Abstract
In 2005, a human bocavirus was discovered in children with respiratory tract illnesses. Attempts to culture this virus on conventional cell lines has failed thus far. We investigated whether the virus can replicate on pseudostratified human airway epithelium. This cell culture system mimics the human airway environment and facilitates culturing of various respiratory agents. The cells were inoculated with human bocavirus-positive nasopharyngeal washes from children, and virus replication was monitored by measuring apical release of the virus via real-time PCR. Furthermore, we identified different viral mRNAs in the infected cells. All mRNAs were transcribed from a single promoter but varied due to alternative splicing and alternative polyadenylation, similar to what has been described for bovine parvovirus and minute virus of canines, the other two members of the Bocavirus genus. Thus, transcription of human bocavirus displays strong homology to the transcription of the other bocaviruses. In conclusion, we report here for the first time that human bocavirus can be propagated in an in vitro culture system and present a detailed map of the set of mRNAs that are produced by the virus.
Parvovirinae, a subfamily of the Parvoviridae family, are nonenveloped, single-stranded DNA (ssDNA) viruses with a viral genome of approximately 4,000 to 6,000 nucleotides (nt) (3). The genera that belong to the Parvovirinae family are parvoviruses, erythroviruses, dependoviruses, amdoviruses, and bocaviruses (3, 9). The virions of the Parvoviridae family are round, exhibit an icosahedral symmetry, and contain a single linear DNA molecule of either negative- or positive-sense orientation (3). The viruses are autonomous and do not require a helper virus for infection, except for the dependoviruses (3). After host cell entry, the viral genomic ssDNA is transported to the nucleus, where it is converted into double-stranded DNA by cellular host proteins (3). Transcription of viral mRNAs occurs in general only during the S phase of the host cell cycle, leading to the synthesis of viral proteins from nonspliced and spliced viral mRNAs (4). Replication of the genomic ssDNA occurs through a so-called rolling-hairpin mechanism, and newly synthesized ssDNA genome products are excised from the replication complex by a process called junction resolution (7). Newly synthesized ssDNA genomes can either serve as template for viral gene expression or be encapsidated in new virions (7).
Presently, the Bocavirus genus includes bovine parvovirus (BPV), minute virus of canines (MVC), and the recently identified human bocavirus (HBoV) and HBoV type 2 (2, 9, 15). HBoV was identified in 2005 within pools of human nasopharyngeal aspirates obtained from individuals with respiratory tract illnesses. The current genomic DNA reference sequence of HBoV is 5,299 nt in length, but sequence information regarding the flanking terminal hairpin structures remains to be determined (2). HBoV has been found worldwide, mainly in respiratory samples, but in some cases, HBoV has also been detected in serum, fecal samples, and urine samples (1, 16, 20, 31). HBoV infections are frequently diagnosed in <2-year-old children with upper or lower respiratory tract illness, often in combination with another respiratory virus (2, 16, 20). One of the most frequently observed clinical symptoms in HBoV-infected patients is acute wheezing (1). Despite the current knowledge regarding HBoV, no in vitro or in vivo model that supports replication of HBoV has been established.
In this study, we investigated whether pseudostratified human airway epithelium cell culture could be utilized as a model for HBoV replication. Pseudostratified epithelium is formed by culturing primary human airway epithelial cells in an air-liquid interface. The morphology and functionality of the cells resemble those of the human airways, and this system has been used previously to culture a wide range of respiratory viruses, e.g., influenza virus (28), parainfluenza virus (33), respiratory syncytial virus (34), adenovirus (21), and severe acute respiratory syndrome coronavirus (26). In this study, we documented HBoV replication upon inoculation with respiratory material from HBoV-infected patients. We observed apical release of virus and analyzed the viral mRNA transcripts in the infected cells.
MATERIALS AND METHODS
Clinical samples.
Clinical patient material (samples Bonn-1, Bonn-2, and Bonn-3) was obtained from three children (from 1 to 1.5 years of age) that were hospitalized due to upper or lower respiratory tract illness. Nasopharyngeal washes were collected two days after onset of symptoms. These symptoms were indistinguishable from the observed symptoms from respiratory syncytial virus infection. Diagnostic PCR and reverse transcription-PCR (RT-PCR) analysis were performed for HBoV, human coronavirus group I and group II, human metapneumovirus, influenza virus types A and B, and respiratory syncytial virus as previous described (18, 25). All nasopharyngeal washes were positive for HBoV, whereas the diagnostic panel was negative for all other viruses. HBoV sequencing revealed that all three strains were type ST2.
