Abstract
Macrophages are an important target cell for infection with cytomegalovirus (CMV). A number of viral genes that either are expressed specifically in this cell type or function to optimize CMV replication in this host cell have now been identified. Among these is the murine CMV (MCMV) US22 gene family member M140, a nonessential early gene whose deletion (RVΔ140) leads to significant impairment in virus replication in differentiated macrophages. We have now determined that the defect in replication is at the stage of viral DNA encapsidation. Although the rate of RVΔ140 genome replication and extent of DNA cleavage were comparable to those for revertant virus, deletion of M140 resulted in a significant reduction in the number of viral capsids in the nucleus, and the viral DNA remained sensitive to DNase treatment. These data are indicative of incomplete virion assembly. Steady-state levels of both the major capsid protein (M86) and tegument protein M25 were reduced in the absence of the M140 protein (pM140). This effect may be related to the localization of pM140 to an aggresome-like, microtubule organizing center-associated structure that is known to target misfolded and overexpressed proteins for degradation. It appears, therefore, that pM140 indirectly influences MCMV capsid formation in differentiated macrophages by regulating the stability of viral structural proteins.
An important feature of cytomegalovirus (CMV) pathogenesis is dissemination of virus to target organs by infected monocytes that harbor CMV DNA (14, 36). Differentiation of these cells into macrophages as they extravasate into tissues triggers production of infectious virus. Infection with human CMV (HCMV) drives monocytes toward a proinflammatory macrophage phenotype with an extended life span (7, 36). This cellular differentiation process is accompanied by numerous physiological changes that could, in theory, negatively affect virus replication. These include increases in reactive oxygen intermediates, phagocytosis, lysosomal compartments, mitochondrial activities, secretory enzymes, and antiviral cytokines, as well as numerous receptors that render these cells highly interactive with the extracellular matrix and immune modulators (7, 14). It is likely, therefore, that CMV encodes products to counteract factors that are adverse to replication or to subvert cellular events to enhance replication. The mouse model has been utilized to identify genes within murine CMV (MCMV) that facilitate replication within differentiated macrophages.
To date, four MCMV genes or gene regions have been identified as determinants of macrophage tropism in that deletion of or insertion within these genes has no impact on virus replication in fibroblasts but significantly depresses replication in macrophages (reviewed in reference 14). Gene M78 encodes a G protein-coupled receptor that facilitates accumulation of immediate-early RNA at low multiplicities (29). The product of gene M45 is virion associated and conveys antiapoptotic functions by binding to RIP1 (4, 21). The US22 gene family member M36 encodes a virion-associated protein that has antiapoptotic function by binding to caspase 8 (24). Finally, the US22 gene family members M139, M140, and M141 optimize MCMV replication in fully differentiated tissue macrophages both in vitro and in vivo (16, 17, 24). Two additional MCMV genes of unknown function are transcribed in infected macrophages but not fibroblasts (38), indicating additional adaptations specific to this target cell.
The M139, M140, and M141 early gene products form at least three stable complexes that colocalize to a perinuclear cis-Golgi region in infected macrophages (18). Sequence analysis of the three genes does not reveal any consensus functional domains, with the exception of a putative nuclear localization sequence within M139 and M141 (15). Deletion of one or more of the three genes does not influence apoptosis (24) or tumor necrosis factor alpha or nitric oxide activities (unpublished data). Therefore, the function of these genes remains unknown. It is apparent that M140 is fundamental to the function(s) of these genes for several reasons. First, the degree of impairment in replication of RVΔ140 is equal to that of a mutant deleted of all three genes (16). Second, pM140 is a component of all three complexes identified: a pM140-pM141 complex and two pM139-pM140-pM141 complexes that differ in size and/or mass (18). Third, pM140 localizes to the nucleus but is redistributed to the cytoplasm when expressed with pM141 (18).
In this study, we took a broad approach to assessing the function of M140 in optimizing MCMV replication in macrophages. The data indicate that impairment in replication of MCMV deleted of M140 is after genome replication but prior to virion assembly. Both the copy numbers and rates of viral genome replication for revertant and mutant virus in macrophages were comparable. However, in the absence of M140, capsids were rarely detected by electron microscopy in the nucleus or cytoplasm. Mutant viral DNA was highly sensitive to DNase, although cleavage of DNA was not compromised. Because cleavage of viral DNA is linked to encapsidation, these data suggest that procapsid stability, rather than formation, was altered by the M140 deletion. In support of this, steady-state levels of the major capsid protein (MCP) and M25 tegument protein in mutant virus-infected cells failed to accumulate to wild-type (WT) levels. The fact that pM140 localized to an aggresome-like structure, which is typically important in regulating viral and cellular protein degradation, implies that pM140 may function to regulate viral protein degradation. These studies indicate that pM140 indirectly regulates virus capsid assembly by regulating the stability of viral structural proteins.
MATERIALS AND METHODS
Cells and viruses.
