO is represented by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001).
Role of Constitutive AHRR Expression in Development
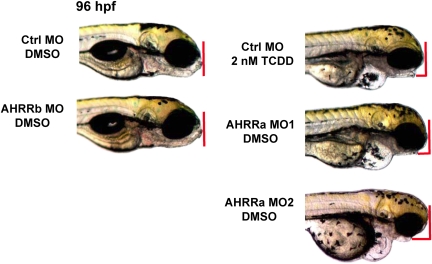
Observations from the AHRR MO experiments suggested that AHRRa knockdown alone (i.e., in the absence of TCDD exposure) could reproduce several of the developmental phenotypes associated with TCDD exposure in embryonic fish. Injection of AHRRa MO1 without subsequent exposure to TCDD caused pericardial edema, yolk sac edema, craniofacial malformations, and cardiac abnormalities (Fig. 7). In contrast to the results seen with AHRRa MO1, AHRRb knockdown had no observable effects on development under control conditions (0.1% DMSO exposure).
FIG. 7.
Recapitulation of TCDD-like developmental phenotypes by AHRRa knockdown in zebrafish embryos. AHRRa or AHRRb translation was blocked by MO injection at the two- to four-cell stage in zebrafish embryos. Common TCDD-induced developmental phenotypes, such as pericardial edema, craniofacial malformations, and cardiac deformities, were phenocopied by AHRRa knockdown in DMSO-treated embryos. Phenotype occurrence was confirmed by microinjection of two separate AHRRa MOs. In contrast, embryos microinjected with AHRRb MO did not display any abnormal developmental phenotypes in DMSO-treated embryos. Results presented are representative of at least three separate experiments.
To assess the specificity of the phenotypes caused by AHRRa knockdown, several attempts were made to rescue them by coinjecting in vitro–transcribed capped mRNA (200 and 400 pg of mRNA) for AHRRa. The coding region of AHRRa was cloned into the pT7TS vector (provided by Krieg PA, University of Arizona) to serve as a template for in vitro transcription of the capped mRNA. The pT7TS vector replaces the 5′ and 3′ UTRs of AHRRa with the UTRs of the Xenopus β-globin transcript, effectively removing the AHRRa MO1 target site from the exogenous AHRRa mRNA. In vitro translation reactions (TNT reactions) were used to confirm that replacement of the 5′ UTR abolishes AHRRa MO1 targeting (Supplementary Fig. 2A). Coinjection of AHRRa mRNA did not block the effect of AHRR knockdown (data not shown). To determine whether exogenous AHRRa protein was expressed in embryos, Western blots were performed on 24 and 48 hpf embryos that were injected with 200 pg of AHRRa mRNA. Additional embryos were microinjected with 100 pg of enhanced green fluorescent protein (eGFP) mRNA to serve as a positive control in which the eGFP protein could be detected by fluorescence microscopy or Western blot using an eGFP-specific antibody. Although eGFP was detectable by Western blot in embryos at 24 and 48 hpf, AHRRa was not detected at either time point (Supplementary Fig. 2B). To further assess the expression of exogenous AHRRa, a construct encoding an AHRRa-eGFP fusion protein (Evans et al., 2008) was used as template to transcribe mRNA. Embryos (one to two cells) were microinjected with either 400 pg of AHRRa-eGFP mRNA or 100 pg of eGFP mRNA. Fluorescence microscopy was used to measure the expression of eGFP or AHRRa-eGFP. At 5 hpf, eGFP fluorescence was detectable by microscopy in both injection groups; however, by 24 hpf, eGFP was no longer detectable in the AHRRa-eGFP group (Supplementary Fig. 2C). In contrast, embryos microinjected with just eGFP mRNA had eGFP expression detectable beyond 24 hpf by fluorescence microscopy. These observations suggest that exogenous AHRRa mRNA or protein has an extremely fast turnover rate in zebrafish embryos, consistent with previous studies suggesting that AHR/ARNT signaling is regulated by a labile repressor (Ma, 2002; Ma and Baldwin, 2002; Ou and Ramos, 1995). Thus, it was not possible to rescue the phenotypes caused by AHRRa knockdown.
As an alternative method (Ekker, 2004) for assessing the specificity of the phenotype observed in AHRRa morphants, we investigated whether a second MO primarily targeting AHRRa (AHRRa MO2) could reproduce the effects of AHRRa MO1. AHRRa MO2 was nearly as effective as AHRRa MO1 at blocking translation of AHRRa (Fig. 2). AHRRa knockdown by AHRRa MO2 caused the same developmental phenotypes seen in AHRRa MO1–injected embryos (Fig. 7). The transcript region targeted by AHRRa MO2 shares strong identity between AHRRa and AHRRb, and the in vitro translation assay suggests that AHRRa MO2 will partially inhibit AHRRb translation. Therefore, to confirm that partial loss of AHRRb played no role in producing the phenotypes, we compared embryos microinjected with AHRRa MO2 and those microinjected with an equal mixture of AHRRa MO1 and AHRRb MO. The dual AHRRa MO1 and AHRRb MO knockdown did not cause any differences in phenotype compared to the AHRRa MO1 or AHRRa MO2 phenotypes (data not shown) in DMSO-exposed embryos. Thus, the phenotypes observed appear to be the result of reduced AHRRa expression.