Human airway epithelial cell culture.
Cryopreserved human trachea epithelial primary cells (HTEpC) were obtained from the European Collection of Cell Cultures. HTEpC were maintained for one serial passage as a monolayer in bronchial/tracheal epithelial cell serum-free growth medium (European Collection of Cell Cultures) supplemented with penicillin-streptomycin. Bronchial/tracheal epithelial cell serum-free growth medium was refreshed every 2 or 3 days. HTEpC cultures were maintained at 37°C in a 5% CO2 incubator. When reaching 75% confluence, cells were dissociated with 2 ml of TrypLE Express enzyme (Invitrogen). HTEpC were diluted in air-liquid interface medium (11), which is a mixture of LHC basal medium (Invitrogen) and Dulbecco's modified Eagle's medium (Invitrogen) supplemented with the required additives (Sigma). A total of 8 × 104 HTEpC were seeded on type IV collagen (Sigma)-coated 12-well ThinCerts with a 0.4-μm pore size (Greiner Bio-One). Medium was renewed every 2 or 3 days. When cultures reached full confluence, the cells were exposed to air. HTEpC cultures on the air-liquid interface were maintained in 12-well deep-well plates (Greiner Bio-One) for 21 days to let the cells differentiate into pseudostratified human airway epithelial cell cultures. Medium from the basolateral compartment was renewed every 6 days, and the apical surface was washed every 2 days with Hanks’ balanced salt solution (HBSS) (Invitrogen).
HBoV infection.
An aliquot of 50 μl clinical patient material was diluted in 200 μl HBSS and centrifuged for 30 min at 4°C with 10,000× relative centrifugal force. Two hundred microliters of diluted clinical sample was directly inoculated upon the apical surface of pseudostratified human airway epithelium and incubated for 2 h at 34°C in a 5% CO2 incubator. After 2 h, 200-μl samples were collected from both the apical and basolateral sides. Inoculated cultures were maintained at 34°C in a 5% CO2 incubator. Samples were collected after 24, 48, 72, and 95 h postinoculation (hpi) from both the apical and basolateral sides. Apical washing and harvesting was performed by adding 200 μl of HBSS to the apical surface and incubation for 10 min at 34°C in a 5% CO2 incubator, followed by the removal and storage of the 200 μl HBSS from the apical surface. Washing is needed to remove the mucus, which will otherwise suffocate the cells. An aliquot of 50 μl apical wash was transferred into 900 μl L6 lysis buffer (6) for HBoV DNA quantification. At the last day of culture, the cells were collected in TRIzol reagent (Invitrogen) for HBoV mRNA analysis. Cultures were transferred to a conventional 12-well plate (Greiner Bio-One) and analyzed by eye with a phase-contrast microscope, prior to cell collection in TRIzol reagent.
HBoV yield.
Viral DNA was isolated from the collected samples of the apical and basolateral harvests by the Boom method for total nucleic acid isolation (6), and elution was performed in 100 μl H2O. Real-time PCRs were performed on the HBoV NS1 gene region. Primer and probe details are available upon request (unpublished data).
DNase protection assay of HBoV DNA from the Bonn-1 culture.
An aliquot of 50 μl apical and basolateral harvest of the Bonn-1 isolate was spiked with 25 μl naked plasmid DNA containing a part of phocid herpesvirus-1 DNA (250 DNA copies/μl) and 5 μl cell culture supernatant of human adenovirus type 5 (1E8 50% tissue culture infective doses/ml). Eighty microliters of the spiked sample was incubated with 18 units of DNase I (Ambion) in a total volume of 100 μl for 45 min at 37°C, followed by total nucleic acid isolation (Boom) as described above. Real-time PCRs for HBoV were performed as described above. The design and characteristics of the phocid herpesvirus-1 and adenovirus real-time PCR are available upon request. In all samples, naked plasmid DNA was degraded (>2-log decrease), whereas adenoviral DNA inside a virus particle was protected from DNase treatment (<1-log decrease) (data not shown).
Full-genome sequencing.