Murine NIH 3T3 fibroblasts (America Type Culture Collection [ATCC] CRL-1658, Manassas, VA) were propagated in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) supplemented with 10% heat-inactivated bovine calf serum (HyClone Laboratories, Logan, UT) and 1% l-glutamine (GIBCO BRL, Grand Island, NY). IC-21 murine macrophages (ATCC TIB 186) were propagated in RPMI medium (Mediatech) supplemented with 10% heat-inactivated fetal calf serum (GIBCO BRL) and 1% l-glutamine. Primary peritoneal exudate macrophages were prepared from adult BALB/c mice (Jackson Laboratories, Bar Harbor, ME) as previously described (17). The viruses used in these studies were a WT MCMV Smith strain (ATCC VR 194), RVΔ140 (16), and the corresponding revertant virus (16). All virus stocks were propagated in NIH 3T3 fibroblasts and quantified by standard plaque assay as previously described (6). Mock virus preparations were supernatants from uninfected NIH 3T3 fibroblasts. Stocks of virus used for infection of macrophages in most experiments were gradient purified to select for single capsid virions, as previously described (17).
Plasmid construction.
The plasmid expressing a fusion protein of M140 with green fluorescent protein (GFP) under the control of the M140 promoter was generated from pGFPM140 (18). First, a HindIII-BamHI fragment (MCMV bases 195847 to 194371) was cloned into pGFPM140 digested with HindIII and BamHI and designated pGFP140J. Next, an AvrII-to-XbaI fragment (MCMV bases 197547 to 199283) was inserted into the NheI site of pGFP140J just upstream of the GFP start site, to produce pIGFP140J. This construct is missing 19 bp from the MCMV sequence upstream of the M140 start codon.
Real-time reverse transcription-PCR.
IC-21 macrophages were infected with gradient-purified mutant or revertant virus at a multiplicity of infection (MOI) of 10. Total RNA was isolated at 48 h postinfection using TRI reagent (Molecular Research Center, Inc.) and RNeasy columns (Qiagen) with DNase I digestion. The RNA (100 ng) was reverse transcribed and tested using the iScript SYBR green reverse transcription-PCR kit (Bio-Rad, Hercules, CA) according to manufacturer's protocols. Primers used were as follows: for m48.2, 5′-GCTAGATCTCTCTACTTTACTC-3′ and 5′-GTCTTATTGTATGACGGTGG-3′; for M86, 5′-GCTCTTCTCGCTGTACTTGGACA-3′ and 5′-TTCAACAACTTTCGCCTGTACTAC-3′. After a 3-min denaturation at 95°, the following PCR conditions were used: for m48.2, 45 cycles of 95° for 1 min, 44° for 30 s, and 72° for 30 s; for M86, 45 cycles of 95° for 1 min, 57° for 30 s, and 72° for 30 s. Real-time PCRs were performed using an iQ iCycler (Bio-Rad). Differences in expression of m48.2 and M86 between WT and mutant virus-infected IC-21 cells were calculated based on comparative threshold cycles, with and without reverse transcriptase, from two independent experiments.
DNA isolation and quantitation of viral genomes.
DNA was harvested at specific times postinfection and extracted using the GenomicPrep DNA isolation kit (Amersham Pharmacia Biotech, Piscataway, NJ). Real-time PCR analysis was performed in triplicate on 1 μg total infected cell DNA using iQ SYBR green supermix (Bio-Rad) and primers specific for MCMV or cellular DNAs. The primer pairs utilized were as follows: ie2, 5′TGGATTGCACTTCGGC-3′ and 5′ TGGGGTATTCCTTGCG-3′; beta interferon (IFN-β), 5′-ATAAGCAGCTCCAGCTCCAA-3′ and 5′-AGTCTCATTCCACCCAGTGC-3′. Real-time PCRs were performed using the iQ iCycler after an initial 3-min denaturation at 95°C. The conditions were 39 cycles of 95° for 30 s, 57° for 30 s, and 72° for 30 s. Copies of MCMV DNA/cell were calculated based on the cycle at which viral DNA crossed the threshold relative to the cellular IFN-β gene, which was present at 2 copies/cell.
DNA cleavage assay.
Two micrograms each of total cell DNA from mock-, revertant-, and RVΔ140-infected cells was subjected to digestion with MluI, and cleavage was analyzed by Southern blot analysis as previously described (17) using as a probe labeled pON4047, which hybridizes to both ends of the cleaved DNA (a generous gift from Michael A. McVoy, Virginia Commonwealth University, Richmond, VA). Band densities were quantitated by scanning with a flat-bed scanner (transmission mode) followed by analysis using UN-SCAN-IT software (Silk Scientific Corporation, Orem, UT).
DNase protection assay.
Two million NIH 3T3 fibroblasts or IC-21 macrophages were infected with RVΔ140 or revertant virus at an MOI of 5. Nuclear extracts were harvested using the NE-PER kit (Pierce, Rockford, IL) according to the manufacturer's instructions. Lysates were divided in half, with one half being treated with 20 U RQ1 DNase I (Promega, Madison, WI) for 4 h and the other half being incubated as a control. After inactivation of the DNase with EDTA, DNA was extracted using the GenomicPrep kit (Amersham Biosciences). Equal volumes from the control and DNase-treated samples were subjected to real-time PCR analysis as described above.
Antibodies.
Polyclonal rabbit antiserum raised against the synthetic peptide RIQQSSQKDLPESQF of the MCMV MCP (M86) (32) was produced at Cocalico Biologicals, Inc. (Reamstown, PA). MCMV M25-specific antibody (5C7) was a generous gift from Carol Wu and John Shanley (University of Connecticut, Farmington, CT). The generation and characterization of rabbit anti-e1 antibody were previously described (8). Mouse anti-α-tubulin (clone B-5-1-2), mouse anti-actin (clone AC-15), and rabbit anti-γ-tubulin were purchased from Sigma (Sigma-Aldrich, St. Louis, MO).