Role of AHRR in Mediation of TCDD Toxicity
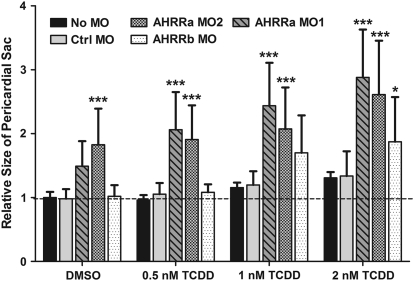
The experiments above suggested that AHRRa regulates endogenous AHR activity, whereas AHRRb modulates changes in CYP1 gene expression following TCDD exposure. To determine the individual roles of AHRRa and AHRRb in mediating TCDD toxicity, embryos were microinjected with a Ctrl MO or individual AHRR MOs and exposed to 0.1% DMSO or one of three concentrations of TCDD (0.5, 1, or 2nM). An increase in pericardial edema, determined by quantification of the size of the pericardial sac at 72 hpf, was used as a measure of the severity of TCDD toxicity. One-way ANOVAs were performed to determine the effect of TCDD treatment on the size of the pericardial sac for each MO treatment. Dunnett's pairwise comparison (p < 0.05) to the DMSO control was used to determine which TCDD doses caused a significant increase in pericardial edema. All No MO and MO-injected embryos displayed TCDD dose-dependent increases in pericardial edema (Fig. 8). The No MO, AHRRb MO–injected, and AHRRa MO1–injected embryos had a significant increase in pericardial edema at both 1 and 2nM TCDD. The Ctrl MO– and AHRRa MO2–injected embryos had a significant increase in pericardial edema only at 2nM TCDD (Supplementary Table 2). A two-way ANOVA, using Bonferroni post-tests to compare all treatments to the Ctrl MO treatment, was performed to determine if the combination of specific MO treatments and TCDD exposure enhanced the severity of pericardial edema. Although both AHRRa MOs caused pericardial edema in the DMSO control, the increase was statistically significant only for AHRRa MO2. However, both AHRRa MOs caused a significant enhancement in pericardial edema at all three TCDD doses compared to the No MO and Ctrl MO–injected embryos. In contrast, AHRRb MO caused significant enhancement of pericardial edema only at 2nM TCDD (Fig. 8).
FIG. 8.
PSS in embryos subjected to AHRR knockdown and exposed to DMSO or TCDD. AHRRa or AHRRb translation was blocked by MO injection at the two- to four-cell stage in zebrafish embryos. Image analysis was used to quantify PSS in 72 hpf embryos after exposure to 0.1% DMSO (carrier) or various concentrations of TCDD. PSS was normalized to the average sac size of noninjected, DMSO-treated embryos and expressed as a ratio of individual/mean of control. Error bars represent one standard deviation; n = 10 individual embryos. A two-way ANOVA was performed to determine the significance of combined TCDD exposure and MO knockdown on the severity of pericardial edema measured by changes in PSS. Statistically significant differences in relative PSS compared to Ctrl MO at each TCDD concentration are represented by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001). Results presented are representative of two separate experiments.
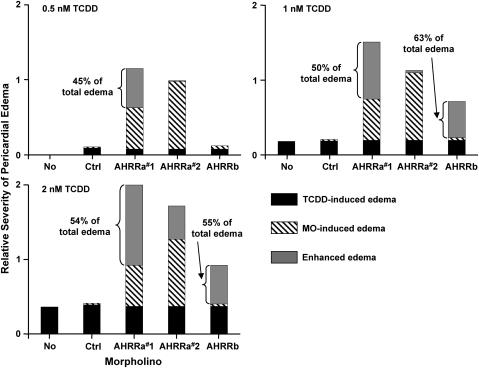
To more clearly illustrate the ability of AHRR knockdown to enhance TCDD toxicity, the amount of pericardial edema (defined as the increase in pericardial sac size [PSS] over the average control value) was divided among specific causative factors (MO injection and TCDD exposure). MO-induced effects on pericardial edema were determined by comparing the average PSS of No MO embryos to the average PSS of Ctrl MO or AHRR MO–injected embryos. Assessments of TCDD-induced pericardial edema in No MO embryos were based on comparison to control embryos (No MO and DMSO treated). Assessments of TCDD-specific pericardial edema for the various AHRR MO treatments were based on the average increase in PSS compared to TCDD-exposed embryos microinjected with Ctrl MO. The sum of these two causative factors was then subtracted from the total amount of pericardial edema in embryos treated with both AHRR MO and TCDD to determine the amount of edema that represents an enhancement (i.e., greater than additive increase) as a result of combined AHRR knockdown and TCDD exposure. The data showing the relative influence of each causative factor on pericardial edema are shown in Figure 9. At the 0.5nM concentration, TCDD exposure alone had negligible effects on pericardial edema in the No MO embryos or any of the MO-injected treatments. However, the two higher doses of TCDD (1 and 2nM) resulted in pericardial edema (20 and 36% increase in PSS), independent of MO treatment. AHRRa MO1 and MO2 knockdowns resulted in pericardial edema (55 and 90% increase in PSS) in the absence of TCDD exposure. In contrast, Ctrl MO and AHRRb MO had negligible effects on pericardial edema. After accounting for TCDD- or MO-specific effects on pericardial edema, it is evident that knockdown of either AHRRa or AHRRb is capable of enhancing the ability of TCDD to cause embryotoxicity, measured as pericardial edema. AHRRa MO1 had the most significant impact on TCDD-induced toxicity; 45, 50, and 54% of the pericardial edema in embryos exposed to TCDD (0.5, 1, and 2nM, respectively) were due to the interaction between TCDD and MO treatment. The AHRRa MO2 produced significant pericardial edema on its own without enhancing the edema caused by 0.5 or 1nM TCDD (Fig. 9). However, in embryos injected with AHRRa MO2 and exposed to 2nM TCDD, 26% of the pericardial edema was due to an interactive effect. AHRRb knockdown did not have a significant effect on pericardial edema in DMSO- or 0.5nM TCDD-treated embryos. However, AHRRb knockdown did enhance TCDD-induced toxicity in the 1 and 2nM TCDD exposures as evidenced by increases in pericardial edema (63 and 55%, respectively) that could not be accounted for by TCDD alone.