The complete genome sequence of the HBoV Stockholm 2 isolate (NC_007455) was used as the template for designing bidirectional primer combinations encompassing 5,299 nt. Primer combinations had an average theoretical fragment length of 600 bp with a minimum overlap of 70 bp with adjacent primer combinations. All primers match identical regions of the most recent available full-genome sequences of different HBoV isolates with the BLAST sequence alignment tool (24). Primer sequences are available upon request. Amplification of the fragment was performed with a thermal cycle profile as follows: 95°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final elongation step at 72°C for 10 min. PCR fragments were visualized upon agarose gel electrophoreses by ethidium bromide staining. Positive PCR fragments were directly sequenced with their respective PCR primers. A sequence reaction was performed without purifying steps, according to the BigDye Terminator version 1.1 cycle sequencing manufacturer protocol (Applied Biosystems) on a GeneAmp PCR system 9700 thermal cycler (Perkin Elmer). Electrophoresis and data collection was performed on a 3100 genetic analyzer (Applied Biosystems). Raw collection data was processed and analyzed with Codoncode Aligner v2.06 software (CodonCode Corporation).
5′ and 3′ RACE of HBoV mRNA.
Cellular mRNA was isolated from whole-cell lysate of the pseudostratified human airway epithelium cell culture in TRIzol reagent, according to the manufacturer's protocol (Invitrogen). Elution was performed in 200 μl of diethyl pyrocarbonate-treated water. Eighty microliters of a whole cellular mRNA fraction was incubated with 18 units of DNase I (Ambion) in a total volume of 100 μl for 45 min at 37°C. The DNase-treated fraction was subsequently phenol-chloroform (Invitrogen) extracted, and this was followed by an overnight ethanol precipitation. The 5′ sequences of the mRNA transcripts were determined with a 5′ system for rapid amplification of cDNA ends (RACE system, version 2.0; Invitrogen), according to the manufacturer's protocol. Gene-specific RT and PCR primers were designed based on the predicted NS1, NP1, VP1, and VP2 and the putative ORFx gene (Table 1). Gene-specific primers for 5′ RACE PCR amplification were designed to flank approximately 100 nt of the 5′ region of the predicted start codon positions of the open reading frames (ORFs). The 3′ ends of HBoV mRNA transcripts were determined with a 3′ RACE system (Invitrogen) according to the manufacturer's protocol with minor modifications. RT was performed with the JZH-OligodT primer (5′-GCTATCATCACAATGGACTTTTTTTTTTTTTTTTTTV-3′), and PCR amplification was performed with the JZH-Nested adaptor primer (5′-GCTATCATCACAATGGAC-3′) and a gene-specific primer (Table 1). The PCR products were excised after agarose electrophoresis and purified with the NucleoSpin Extract II kit (Machery-Nagel) according to the manufacturer's protocol. Purified PCR products were cloned into the pCR2.1-TOPO TA vector (Invitrogen) and transformed in chemically competent TOP10 Escherichia coli (Invitrogen), according to manufacturer protocol. Transformants were directly analyzed via colony PCR with T7 and M13RP primers. PCR products were sequenced as described above.
TABLE 1.
Primers for HBoV mRNA analysis
| Target | Assay | Primer | Position (nt) | Orientation | Sequence (5′→3′) |
|---|---|---|---|---|---|
| NS1 | 5′ RACE | NS1_End | 2172 | Reverse | TTACTTACTTGGTGTGATGTCTCCTG |
| NS1_5RACE1 | 326 | Reverse | GGATATGGAAATTTGAAGACATAAG | ||
| NP1 | 5′ RACE | NP1_End | 3069 | Reverse | TTAATTGGAGGCATCTGCTTCCATG |
| BOCA_R12 | 2625 | Reverse | TTGGTCTGAGGTCTTCGAAGCAGTG | ||
| VP1 | 5′ RACE | VP1_End | 3442 | Reverse | TTTTGAGGTTCCTGGTTTAGGTTCAC |
| BOCA_VP1 | 3167 | Reverse | CAGCGCGATCAGCGTTATTTAC | ||
| NS1 | 3′ RACE | BOCA_F07 | 1505 | Forward | CTGTTTGCTTTTACGGGCCTGC |
| BOCA_F09 | 2000 | Forward | CAAACTCATTTCCTCTTGGGGTC | ||
| NP1 | 3′ RACE | BOCA_NP1_2410 | 2410 | Forward | ATGAGCTCAGGGAATATGAAAGAC |
| BOCA_F11 | 2498 | Forward | CAACTCATCACAGGAGCAGGAG | ||
| VP2 | 3′ RACE | BOCA_F19 | 4490 | Forward | CCAACTAACCTAGAATACAAACTTC |
| BOCA_F21 | 4989 | Forward | TGGATCCAACAGGAGCATACATC | ||
| Leader | Spliced messenger PCR | BOCA_5UTR1 | 186 | Forward | AGACTGCATCCGGTCTCCGGCGAGTGAAC |
| NP1 | Spliced messenger PCR | BOCA_R12 | 2625 | Reverse | TTGGTCTGAGGTCTTCGAAGCAGTG |
| VP1 | Spliced messenger PCR | VP1_End | 3442 | Reverse | TTTTGAGGTTCCTGGTTTAGGTTCAC |
| VP2 | Spliced messenger PCR | BOCA_R16 | 3605 | Reverse | CCACATATTTGTCTGAAAAGTGAG |
| ORFx | Spliced messenger PCR | BOCA_R18 | 4108 | Reverse | AAAAGGCATCTGATACAGAAGTG |
| NS1 | Spliced messenger PCR | BOCA_R06 | 1128 | Reverse | CTGATCTCCTACCTCAGGAAGATG |
In silico analysis.