Western blot analysis.
Expression of the MCP in mutant and revertant virus infected cells was monitored by Western blot analysis as previously described (16). An MOI of 2 was used for NIH 3T3 fibroblasts, and an MOI of 4 was used to infect IC-21 macrophages. For intracellular localization, cell fractionation was performed using the NE-PER kit (Pierce) according to the manufacturer's instructions. Volumes of lysate from the cytoplasmic and nuclear fractions representing equal numbers of infected cells were run on 12.5% acrylamide gels and blotted as described above. For the anisomycin block experiments, cells were infected as described above and anisomycin (100 μM final concentration) was added at 40 h postinfection. Cells were harvested at 0, 8, or 16 h after anisomycin treatment for Western blot analysis. Protein bands were quantitated using the Odyssey infrared imaging system (Li-Cor Biotechnology, Lincoln, NE) with rabbit anti-MCP and mouse anti-actin, followed by goat anti-rabbit infrared dye 800CW and goat anti-mouse infrared dye 680 (Li-Cor). The half-life of the MCP was calculated from the intensities of the MCP bands normalized to actin, which has a half-life of 60 h.
Electron microscopy.
IC-21 macrophages were infected at an MOI of 5 with sucrose gradient-purified virus for 48 h before fixation with 1% glutaraldehyde in phosphate-buffered saline (PBS). Fixed cells were embedded in plastic (Epon), and 80-nm thin sections were cut and mounted on standard electron microscopy grids before staining with uranyl acetate and lead citrate. Images were obtained using a Phillips 400T transmission electron microscope operated at 80 keV. The numbers of virion-like particles within an area representing 12 to 40 μm2 in seven mutant virus- and seven revertant virus-infected nuclei were quantitated, and results were expressed as virions/μm2. A one-tailed Student t test was used to calculate the probability that the mean values were significantly different.
Indirect immunofluorescence.
IC-21 macrophages were transfected with pIGFP140J using Metafectene (Biontex, Planegg, Germany). Twenty-four hours later, the cells were harvested and seeded into chambered slides. The following day, the cells were infected with WT or RVΔ140 MCMV at an MOI of 5. After 24 h of incubation, the cells were washed three times with sterile PBS, fixed with ice-cold 100% methanol, and washed again with PBS. After blocking with 5% normal goat serum in PBS, cells were stained with rabbit antiserum specific for e1 (M112/113) protein and mouse anti-α-tubulin antibody followed by goat anti mouse-tetramethyl rhodamine isocyanate (Sigma-Aldrich) and Alexa Fluor 647 goat anti-rabbit immunoglobulin G (Invitrogen). Cells were mounted with Slowfade mounting medium (Invitrogen) and examined using a Zeiss 510 confocal microscope at a magnification of ×40 with 4× zoom.
RESULTS
Deletion of M140 has no impact on the rate and level of MCMV DNA replication or the extent of DNA cleavage.
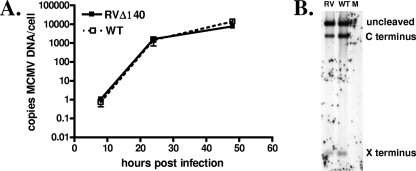
The growth defect in RVΔ140 in macrophages is evident from one-step growth kinetics, indicating that M140 regulates some aspect of viral replication (17). To define where in the replication cycle the defect occurs, we began by comparing genome copy numbers in RVΔ140-infected macrophages and revertant virus-infected cells. This parameter of viral replication provided a quantitative assessment as to whether the defect was at an early or late stage. Real-time PCR analysis of viral DNA levels revealed that genome replication was unimpaired in RVΔ140-infected macrophages (Fig. 1A). No significant differences in either the rate of replication or levels of viral DNA were detected. These data indicated that pM140 regulates some aspect of late-phase replication.
FIG. 1.
Viral DNA replication and cleavage are unimpaired in RVΔ140-infected macrophages. (A) Viral DNA replication. IC-21 macrophages were infected with sucrose gradient purified RVΔ140 or revertant virus (WT) at an MOI of 5 or mock infected with an equal volume of uninfected cell supernatant. At 8, 24, and 48 h postinfection, total DNA was harvested. Real-time PCR was performed in triplicate on 1 μg total infected cell DNA using primers specific for MCMV or cellular (IFN-β) DNA. Viral DNA levels were normalized to cellular gene levels. The experiment was repeated three times, and the mean results are shown. Error bars represent standard deviations. (B) Viral DNA cleavage. Two micrograms each of total cell DNA from mock (M)-, revertant virus (WT)-, and RVΔ140 (RV)-infected cells, prepared as described above, were subjected to digestion with MluI, and cleavage was analyzed by Southern blot analysis using a probe (pON4047) that hybridizes to both ends of the cleaved DNA. The experiment was repeated three times, and representative data are shown. The X and C termini are named as such based on EcoRI X and C fragments at the termini of the MCMV genome (22).