FIG. 9.
Enhancement of TCDD toxicity by AHRR knockdown as determined by severity of pericardial edema. Assessment of TCDD-induced pericardial edema for the various AHRR MO treatments are assumed to be the same as the average increase in PSS from TCDD-exposed embryos microinjected with Ctrl MO. Assessments of TCDD-induced pericardial edema in noninjected embryos (No MO) were based on comparison to control embryos (noninjected and DMSO treated). MO-induced changes in PSS were determined from DMSO-treated embryos. The sum of these two causative factors (MO and TCDD treatment) was used to determine the magnitude of TCDD-induced edema that is specifically enhanced (i.e., more than additive) as a result of AHRR knockdown. AHRRa knockdown alone resulted in significant pericardial edema that was further enhanced by TCDD exposure. In contrast, AHRRb knockdown alone caused minimal pericardial edema in control embryos but significantly enhanced the TCDD-induced pericardial edema.
Because of the partial AHRRb knockdown by AHRRa MO2, a comparison between AHRRa MO1, AHRRb MO, and dual microinjection of AHRRa MO1 and AHRRb MO was performed to determine if the phenotypic changes associated with AHRRa knockdown were enhanced by coinhibition of AHRRb. There were no differences in phenotype severity between embryos injected with AHRRa MO1 and those injected with AHRRa MO1 and AHRRb MO together (Supplementary Fig. 3). Thus, the results of AHRRa MO2 can be considered specific for AHRRa knockdowns even though there may be minor inhibition of AHRRb translation.
To track the development of pericardial edema over time in embryos injected with MOs and exposed to TCDD, additional experiments were performed with a single TCDD concentration. Because of the severe morbidity and higher incidence of mortality at later time points in preliminary studies in which embryos were subjected to AHRR knockdown and TCDD concentrations of 2nM (data not shown), a TCDD concentration of 0.5nM was used for exposure. Embryos were microinjected with a Ctrl MO or individual AHRR MOs and exposed to 0.1% DMSO or 0.5nM TCDD; pericardial sac measurements were taken at 72, 96, and 120 hpf and used to calculate the mean PSS and the prevalence of pericardial edema (percentage of fish exhibiting PSS > mean PSS + 2 × standard deviation). There was no increase in PSS at any time point in either the DMSO or 0.5nM TCDD treatments in No MO embryos or embryos microinjected with Ctrl MO or AHRRb MO (Supplementary Fig. 4). In contrast, both AHRRa MOs caused a progressive increase in pericardial edema in both the DMSO and 0.5nM TCDD exposure groups compared to Ctrl MO treatments at each respective time point (Supplementary Fig. 4). There were no significant differences in pericardial edema between the DMSO and 0.5nM TCDD exposure groups in the AHRRa MO1 or AHRRa MO2 treatments at any time point. The prevalence of pericardial edema in the No MO embryos or embryos injected with Ctrl MO or AHRRb MO remained at 10% or below over the course of the experiment in both the DMSO and 0.5nM TCDD exposure groups (Supplementary Fig. 5). In contrast, both AHRRa MOs caused pericardial edema in 50–100% of the embryos regardless of TCDD exposure.
DISCUSSION
The AHRR is an AHR-regulated protein that forms a negative feedback loop with the AHR by repressing AHR signaling. Most of the current knowledge of AHRR function comes from in vitro studies used to characterize AHRR expression or the role of this protein in transcriptional regulation of AHR signaling. Although several of the key components of the AHR pathway, including AHR, ARNT, and ARA9, have been shown to play important roles in basic biological processes, very little is known regarding the role that AHRR plays in development or AHR signaling in vivo. We report here the functional characterization of two AHRR isoforms, AHRRa and AHRRb, in zebrafish embryos. We demonstrate that AHR2 regulates the TCDD-induced expression of both AHRRs and that AHRRa and AHRRb have distinct roles in developing embryos. Our results show that AHRRb represses CYP1 expression in response to TCDD exposure, while AHRRa plays a separate role in controlling constitutive AHR signaling. We also present data suggesting that both AHRRa and AHRRb are able to modulate TCDD toxicity.