The obtained HBoV genomic sequence was used for prediction of ORFs with ORFfinder (24). Identification of the putative internal ribosomal entry site (IRES) was performed with RegRNA V1.0 prediction software on in silico-derived mature mRNA sequences (13). The NetPicoRNA, NetCorona, ProP, and PeptideCutter software packages were used for identification of putative proteases that specifically cleave at the VP1/VP2 boundary (5, 8, 12, 17).
Nucleotide sequence accession number.
The genome sequence of the Bonn-1 isolate has been submitted to GenBank under accession number FJ858259.
RESULTS
HBoV infection of human airway epithelial cultures.
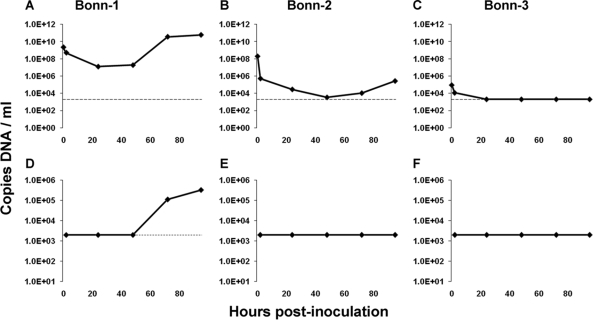
Currently, there is no culture system that supports HBoV replication. Pseudostratified human airway epithelium, which is formed after exposure of human tracheal epithelial cells to air, is the best imitation of the human trachea. We tested whether this system allows propagation of HBoV. Clinical material from three HBoV-infected patients was inoculated at the apical side of pseudostratified human airway epithelium and incubated for 2 h at 34°C. The culture was maintained, and 200-μl harvests were collected from the apical and basolateral sides at 2, 24, 48, 72, and 95 hpi. The concentration of the input virus in the three clinical samples ranged from 9.2E4 HBoV DNA copies/ml (isolate Bonn-3) to 2.2E9 HBoV DNA copies/ml (isolate Bonn-1), as determined by quantitative real-time PCR. A steep decrease in HBoV DNA concentration was noted for all three infections over the first 24 h (Fig. 1A to C), and this decrease correlates with entry of the virus and subsequent washing of the apical side of the cells. A 3-log increase of the HBoV yield was observed for the Bonn-1 isolate at 72 hpi (Fig. 1A). This increase in viral DNA concentration strongly suggests that there is active HBoV replication. The fact that the Bonn-1 virus yields at 72 and 95 hpi (3.6E10 and 6.0E10 HBoV DNA copies/ml, respectively) are higher than the concentration of the input virus (2.2E9 HBoV DNA copies/ml) provides evidence that active replication occurs in the cell culture system. For the Bonn-2 and Bonn-3 isolates, no clear sign of replication was noticed, but this may relate to the lower concentrations of virus in these samples (Fig. 1B and C, respectively). We also monitored the HBoV DNA yield at the basolateral side to investigate whether the viral secretion occurs exclusively at the apical surface. We detected HBoV Bonn-1 DNA at the basolateral side starting at 48 hpi, with an increase over time (Fig. 1D; see Fig. 1E and F for Bonn-2 and Bonn-3 isolates). To investigate whether the secreted HBoV DNA in the Bonn-1 harvests was from virus particles or naked DNA from broken cells, a DNase protection assay was conducted. The apical HBoV DNA was insensitive to DNase (<1-log decrease; Fig. 2A), while the basolateral harvest was sensitive to DNase (>2-log decrease; Fig. 2B). Thus, the HBoV DNA from the basolateral side was most likely the result of leakage from dying cells, but the apical HBoV DNA was protected by a viral capsid.