We next assessed viral DNA cleavage, which is commensurate with packaging of viral genomes. CMV genomes are replicated to form head-to-tail-linked concatemeric genomes, which are packaged into preformed procapsids, concomitant with cleavage at specific recognition sites into unit-length genomes (reviewed in reference 25). The viral DNA was therefore subjected to Southern blot analysis with probes specific for the genomic ends to evaluate DNA cleavage (22) (Fig. 1B). When the intensities of the bands were analyzed, no more than a twofold difference in the intensity of each band between revertant and mutant virus-infected cells was detected. These data indicate that replication of RVΔ140 in macrophages proceeded like that of WT virus through genome replication to the DNA cleavage stage commensurate with packaging into procapsids.
Infection with RVΔ140 results in a reduction in viral particles.
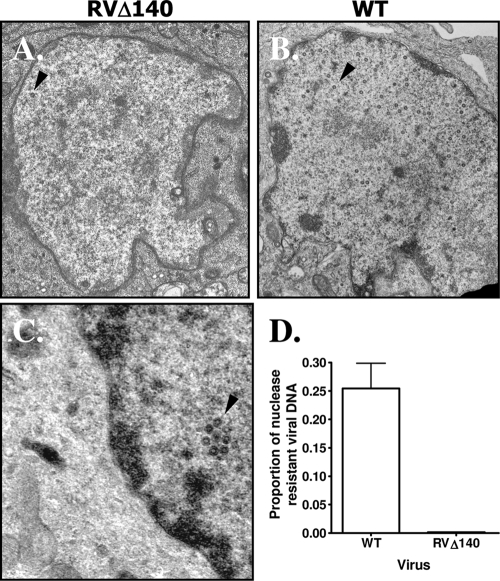
Because the DNA replication and cleavage analyses indicated that the defect in mutant virus-infected macrophages was at a late stage of viral assembly or even egress, we performed electron microscopic examination of revertant virus- and RVΔ140-infected IC-21 macrophages (Fig. 2). Viral capsid-like particles were rarely observed in the nuclei of mutant virus-infected cells (Fig. 2A) but were readily detected in nuclei of revertant-infected macrophages (Fig. 2B). The average number of capsids per area was 2.8 ± 1.5/μm2 in revertant virus-infected macrophages and 0.4 ± 0.4/μm2 in RVΔ140-infected cells (P = 0.0009). When capsid-like structures were evident in the RVΔ140 infected nuclei, they were predominantly empty (Fig. 2C). These data indicate that in macrophages, MCMV deleted of M140 is defective in assembly of mature capsids at a stage between formation of the procapsid and formation of the mature capsid.
FIG. 2.
Infection of macrophages with RVΔ140 produces a limited number of viral capsids. (A to C) Electron micrographs of mutant or revertant virus-infected macrophages. IC-21 macrophages were infected at an MOI of 5 with sucrose gradient-purified revertant virus (WT) (B) or RVΔ140 virus (A and C) for 48 h before fixation and staining for transmission electron micrographic analysis. Infected cell nuclei are shown, with arrowheads pointing to representative virions. (D) Mutant viral DNA remains sensitive to DNase. Two million IC-21 macrophages were infected with sucrose gradient-purified revertant virus (WT) or RVΔ140 at an MOI of 5, and nuclear extracts were harvested at 48 h postinfection. Half of each lysate was treated with DNase; the other half was incubated as a control without enzyme. After inactivation of the DNase, DNA was extracted, equal volumes from the control and DNase-treated samples were subjected to real-time PCR analysis, and the relative proportion of viral DNA remaining after treatment was determined. The results shown are the means and standard deviations for two independent experiments.
To further document a defect in capsid assembly, we assessed the degree to which the viral genome was protected from DNase. If the viral genomes have been properly encapsidated, they will be protected from DNase digestion. Consistent with the electron microscopy results, we found a 2-log-unit reduction in the proportion of viral DNA that was resistant to DNase treatment in the mutant virus-infected nuclei compared to revertant virus-infected nuclei (Fig. 2D). Approximately 25% of the revertant viral DNA was resistant to DNase treatment at 48 h postinfection. In contrast, only 0.2% of the RVΔ140 DNA was resistant to DNase. Collectively, the data reveal a defect in formation of mature capsids with DNase-resistant viral genomes. Because DNA cleavage was not significantly compromised, it appears that viral capsids were formed but were structurally defective and therefore unstable. In support of this conclusion, previous studies revealed that transcription of viral structural genes was unaltered in the absence of M140. Specifically, microarray analyses to assess MCMV gene expression in the presence or absence of M140 indicated that viral late mRNA was not compromised by deletion of M140 (unpublished data).
The M140 protein colocalizes with nuclear replication compartments and cytoplasmic aggresome-like structures in infected macrophages.