AHRR Expression and Regulation by AHR2
Studies in mammals have demonstrated that AHRR is expressed in a variety of organs and is inducible in a tissue-specific manner by different AHR agonists (Bernshausen et al., 2005; Mimura et al., 1999). In a previous study, we demonstrated that zebrafish AHRRa and AHRRb are inducible in embryos by TCDD exposure at 24 hpf (Evans et al., 2005). We performed new dose-response experiments to determine the effects of early (6 hpf) TCDD exposure on AHRR expression (Fig. 1). The timing and magnitude of CYP1A induction in response to TCDD were consistent with previous studies (Andreasen, Spitsbergen, et al., 2002; Jönsson, Jenny, et al., 2007). Although both AHRRa and AHRRb were very responsive to TCDD at the later (48 and 72 hpf) time points, neither AHRR displayed any signs of induction by 24 hpf at either a moderate or high TCDD concentration (Fig. 1). This delay in AHRR induction is contrasted by the strong induction of CYP1A (Andreasen, Spitsbergen, et al., 2002; Prasch et al., 2003), CYP1B1, 1C1, and 1C2 (Jönsson, Jenny, et al., 2007) by various AHR agonists at this earlier developmental time point. Whether such differences in induction kinetics reflect differential RNA turnover or other mechanisms remains to be determined.
AHR-deficient adult mice display a significant decrease (> 100-fold decrease) in constitutive expression of AHRR compared to wild-type animals (Bernshausen et al., 2005), whereas mice overexpressing a constitutively active AHR mutant display increased AHRR transcription (Andersson et al., 2002). In addition, AHR agonists induce the expression of the AHRR, and in vitro promoter assays have demonstrated the functionality of the AHREs found in the proximal promoter of AHRR in a variety of vertebrate species (Baba et al., 2001; Haarmann-Stemmann et al., 2007; Karchner et al., 2002; Mimura et al., 1999). These observations clearly establish a role for AHR in regulating constitutive AHRR expression and support the idea that AHR and AHRR form a regulatory loop. However, the AHR dependence of AHRR induction in vivo has not been demonstrated. Considering the primary role of AHR2 in regulating CYP1 expression in zebrafish (Jönsson, Jenny, et al., 2007; Prasch et al., 2003), we hypothesized that AHR2 might also control the induction of AHRRa and AHRRb. We show here that AHR2 knockdown via MO severely inhibited the TCDD-dependent induction of both AHRRa and AHRRb, providing strong evidence for the role of AHR2 in the regulation of AHRR induction by TCDD in vivo (Fig. 3). Although our data demonstrate that AHR2 is primarily responsible for regulating the TCDD-dependent induction of both zebrafish AHRRs in whole embryos, future studies are required to establish any potential roles for either AHR1a or AHR1b in regulating AHRRa or AHRRb expression in response to other AHR agonists or in a tissue-specific manner.
Distinct Roles of AHRRa and AHRRb in Modulation of CYP1 Gene Expression
Previous studies have demonstrated a regulatory role for AHRR in the constitutive expression of CYP1A1, as well as its induction by AHR agonists (Haarmann-Stemmann et al., 2007; Hosoya et al., 2008). Hosoya et al. (2008) demonstrated that AHRR-deficient mice exhibited enhanced CYP1A1 induction in a tissue-specific manner after treatment with 3-methylcholanthrene. Similarly, AHRR mRNA silencing enhanced the basal expression of CYP1A1 in HeLa cells (Haarmann-Stemmann et al., 2007). Consistent with these observations, we demonstrated that dual AHRRa/AHRRb knockdown enhanced the induction of CYP1A in ZF-L cells (Fig. 4) as well as whole embryos (Supplementary Fig. 1B). Surprisingly, even though zebrafish CYP1A, 1B1, 1C1, and 1C2 have all been shown to be induced by TCDD in an AHR2-dependent manner (Jönsson, Jenny, et al., 2007), only CYP1A induction was significantly enhanced by the dual AHRR knockdown in ZF-L cells (Fig. 4). The results suggest the possibility that AHRR does not repress all AHR target genes or that there are differences between cultured cells and in vivo systems in the ability of AHRR to repress AHR transactivation. Consistent with this, it has long been known that AHR target genes are not necessarily coordinately regulated (Dunn et al., 1988; Fagan et al., 1986; Zhang et al., 2006). Thus, AHRR could contribute to gene-specific differences in responsiveness to TCDD exposure.
Previously, we provided evidence that AHRRa and AHRRb are co-orthologs of the human AHRR, originating as part of the fish-specific whole-genome duplication (Evans et al., 2005). Here, we used zebrafish embryos to assess AHRR function during embryonic development. To determine if the AHRR duplication has led to subfunction partitioning, we performed individual knockdowns of AHRRa and AHRRb and assessed the effects on CYP1 induction in whole embryos. AHRRa knockdown did not have a significant effect on CYP1 expression or inducibility, whereas AHRRb knockdown caused a significant enhancement (∼50% increase) in CYP1A, 1B1, and 1C1 induction by TCDD (Fig. 5). These observations were somewhat unexpected because in vitro promoter experiments showed that both AHRRa and AHRRb are capable of repressing AHR2-dependent transactivation to a similar extent (Evans et al., 2005). Despite the evidence for conservation of repressor function in vitro, AHRRa and AHRRb appear to differ in their roles in controlling AHR2 signaling in response to TCDD in vivo. However, the most striking observation from the individual AHRR knockdowns was that, in the absence of TCDD exposure, AHRRa morphants displayed several of the classic signs of TCDD embryotoxicity, such as pericardial edema, craniofacial malformations, and cardiac abnormalities (Fig. 7). In contrast, AHRRb morphants showed no sign of any developmental abnormalities. Together, these results suggest that AHRRa and AHRRb have distinct functions in the zebrafish embryo that may reflect partitioning of subfunctions of the putative single AHRR that existed in the most recent common ancestor of mammals and fish.