FIG. 1.
HBoV propagation on human airway epithelium cell culture. HBoV DNA concentration (copies of DNA/ml; y axis) in the apical washings at different hours postinoculation (x axis) for the Bonn-1 (A), Bonn-2 (B), and Bonn-3 (C) HBoV-inoculated human airway epithelium cell cultures. HBoV DNA quantification at the basolateral side is shown in panels D (Bonn-1), E (Bonn-2), and F (Bonn-3). The horizontal dashed line represents the detection threshold of the HBoV DNA real-time PCR assay.
FIG. 2.
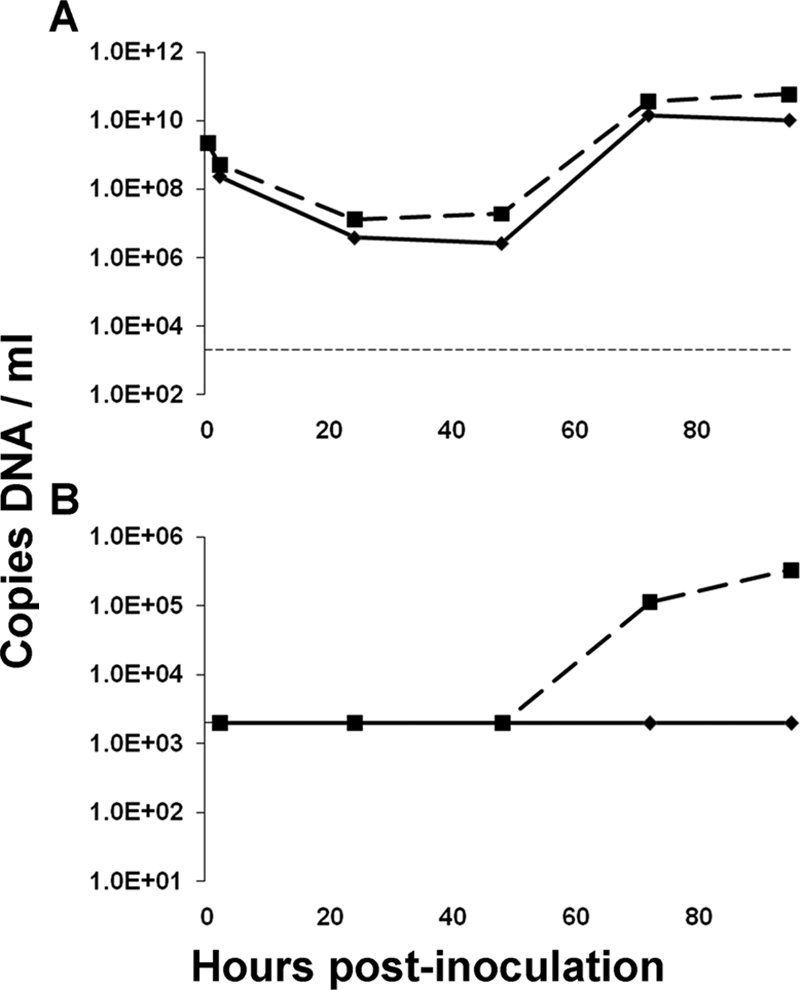
DNase protection assay of HBoV DNA from the Bonn-1 culture. HBoV DNA concentration (copies of DNA/ml; y axis) in DNase-treated (continuous line) and non-DNase-treated (dashed line) apical harvests (A) and basolateral harvests (B). The horizontal dashed line in panel A represents the detection threshold of the assay.
To investigate the cytopathic effect of HBoV infection, we monitored the morphological changes of the cells at 95 hpi with the Bonn-1 isolate by phase-contrast microscopy. Minor changes in the culture were noticed. Stretching of cells (which was absent in the control culture) was observed, but there was no disruption of the pseudostratified epithelium layer and the ciliated cell density. Furthermore, the ciliary movement and the mucosal secretion of the Bonn-1 virus-inoculated culture were similar to those of the control culture.