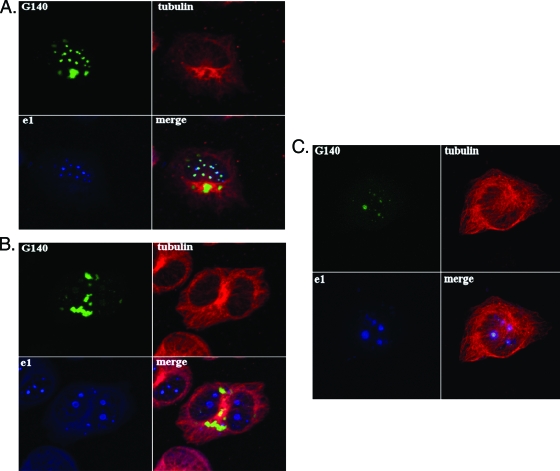
The above data indicating that pM140 regulates nuclear events related to capsid formation are not inconsistent with previous data indicating that pM140 localizes to both nuclear and cytoplasmic fractions of infected cells (16). In fact, when transiently expressed alone under the control of the HCMV major immediate-early promoter, pM140 is distributed diffusely throughout the nucleus (18). However, it relocalizes to a perinuclear cytoplasmic site when coexpressed with one of its binding partners, pM141 (18). Under these conditions, pM140 is likely to be highly overexpressed and with improper kinetics for an early gene. In order to validate this cellular localization under more natural conditions, we generated a plasmid expressing a full-length GFP-M140 fusion protein (pGFPM140) under the control of the native M140 promoter. The GFP tag was necessary because polyclonal antiserum specific for pM140 fails to detect the protein in infected cells by immunofluorescence (data not shown). The GFP-M140 fusion protein was not expressed in the absence of MCMV superinfection (data not shown), as expected of an early gene product, but localized to both the nucleus and cytoplasm in infected cells (Fig. 3). Interestingly, at 24 h postinfection (considered an early time point for macrophages), pGFPM140 in the nucleus largely colocalized with viral replication compartments. This was evidenced by colocalization with pM112/113 (e1), which is required for viral DNA replication and transcription (Fig. 3A) (8). Because this colocalization was also evident at early times in fibroblasts (12 h postinfection [Fig. 3C]), the association of pM140 with e1 in the nucleus is likely unrelated to the phenotype of RVΔ140 in macrophages.
FIG. 3.
The M140 protein colocalizes with viral replication compartments and adjacent to condensed MTOCs in infected macrophages. (A and B) IC-21 macrophages were transfected with a plasmid expressing a GFP-M140 fusion protein under the control of the M140 promoter. The transfected cells were infected with WT MCMV 48 h later and processed for confocal microscopy at 24 (A) or 48 (B) h postinfection. Mouse anti-tubulin and rabbit anti-e1 antibodies were used, followed by goat anti-mouse-tetramethyl rhodamine isocyanate or -fluorescein isothiocyanate and Alexa Fluor 647 goat anti-rabbit conjugates before analysis on a Zeiss 510 confocal microscope. Results were similar when transfected cells were superinfected with RVΔ140 instead of WT virus. (C) NIH 3T3 fibroblasts were transfected and infected as described above for 12 h before processing for confocal microscopy using the antibodies described above.
In the cytoplasm of macrophages at both 24 and 48 h postinfection, pGFPM140 localized to a perinuclear site as previously reported (Fig. 3A and B) (18). The use of anti-α-tubulin antibodies in the current study revealed that this site is adjacent to the microtubule organizing center (MTOC). Additional evidence that this structure is the MTOC was provided by γ-tubulin staining of centrioles at the center and sensitivity of the structure to nocodazole (41) (data not shown). We had previously identified the presence of cis-Golgi markers at this site (18). This suggested that this structure may be an agressome, as aggresomes are located at the MTOC in association with cis-Golgi markers (13). They are sites of aggregated and/or misfolded proteins destined for degradation by either the proteasome or autophagy (11, 13, 41). In virally infected cells, they may function to degrade viral proteins as a defense mechanism or may be sites of concentrated viral proteins in an assembly compartment (41). Interestingly, the enlarged aggresome-like structure was present in MCMV-infected macrophages but not in fibroblasts at early (Fig. 3C) or late (data not shown) times. We considered, therefore, that M140 might influence viral protein stability in macrophages.
Viral capsid protein levels are reduced in mutant virus-infected macrophages.
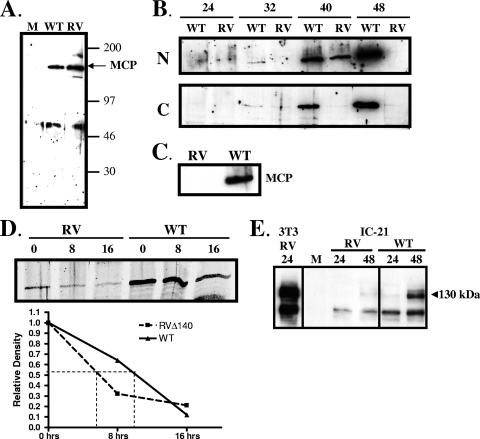
Based on the above findings, we assessed steady-state levels of late viral proteins associated with capsid maturation. Microarray analyses indicated no defect in expression of mRNAs for any of the viral structural components (unpublished data). This was confirmed by real-time reverse transcription-PCR for the small capsid protein m48.2 and the MCP (M86). The ratios of RNA from mutant virus-infected IC-21 cells to that from WT virus-infected cells were 1.0 for m48.2 and 1.3 for M86, thus confirming that the M140 deletion did not compromise RNA levels of these structural proteins. We therefore turned our attention to steady-state levels of the MCP. We generated peptide-specific antisera, which recognized a viral protein of appropriate size, approximately 150 kDa (Fig. 4A). In nuclear fractions of revertant virus-infected macrophages, MCP was expressed at low levels at 24 and 32 h postinfection, with steady-state levels increasing significantly at 40 and 48 h postinfection (Fig. 4B). These kinetics reflect the protracted rate of MCMV gene expression in macrophages compared to fibroblasts (14). However, in nuclear fractions of macrophages infected with RVΔ140, MCP was clearly evident only at 40 h postinfection, and it was undetectable at 48 h (Fig. 4B). Protein levels in the cytoplasm of revertant-virus infected cells followed a pattern similar to that seen in nuclear fractions. However, levels of MCP in the cytoplasm of RVΔ140-infected cells were barely detectable throughout this time course. The fact that levels of MCP in mutant virus-infected cells were undetectable at 48 h postinfection in both the nuclear and cytoplasmic fractions indicated that MCP trafficking was not defective, and it more likely reflects a lack of protein stability. Decreased steady-state levels of MCP were selective for macrophages, as seen by comparing results for NIH 3T3 fibroblasts (Fig. 4A) with those for IC-21 macrophages (Fig. 4B). This difference corresponds to the cell type-specific defect in virion production. This reduction in steady-state levels of the MCP was also seen in infected primary peritoneal macrophages (Fig. 4C), confirming that this is not a cell line-specific phenomenon.