Some of the apparent functional differences between AHRRa and AHRRb may be due to tissue-specific expression of one or both repressors; whether these AHRRs are expressed in a tissue-specific manner during zebrafish development is not yet known. However, it is possible that tissue specificity alone may not explain all the differences in developmental phenotype or enhanced CYP1 induction. Even when coexpressed in vivo, paralogous proteins may exhibit distinct functions. For example, ARNT2 is generally coexpressed with AHR2 in zebrafish embryos (Andreasen, Spitsbergen, et al., 2002) and is more highly expressed than ARNT1 in whole embryos (Prasch et al., 2006). Despite this and although ARNT2 is capable of partnering with AHR2 to drive the transcription of reporter constructs in vitro (Prasch et al., 2006), it is ARNT1 rather than ARNT2 that supports AHR2-dependent induction of CYP1A and the TCDD-induced developmental phenotypes in vivo (Prasch, Heideman, et al., 2004; Prasch et al., 2006). The functional differences between AHRR paralogs identified in the present manuscript resemble those described for ARNT1 and ARNT2. AHRRa has a higher transcript abundance in whole embryos compared to AHRRb levels (Evans et al., 2005; data not shown) but does not appear to play a role in regulating the induced expression of CYP1 genes in response to TCDD. In contrast to these in vivo observations, both AHRRa and AHRRb are capable of repressing AHR2 transactivation in transient transfection assays (Evans et al., 2005), indicating conserved functional capabilities. These observations highlight the need for comprehensive in situ hybridization and immunohistochemistry experiments to determine the tissue specificity of the AHRs and AHRRs in zebrafish embryos. The results also demonstrate that in vitro assays do not always predict in vivo functionality.
Distinct Roles of AHRRa and AHRRb in Development
The AHR regulates basic biological processes including cellular differentiation, cell cycle progression, and apoptosis (reviewed in Hahn et al., 2009; Ma et al., 2009). The role of AHR in these processes may involve cross-talk between AHR and variety of other cell signaling proteins, including estrogen receptor, hypoxia-inducible factors (HIFs), RelA NF-κB subunit, peroxisome proliferator–activated receptors, and Wnt/β-catenin signaling pathways (Fallone et al., 2005; Hanlon et al., 2003; Kharat and Saatcioglu, 1996; Lee et al., 2006; Mathew et al., 2008; Prasch, Andreasen, et al., 2004; Vogel et al., 2007; Zhang and Walker, 2007). Virtually, nothing is known regarding the role of AHRR in any of these processes, but recent studies suggest that AHRR, like AHR, interacts with other signaling systems. For example, Kanno et al. (2008) showed that overexpression of human AHRR in breast cancer–derived cells repressed both endogenous expression of estrogen-responsive genes and estradiol-stimulated cell proliferation (Kanno et al., 2008). Additional studies from our laboratory have demonstrated that human AHRR is capable of repressing HIF-dependent signaling (Karchner et al., 2009). These two studies show that AHRR plays a broader role in transcriptional regulation that extends beyond its role as a repressor of AHR.
The results presented here suggest a potential role of AHRR also in regulating the Wnt/β-catenin signaling pathway, as demonstrated by the suppression of Sox9b expression by AHRRb (Fig. 6). Previous studies have identified Sox9b as a gene that is repressed in some tissues by TCDD exposure (Andreasen et al., 2006; Xiong et al., 2008) and plays a role in mediating the craniofacial malformations associated with TCDD developmental toxicity in fish (Xiong et al., 2008). In the present study, TCDD exposure suppressed Sox9b expression in whole embryos (Fig. 6). However, neither the AHRRa MO nor the AHRRb MO affected Sox9b suppression by TCDD. With the prevalence of pericardial edema and craniofacial malformations in AHRRa morphants, we were surprised that AHRRa knockdown did not affect Sox9b expression in DMSO-treated embryos, whereas AHRRb knockdown caused a significant decrease in Sox9b expression (Fig. 6). As previously mentioned (Andreasen et al., 2006; Xiong et al., 2008), the Sox9b morphants (Yan et al., 2005) display phenotypes very similar to those induced by TCDD. The phenotype of Sox9b morphants is also similar to that of AHRRa morphants. Thus, it is possible that AHRRa regulates Sox9b in a tissue-specific manner to cause the developmental phenotypes. However, our studies do not directly address the possible role of Sox9b in the phenotypes of AHRRa morphants. Nevertheless, the decrease in constitutive expression of Sox9b and enhanced CYP1A induction in response to TCDD in AHRRb morphants may suggest a role for AHRRb in modulating both positive and negative regulation of transcription by AHR.
With respect to aromatic hydrocarbon exposure, the AHR plays a dual role in regulating the enzymatic metabolism of these compounds and mediating their toxic effects (Prasch et al., 2003; Walisser et al., 2005). However, in the absence of toxicant exposure, AHR and several components of the AHR signaling pathway also play important roles in development, especially cardiovascular development (Kozak et al., 1997; Lahvis et al., 2000; Lin et al., 2007). Given these observations, it is reasonable to suggest that AHRR may also play a role both in basic biological processes and in the response to toxicants.