HBoV transcription.
The apical release of Bonn-1 virus particles demonstrates viral replication, yet further evidence of productive infection can be obtained by measuring intracellular viral mRNA transcripts. This can be done by Northern blotting; however, this was not possible, due to the low number of cells in this culture system. Alternatively, the mRNAs can be investigated by 5′ RACE and 3′ RACE. For primer design, we first established the viral genome sequence; more specifically, we obtained this information for the ORFs in the Bonn-1 isolate. As a template for sequencing, we used the apical washing of Bonn-1 at 95 hpi. The acquired genomic sequence encompasses 5,299 nt with an overall similarity of 99.8% with the HBoV ST2 reference sequence (NC_007455) and 97.9% with the HBoV ST1 reference sequence (DQ000495). In silico analysis of the putative ORFs within the Bonn-1 genomic sequence resulted in four hits: the NS1, NP1, and VP1/VP2 genes and one additional putative gene (ORFx) encoding a 120 amino acid protein that has not been described previously. The deduced amino acid sequence of the ORFx protein has no viral or cellular homologue. The deduced amino acid sequences of the putative NS1 and NP1 proteins of Bonn-1 shared 100% homology with those of the HBoV ST2, and for the VP1/VP2 protein, the shared homology is 99%.
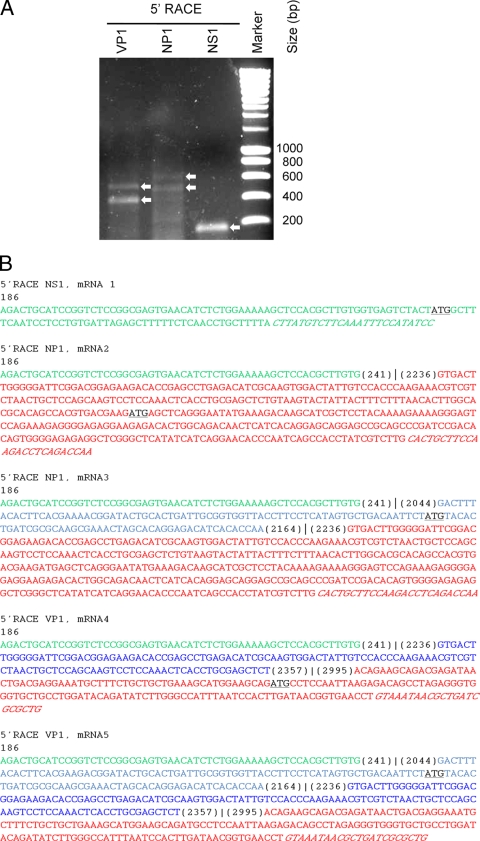
The 5′ terminal sequence of the various mRNAs was determined by 5′ RACE with primers annealing in NS1, NP1, VP1, VP2, and ORFx (Table 1). Only with NS1, NP1, and VP1 primers was the 5′ RACE positive (Fig. 3A). The NP1 and VP1 5′ RACE products display two fragments, and the NS1 product displays only one. Sequencing of the fragments revealed that all three transcripts start at position 186 (Fig. 3B). Prediction of promoter regions with the neural network promoter prediction software (version 2.2) indeed shows that at positions 146 to 196, a promoter region (5′-TATTAAACCTATATAAGCTGCTGCACTTCCTGATTCAATCAGACTGCATC-3′ [the putative TATA box is underlined and the starting nucleotide is in boldface) is present, and our 5′ RACE experiments suggest that it is the only promoter used (23). The 5′ RACE experiment further revealed that the transcript containing the NS1 gene was not spliced at the 5′ terminus (mRNA 1; Fig. 3B and 4). This was not the case for the mRNAs that contained the NP1 gene and the VP1 gene, which are the products of alternative splicing. There are two transcripts that contain the NP1 gene (mRNA 2 and mRNA 3; Fig. 3B and 4). In mRNA 2, only one splicing event occurred (donor 241 and acceptor 2236), whereas in the other mRNA (mRNA 3), splicing occurred twice (donor 241 and acceptor 2044 and donor 2164 and acceptor 2236) (Fig. 3B and 4). In mRNA 3, the sequence between positions 2044 and 2164 is retained in the mRNA.