FIG. 4.
Steady-state levels of the MCP and a tegument protein are reduced in RVΔ140-infected macrophages. (A) Confirmation of the specificity of the rabbit antiserum. Two million NIH 3T3 fibroblasts were infected with RVΔ140 (RV) or revertant virus (WT) at an MOI of 2 or were mock (M) infected. Cell lysates were harvested at 24 h postinfection and subjected to Western blot analysis using rabbit antiserum raised against an M86 peptide. (B) Steady-state levels of MCP in nuclear (N) and cytoplasmic (C) fractions of RVΔ140 (RV) and revertant (WT) virus-infected macrophages. IC-21 macrophages were infected at an MOI of 4 with sucrose gradient-purified viruses. Cell lysates were harvested at the indicated hours postinfection, and fractionation was performed using the NE-PER kit. Western blotting utilized the antisera described above. (C) Steady-state levels of MCP in infected primary macrophages. Peritoneal exudate cells from BALB/c mice were infected with sucrose gradient purified revertant virus (WT) or RVΔ140 (RV) at an MOI of 5. Cell lysates were harvested at 48 h postinfection and processed for Western blot analysis using antisera to the MCP as described above. (D) Protein synthesis block to assess stability of MCP expression in mutant and revertant virus-infected macrophages. IC-21 macrophages infected as described above were treated with 100 μM anisomycin starting at 40 h postinfection. Total cell lysates were harvested at the indicated times posttreatment (hours) and subjected to Western blot analysis for the MCP and actin (not shown). Protein bands were quantitated using the Odyssey infrared imaging system, normalized to actin levels, and graphed as a function of time postblock. (E) Levels of tegument protein M25 are reduced in mutant virus-infected IC-21 macrophages. NIH 3T3 fibroblasts (3T3) or IC-21 macrophages were mock infected (M) or infected at an MOI of 2 with revertant (WT) or mutant (RV) virus. Total cell lysates were harvested at the indicated hours postinfection for Western blot analysis using a monoclonal antibody specific for MCMV tegument protein M25.
In order to further investigate this finding, we subjected revertant and mutant virus-infected IC-21 macrophages to a protein synthesis block using the inhibitor anisomycin. The time of addition of anisomycin (40 h postinfection) was based on the optimal MCP expression levels in mutant virus-infected cells. As shown in Fig. 4D, levels of MCP at 40 h postinfection (0-h block) were reduced substantially in RVΔ140-infected cells compared to revertant virus-infected macrophages, and steady-state levels declined at a higher rate in mutant virus-infected cells. When band intensities were quantitated and normalized to actin levels, the half-life of MCP was calculated to be 10 h in revertant virus-infected macrophages but only 6 h in mutant virus-infected cells (Fig. 4D). These data confirm the reduction in stability of the MCP.
We also evaluated levels of one tegument protein, M25. We were particularly interested in this protein because it is abundant in nuclear aggregates with MCP in fibroblasts infected with an HCMV UL97 deletion mutant (30), and some HCMV UL97 mutants exhibit viral DNA cleavage with poor encapsidation (42). Interestingly, we saw a pronounced reduction in levels of the 130-kDa M25 protein, which is the virion-associated form (43), in mutant virus-infected macrophages compared to revertant virus-infected cells (Fig. 4E). Infection of NIH 3T3 fibroblasts with mutant virus deleted of M140 produced abundant levels of M25 (Fig. 4E). These data indicating that steady-state levels of two virion-associated proteins are significantly reduced in the absence of M140 are likely related to the observed scarcity of virions in macrophages infected with the mutant virus.
DISCUSSION
Previously we and others reported that the MCMV M140 protein is necessary for efficient viral replication in macrophages (16, 24). Our results indicate that this protein functions late in infection, after the stage of viral DNA replication. The reduction in both the number of viral capsids and the percentage of DNase-resistant viral genomes in the presence of efficient cleavage of viral DNA indicates that pM140 is necessary, directly or indirectly, for packaging of the viral genomes into stable virions.
Reduced steady-state levels of the MCP and M25 tegument protein are likely related to capsid instability. However, this reduction could be either a cause or an effect of such instability. Reduced levels of MCP may lead to altered stoichiometry of capsid components that provide sufficient structure for cleavage of viral DNA but incorrect assembly, leading to unstable virions. Currently, there is no direct evidence for colocalization or direct interactions between pM140 and either the MCP or M25 protein. At least under stringent conditions, neither structural protein coprecipitates with pM140 using antibody or tandem affinity purification (unpublished data), and thus they are not likely components of the pM139-pM140-pM141 complex(es) that we previously identified (18). Indirect effects of pM140 on MCP or M25 trafficking or posttranslational modifications could alter capsid stability, and either could result in increased degradation. However, effects on MCP trafficking are not likely, as the ratio of nuclear to cytoplasmic protein was unaltered by the M140 deletion.