It is currently thought that many of the toxic end points induced by TCDD or related pollutants may be the result of hyperactivation of the AHR resulting in misregulation of target genes involved in basic biological processes. Consistent with this idea, the recapitulation of TCDD-like developmental phenotypes by AHRRa knockdown in DMSO-treated embryos (Fig. 7) suggests that AHR is constitutively active during development and that AHRRa has an important role in controlling that activity. The observation that AHRRa knockdown has no effect on CYP1A expression but can phenocopy TCDD toxicity, presumably by hyperactivating the AHR, is consistent with the idea that at least some TCDD-induced phenotypes are independent of CYP1A expression (Carney et al., 2004).
Distinct Roles of AHRRa and AHRRb in Enhancement of TCDD Toxicity
We have presented data that suggest separate roles for AHRRa and AHRRb in preventing TCDD-like developmental defects that may be caused by constitutive activation of the AHR or regulating AHR signaling by altering the expression of AHR-responsive genes (CYP1 and Sox9b), respectively. To determine whether AHRR knockdown affects TCDD-induced toxicity, we monitored the severity of pericardial edema in AHRRa and AHRRb morphants after exposure to a series of TCDD concentrations. Knockdown of AHRRa or of AHRRb enhanced the toxic effects of TCDD exposure in vivo (Figs. 8 and 9; Supplementary Table 2). Interestingly, the opposite relationship was observed in AHRR-deficient mice, which displayed some resistance to B[a]P-induced chemical carcinogenesis, presumably by “superinduction” of CYP1A1 and accelerating the rate of B[a]P clearance (Hosoya et al., 2008). However, this difference in observations is likely due to the different end points measured as well as the metabolic differences between these two AHR agonists, including the well-established persistence of TCDD even in the face of induced CYP1A.
CONCLUSION
The results presented here provide new information concerning the regulation and function of both isoforms of AHRR (AHRRa and AHRRb) in zebrafish. Using antisense MOs, we demonstrate an AHR2-mediated role in the induction of both AHRRs by TCDD. We also demonstrate separate roles for AHRRa and AHRRb in regulating developmental processes, AHR signaling in response to TCDD exposure, and TCDD-induced embryotoxicity. AHRRa appears to play a primary role in regulating constitutive AHR signaling during development, whereas AHRRb regulates the TCDD-induced expression of CYP1 genes. However, both AHRRs play a role in modulating TCDD embryotoxicity. Our results suggest that the zebrafish AHRRs may have partitioned the multiple functions of the single AHRR found in mammals and thus will be useful in future studies to further elucidate AHRR function in vivo.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Health grants (R01ES006272 to M.E.H., R01ES015912 to J.J.S., K99ES017044-01 to M.J.J.); Superfund Basic Research Program (P42ES007381).
Acknowledgments
The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
References
- Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang Y-L, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Andersson P, McGuire J, Rubio C, Gradin K, Whitelaw ML, Pettersson S, Hanberg A, Poellinger L. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9990–9995. doi: 10.1073/pnas.152706299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen EA, Hahn ME, Heideman W, Peterson RE, Tanguay RL. The zebrafish (Danio rerio) aryl hydrocarbon receptor type 1 is a novel vertebrate receptor. Mol. Pharmacol. 2002;62:234–249. doi: 10.1124/mol.62.2.234. [DOI] [PubMed] [Google Scholar]
- Andreasen EA, Mathew LK, Lohr CV, Hasson R, Tanguay RL. Aryl hydrocarbon receptor activation impairs extracellular matrix remodeling during zebrafish fin regeneration. Toxicol. Sci. 2007;95:215–226. doi: 10.1093/toxsci/kfl119. [DOI] [PubMed] [Google Scholar]
- Andreasen EA, Mathew LK, Tanguay RL. Regenerative growth is impacted by TCDD: Gene expression analysis reveals extracellular matrix modulation. Toxicol. Sci. 2006;92:254–269. doi: 10.1093/toxsci/kfj118. [DOI] [PubMed] [Google Scholar]
- Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: Effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol. Sci. 2002;68:403–419. doi: 10.1093/toxsci/68.2.403. [DOI] [PubMed] [Google Scholar]
- Baba T, Mimura J, Gradin K, Kuroiwa A, Watanabe T, Matsuda Y, Inazawa J, Sogawa K, Fujii-Kuriyama Y. Structure and expression of the Ah receptor repressor gene. J. Biol. Chem. 2001;276:33101–33110. doi: 10.1074/jbc.M011497200. [DOI] [PubMed] [Google Scholar]
- Bernshausen T, Jux B, Esser C, Abel J, Fritsche E. Tissue distribution and function of the aryl hydrocarbon receptor repressor (AhRR) in C57BL/6 and aryl hydrocarbon receptor deficient mice. Arch. Toxicol. 2005;80:206–211. doi: 10.1007/s00204-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, Bradfield CA. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- Carney SA, Peterson RE, Heideman W. 2,3,7,8-Tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol. Pharmacol. 2004;66:512–521. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Carney SA, Prasch AL, Heideman W, Peterson RE. Understanding dioxin developmental toxicity using the zebrafish model. Birth Defects Res. A. 2006;76:7–18. doi: 10.1002/bdra.20216. [DOI] [PubMed] [Google Scholar]