The condensed MTOC observed in MCMV-infected macrophages is an aggresome-like structure, possibly signifying the importance of regulating protein turnover in this cell type. Although M140 is not required for generating this perinuclear structure (unpublished data), localization of pM140 at this site indicates a possible role for pM140 in regulating proteolysis in differentiated macrophages. We also considered that the pronounced MTOC may represent an assembly compartment. However, the MCP did not consistently localize to this site (unpublished data), and unlike the assembly compartment for HCMV (33), this structure colocalizes with cis-Golgi markers (18). Golgi-derived vesicles are, however, components of aggresome-like virus assembly compartments for herpes simplex virus (HSV) (27).
Our study is not the first to show that viral DNA cleavage and packaging into mature capsids are not inextricably connected. The use of herpesvirus deletion mutants has demonstrated that efficient DNA cleavage is not always associated with mature, DNA-containing C capsids. For example, deletion of the HSV type 1 or bovine herpesvirus UL25 gene or of HCMV gene UL97 or TRS1 results in reduced C capsids and/or DNase-resistant genomes in spite of efficient viral DNA cleavage (1, 12, 23, 37, 42). The exact role for any of these viral gene products in mature capsid formation has not been elucidated. However, for UL25 it has recently been demonstrated that certain mutants result in aberrant cleavage of the DNA, leading to truncated genomes which are not retained in the capsids while capsids containing full-length genomes are stabilized (9). In these studies, Cockrell et al. (9) found that one of the two termini expected after DNA cleavage was absent, significantly reduced, or altered in size from that of WT virus. Since we found no difference in relative levels or size of either terminus for RVΔ140 DNA, it is unlikely that M140 functions similarly to HSV UL25.
There are several additional differences between the above-described herpesvirus proteins that affect viral DNA encapsidation (or subsequent virion stability) and pM140. First, all of the other proteins are virion associated (2, 10, 28, 31, 35, 39, 40), whereas pM140 is not detected in purified virion preparations (19). Thus, unlike HSV UL25 and HCMV UL97 or TRS1, either M140 exerts an indirect effect upon viral DNA packaging or there are transient interactions during viral assembly. Second, UL25 and UL97 deletion mutants exhibit no reduction in the overall number of capsids; however, the proportion of C capsids is reduced (5, 12, 23, 37, 42). Only with the TRS1 mutation in HCMV was a reduction in total capsids detected, despite viral DNA cleavage (1). Thus, M140 conveys a phenotype most similar to that of this other member of the US22 gene family.
It is interesting that infection of macrophages with RVΔ140 resulted in a reduction in viral capsids, as previously reported for human fibroblasts infected with the HCMV TRS1 deletion mutant (3), but also in a reduction in steady-state levels of the MCP. We hypothesize that this may be due to differences between fibroblasts and macrophages. As a professional antigen-presenting cell, macrophages have active protein degradative systems, including the aggresome-associated proteasome and a constitutively active autophagy system (34). It is possible that in this cell type, protein products are subject to more rapid degradation. Therefore, in RVΔ140-infected macrophages, the reduction in the number of capsids detected is accompanied by reduced steady-state levels of the MCP. The M140 gene may be required in macrophages to prevent degradation of viral proteins that would otherwise be targeted by aggresome-associated degradative systems.
There is evidence that at least the HCMV MCP is susceptible to proteolytic cleavage or additional processing. Specific mutations in the capsid scaffolding components UL80a and UL80.5 that delay transport of the MCP to the nucleus result in an increase in a specific proteolytic cleavage of the MCP, likely due to the activity of other viral proteins (20, 26). The MCPs of HCMV and MCMV may therefore be susceptible to proteolytic processing under specific circumstances.
The reduction in levels of the 130-kDa virion-associated M25 protein is also intriguing in this light, since the MCP and M25 proteins are two of the most abundant proteins in intranuclear aggregates in an HCMV UL97 mutant (30). Such aggregates would be prime targets for degradation by the aggresome. An MCMV mutant deleted of the UL97 homologue M97 exhibits a decrease in mature C capsid production (5). It is possible that in RVΔ140-infected cells, insoluble MCP aggregates are formed, resulting in an apparent reduction in protein levels. Whether M140 influences M97 function has yet to be determined.
Based on these results, it is likely that the levels of other capsid and tegument components are also reduced; however, these analyses are limited by the dearth of antibodies to MCMV structural proteins. Viral particles released into the supernatant from revertant virus- and RVΔ140-infected IC-21 macrophages exhibited no difference in sensitivity to degradation by incubation for up to 8 h at 37°C (unpublished data), which supports the hypothesis that the defect is specifically in some aspect of procapsid-to-capsid maturation.
In conclusion, the data indicate that M140 is required for efficient production of mature capsids in macrophages. The reduced steady-state levels of the MCP and M25 suggest that capsid formation is defective in the absence of M140. However, efficient viral DNA cleavage signifies that at least some form of procapsids are assembled, although they are apparently unstable and do not progress to mature capsids. Thus, MCMV mutants deleted of M140 could provide a useful tool for clarifying how this poorly understood stage of virion production is controlled.
Acknowledgments
This work was supported by National Cancer Institute grant CA41451 awarded to A.E.C., National Institute of Allergy and Infectious Diseases (NIAID) grant AI41136 and a grant from the Mathers Foundation awarded to G.G.M., and NIAID grant AI041644 awarded to J.C.B.
We gratefully acknowledge helpful discussions with Michael A. McVoy, Virginia Commonwealth University, regarding the DNA cleavage assays. We thank Julie A. Kerry for assistance with figures and critical reviews of this paper.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Adamo, J. E., J. Schroer, and T. Shenk. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 7810221-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216278-283. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship, C. A., and T. Shenk. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J. Virol. 7612290-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291303-305. [DOI] [PubMed] [Google Scholar]
- 5.Buser, C., F. Fleischer, T. Mertens, D. Michel, V. Schmidt, and P. Walther. 2007. Quantitative investigation of murine cytomegalovirus nucleocapsid interaction. J. Microsc. 22878-87. [DOI] [PubMed] [Google Scholar]
- 6.Cavanaugh, V. J., R. M. Stenberg, T. L. Staley, H. W. t. Virgin, M. R. MacDonald, S. Paetzold, H. E. Farrell, W. D. Rawlinson, and A. E. Campbell. 1996. Murine cytomegalovirus with a deletion of genes spanning HindIII-J and -I displays altered cell and tissue tropism. J. Virol. 701365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, G., E. R. Bivins-Smith, M. S. Smith, P. M. Smith, and A. D. Yurochko. 2008. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J. Immunol. 181698-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciocco-Schmitt, G. M., Z. Karabekian, E. W. Godfrey, R. M. Stenberg, A. E. Campbell, and J. A. Kerry. 2002. Identification and characterization of novel murine cytomegalovirus M112-113 (e1) gene products. Virology 294199-208. [DOI] [PubMed] [Google Scholar]
- 9.Cockrell, S. K., M. E. Sanchez, A. Erazo, and F. L. Homa. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 8347-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corboy, M. J., P. J. Thomas, and W. C. Wigley. 2005. Aggresome formation. Methods Mol. Biol. 301305-327. [DOI] [PubMed] [Google Scholar]
- 12.Desloges, N., and C. Simard. 2003. Implication of the product of the bovine herpesvirus type 1 UL25 gene in capsid assembly. J. Gen. Virol. 842485-2490. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1461239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson, L. K., and A. E. Campbell. 2006. Determinants of macrophage tropism, p. 419-443. In M. J. Reddehase (ed.), Cytomegaloviruses. Molecular biology and immunology. Caister Academic Press, Norfolk, United Kingdom.
- 15.Hanson, L. K., B. L. Dalton, Z. Karabekian, H. E. Farrell, W. D. Rawlinson, R. M. Stenberg, and A. E. Campbell. 1999. Transcriptional analysis of the murine cytomegalovirus HindIII-I region: identification of a novel immediate-early gene region. Virology 260156-164. [DOI] [PubMed] [Google Scholar]
- 16.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 756292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. t. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 735970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karabekian, Z., L. K. Hanson, J. S. Slater, N. K. Krishna, L. L. Bolin, J. A. Kerry, and A. E. Campbell. 2005. Complex formation among murine cytomegalovirus US22 proteins encoded by genes M139, M140, and M141. J. Virol. 793525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 7811187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loveland, A. N., N. L. Nguyen, E. J. Brignole, and W. Gibson. 2007. The amino-conserved domain of human cytomegalovirus UL80a proteins is required for key interactions during early stages of capsid formation and virus production. J. Virol. 81620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack, C., A. Sickmann, D. Lembo, and W. Brune. 2008. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. USA 1053094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks, J. R., and D. H. Spector. 1988. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology 16298-107. [DOI] [PubMed] [Google Scholar]
- 23.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 775557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 26.Nguyen, N. L., A. N. Loveland, and W. Gibson. 2008. Nuclear localization sequences in cytomegalovirus capsid assembly proteins (UL80 proteins) are required for virus production: inactivating NLS1, NLS2, or both affects replication to strikingly different extents. J. Virol. 825381-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozawa, N., Y. Yamauchi, K. Ohtsuka, Y. Kawaguchi, and Y. Nishiyama. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299486-497. [DOI] [PubMed] [Google Scholar]
- 28.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 751427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 983237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 7915494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 715703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupp, B., Z. Ruzsics, T. Sacher, and U. H. Koszinowski. 2005. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 79486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid, D., M. Pypaert, and C. Munz. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2679-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, M. S., G. L. Bentz, J. S. Alexander, and A. D. Yurochko. 2004. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 784444-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, Q., E. A. Murphy, and G. G. Maul. 2006. Experimental confirmation of global murine cytomegalovirus open reading frames by transcriptional detection and partial characterization of newly described gene products. J. Virol. 806873-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Zeijl, M., J. Fairhurst, E. Z. Baum, L. Sun, and T. R. Jones. 1997. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology 23172-80. [DOI] [PubMed] [Google Scholar]
- 41.Wileman, T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61149-167. [DOI] [PubMed] [Google Scholar]
- 42.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 981895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, C. A., M. E. Carlson, S. C. Henry, and J. D. Shanley. 1999. The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology 262265-276. [DOI] [PubMed] [Google Scholar]