- Dunn TJ, Lindahl R, Pitot HC. Differential gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Noncoordinate regulation of a TCDD-induced aldehyde dehydrogenase and cytochrome P-450c in the rat. J. Biol. Chem. 1988;263:10878–10886. [PubMed] [Google Scholar]
- Ekker SC. Nonconventional antisense in zebrafish for functional genomics applications. Methods Cell Biol. 2004;77:121–136. doi: 10.1016/s0091-679x(04)77007-7. [DOI] [PubMed] [Google Scholar]
- Evans BR, Karchner SI, Allan LL, Pollenz RS, Tanguay RL, Jenny MJ, Sherr DH, Hahn ME. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: Role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 2008;73:387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BR, Karchner SI, Franks DG, Hahn ME. Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: Structure, function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 2005;441:151–167. doi: 10.1016/j.abb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Fagan JB, Pastewka JV, Chalberg SC, Gozukara E, Guengerich FP, Gelboin HV. Noncoordinate regulation of the mRNAs encoding cytochromes P-450 BNF/MC-3 and ISF/BNF-G. Arch. Biochem. Biophys. 1986;244:261–272. doi: 10.1016/0003-9861(86)90116-5. [DOI] [PubMed] [Google Scholar]
- Fallone F, Villard P-H, Decome L, Seree E, Meo M d, Chacon C, Durand A, Barra Y, Lacarelle B. PPARalpha activation potentiates AhR-induced CYP1A1 expression. Toxicology. 2005;216:122–128. doi: 10.1016/j.tox.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T, Abel J. The aryl hydrocarbon receptor repressor (AhRR): Structure, expression, and function. Biol. Chem. 2006;387:1195–1199. doi: 10.1515/BC.2006.147. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T, Bothe H, Kohli A, Sydlik U, Abel J, Fritsche E. Analysis of the transcriptional regulation and molecular function of the aryl hydrocarbon receptor repressor in human cell lines. Drug Metab. Dispos. 2007;35:2262–2269. doi: 10.1124/dmd.107.016253. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling: Complex interactions involving the AHR repressor. Biochem. Pharmacol. 2009;77:485–497. doi: 10.1016/j.bcp.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Evans BR, Franks DG, Merson RR, Lapseritis JM. Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: Insights from comparative genomics. J. Exp. Zool. Part A. 2006;305:693–706. doi: 10.1002/jez.a.323. [DOI] [PubMed] [Google Scholar]
- Hanlon PR, Ganem LG, Cho YC, Yamamoto M, Jefcoate CR. AhR- and ERK-dependent pathways function synergistically to mediate 2,3,7,8-tetrachlorodibenzo-p-dioxin suppression of peroxisome proliferator-activated receptor-y1 expression and subsequent adipocyte differentiation. Toxicol. Appl. Pharmacol. 2003;189:11–27. doi: 10.1016/s0041-008x(03)00083-8. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Teraoka H, Heideman W, Peterson R. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 2005;86:6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Harada N, Mimura J, Motohashi H, Takahashi S, Nakajima O, Morita M, Kawauchi S, Yamamoto M, Fujii-Kuriyama Y. Inducibility of cytochrome P450 1A1 and chemical carcinogenesis by benzo[a]pyrene in AhR repressor-deficient mice. Biochem. Biophys. Res. Commun. 2008;365:562–567. doi: 10.1016/j.bbrc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebrafish exposed to 3,3′,4,4′,5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2007;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3′,4,4′,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B, and 1C genes in zebrafish. Toxicol. Appl. Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Takane Y, Takizawa Y, Inouye Y. Suppressive effect of aryl hydrocarbon receptor repressor on transcriptional activity of estrogen receptor alpha by protein-protein interaction in stably and transiently expressing cell lines. Mol. Cell. Endocrinol. 2008;291:87–94. doi: 10.1016/j.mce.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Hahn ME. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: Tandem arrangement of ahr1b and ahr2b genes. Biochem. J. 2005;392:153–161. doi: 10.1042/BJ20050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Powell WH, Hahn ME. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR repressor, AHR1, and AHR2. J. Biol. Chem. 2002;277:6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, Sherr DH, Hahn ME. The active form of human aryl hydrocarbon receptor repressor lacks exon 8 and its Pro185 and Ala185 variants repress both AHR and HIF. Mol. Cell. Biol. 2009;29:3465–3477. doi: 10.1128/MCB.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharat I, Saatcioglu F. Antiestrogenic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin are mediated by direct transcriptional interference with the liganded estrogen receptor. J. Biol. Chem. 1996;271:10533–10537. doi: 10.1074/jbc.271.18.10533. [DOI] [PubMed] [Google Scholar]
- Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev. Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPres JJ, Glover E, Dunham EE, Bunger MK, Bradfield CA. ARA9 modifies agonist signaling through an increase in cytosolic aryl hydrocarbon receptor. J. Biol. Chem. 2000;275:6153–6159. doi: 10.1074/jbc.275.9.6153. [DOI] [PubMed] [Google Scholar]
- Lee K, Burgoon LD, Lamb L, Dere E, Zacharewski TR, Hogenesch JB, LaPres JJ. Identification and characterization of genes susceptible to transcriptional cross-talk between the hypoxia and dioxin signaling cascades. Chem. Res. Toxicol. 2006;19:1284–1293. doi: 10.1021/tx060068d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BC, Nguyen LP, Walisser JA, Bradfield CA. A hypomorphic allele of aryl hydrocarbon receptor-associated protein-9 produces a phenocopy of the Ahr-null mouse. Mol. Pharmacol. 2008;74:1367–1371. doi: 10.1124/mol.108.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BC, Sullivan R, Lee Y, Moran S, Glover E, Bradfield CA. Deletion of the aryl hydrocarbon receptor-associated protein 9 leads to cardiac malformation and embryonic lethality. J. Biol. Chem. 2007;282:35924–35932. doi: 10.1074/jbc.M705471200. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma C, Marlowe JL, Puga A. The aryl hydrocarbon receptor at the crossroads of multiple signaling pathways. EXS. 2009;99:231–257. doi: 10.1007/978-3-7643-8336-7_9. [DOI] [PubMed] [Google Scholar]
- Ma Q. Induction and superinduction of 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible poly(ADP-ribose) polymerase: Role of the aryl hydrocarbon receptor/aryl hydrocarbon receptor translocator transcription activation domains and a labile transcription repressor. Arch. Biochem. Biophys. 2002;404:309–316. doi: 10.1016/s0003-9861(02)00339-9. [DOI] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT. A cycloheximide-sensitive factor regulates TCDD-induced degradation of the aryl hydrocarbon receptor. Chemosphere. 2002;46:1491–1500. doi: 10.1016/s0045-6535(01)00270-3. [DOI] [PubMed] [Google Scholar]
- Mathew LK, Sengupta SS, LaDu J, Andreasen EA, Tanguay RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22:3087–3096. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 1999;13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okey A B. An aryl hydrocarbon receptor odyssey to the shores of toxicology: The Deichmann Lecture, International Congress of Toxicology-XI. Toxicol. Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- Ou X, Ramos KS. Regulation of cytochrome P4501A1 gene expression in vascular smooth muscle cells through aryl hydrocarbon receptor-mediated signal transduction requires a protein synthesis inhibitor. Arch. Biochem. Biophys. 1995;316:116–122. doi: 10.1006/abbi.1995.1017. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan Y-L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Andreasen EA, Peterson RE, Heideman W. Interactions between 2,3,7,8-tetrachlorodibenzon-p-dioxin (TCDD) and hypoxia signaling pathways in zebrafish: Hypoxia decreases responses to TCDD in zebrafish embryos. Toxicol. Sci. 2004;78:68–77. doi: 10.1093/toxsci/kfh053. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Heideman W, Peterson RE. ARNT2 is not required for TCDD developmental toxicity in zebrafish. Toxicol. Sci. 2004;82:250–258. doi: 10.1093/toxsci/kfh235. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Tanguay RL, Mehta V, Heideman W, Peterson RE. Identification of zebrafish ARNT1 homologs: 2,3,7,8-Tetrachlorodibenzo-p-dioxin toxicity in the developing zebrafish requires ARNT1. Mol. Pharmacol. 2006;69:776–787. doi: 10.1124/mol.105.016873. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol. Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Tanguay RL, Abnet CC, Heideman W, Peterson RE. Cloning and characterization of the zebrafish (Danio rerio) aryl hydrocarbon receptor. Biochim. Biophys. Acta. 1999;1444:35–48. doi: 10.1016/s0167-4781(98)00252-8. [DOI] [PubMed] [Google Scholar]
- Tanguay RL, Andreasen E, Heideman W, Peterson RE. Identification and expression of alternatively spliced aryl hydrocarbon nuclear translocator 2 (ARNT2) cDNAs from zebrafish with distinct functions. Biochim. Biophys. Acta. 2000;1494:117–128. doi: 10.1016/s0167-4781(00)00225-6. [DOI] [PubMed] [Google Scholar]
- Tyson-Capper AJ, Europe-Finner GN. Novel targeting of cyclooxygenase-2 (COX-2) pre-mRNA using antisense morpholino oligonucleotides directed to the 3’ acceptor and 5’ donor splice sites of exon 4: Suppression of COX-2 activity in human amnion-derived WISH and myometrial cells. Mol. Pharmacol. 2006;69:796–804. doi: 10.1124/mol.105.020529. [DOI] [PubMed] [Google Scholar]
- Vogel CFA, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol. Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisser JA, Bunger MK, Glover E, Harstad EB, Bradfield CA. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J. Biol. Chem. 2004;279:16326–16331. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor-mediated down-regulation of Sox9b causes jaw malformation in zebrafish embryos. Mol. Pharmacol. 2008;74:1544–1553. doi: 10.1124/mol.108.050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y-L, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: Distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Zhang N, Walker MK. Crosstalk between the aryl hydrocarbon receptor and hypoxia on the constitutive expression of cytochrome P4501A1 mRNA. Cardiovasc. Toxicol. 2007;7:282–290. doi: 10.1007/s12012-007-9007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-Y, Pelletier RD, Wong YN, Sugawara M, Zhao N, Littlefield BA. Preferential inducibility of CYP1A1 and CYP1A2 by TCDD: Differential regulation in primary human hepatocytes versus transformed human cells. Biochem. Biophys. Res. Commun. 2006;341:399–407. doi: 10.1016/j.bbrc.2005.12.203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.