FIG. 3.
Identification of mRNA transcripts from Bonn-1. (A) Agarose gel with ethidium bromide-stained 5′ RACE PCR products of NS1, NP1, and VP1 mRNAs of the Bonn-1 isolate at 95 hpi. The arrows indicate 5′ RACE-amplified PCR products. (B) The determined cDNA nucleotide sequences of the NS1, NP1, and VP1 5′ RACE products. The start position of each transcript is indicated at the beginning of the nucleotide sequence. The spectrum of color highlights the nucleotide sequences corresponding with different regions along the genome. The genome positions at the splice donor and acceptor junction sites are shown in parentheses. The first start codon triplets along each fragment are indicated in underlined black letters. In mRNA 1, the start codon is for NS1; for mRNA 2, the start codon is for NP1; for mRNA 3, the start codon is for UP1; for mRNA 4, the start codon is for VP1; and for mRNA5, the start codon is for UP2. The binding region of the reverse 5′ RACE primer is shown in italics. (C) Agarose gel with ethidium bromide-stained 3′ RACE products of the NP1 and VP2 mRNAs of the Bonn-1 isolate at 95 hpi. The arrows indicate 3′ RACE-amplified PCR products that were properly primed on the polyadenosine tails. The additional fragments were generated by nonspecific priming of the RT primer at polyadenosine stretches along the HBoV genome. (D) The determined cDNA nucleotide sequences of the VP2 and NP1 3′ RACE products. The forward 3′ RACE primer is shown in italics. The position of the stop codon of the NP1 and VP2 transcripts is indicated with underlined black letters, and the proximal polyadenylation [(pA)p] (NP1 3′RACE products) and distal pA [(pA)d] (VP2 3′RACE product) are underlined. The genome position of the stop codon and the pA sites are indicated above the sequence.
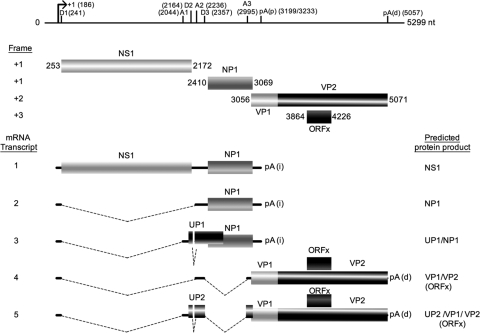
FIG. 4.
Schematic representation of the genomic and transcriptional map of HBoV Bonn-1 isolate. The transcriptional start (arrow), the splice donor (D numbers) and acceptor (A numbers) sites, the proximal and distal polyadenylation signals [pA(p) and pA(d), respectively], and the predicted ORFs are positioned along the HBoV genome of the Bonn-1 isolate. In the lower panel, a schematic overview of the identified mRNA transcripts of Bonn-1 HBoV and their suggested protein products is presented.
The 5′ RACE with a VP1-specific primer showed that there are two different transcripts that contain the VP1 gene (Fig. 3A). One of the transcripts is spliced twice (mRNA 4, donor 241 and acceptor 2236 and donor 2357 and acceptor 2995), whereas the other (mRNA 5) is spliced three times (donor 241 and acceptor 2044, donor 2164 and acceptor 2236, and donor 2357 and acceptor 2995) (Fig. 3B and 4). Again, there were mRNA variants that retained the sequence between positions 2044 and 2164. We searched for mRNAs that are specific for VP2 and ORFx by RT-PCR experiments with the 5′ untranslated region and VP2- and ORFx-specific primers. In none of these experiments could we identify VP2- or ORFx-specific mRNAs; in all cases, the VP1 gene was included in the transcripts. This finding suggests that mRNA 4 and 5 are polycistronic mRNAs that can encode different proteins (VP1/VP2 and ORFx). A confirmation of the splicing that we observed in 5′ RACE of the various mRNAs was obtained by using a PCR with 5′ untranslated region primers and 3′ primers at the 3′ regions of the NS1, NP1, and VP1 genes. The 3′ primers were selected on specificity, so they could not amplify transcripts containing other ORFs (Table 1). The same splicing pattern noticed with 5′ RACE was observed (data not shown).
Subsequently, we determined the 3′ terminal sequence of the viral mRNA transcripts to investigate whether splicing occurs downstream of the ORFs and to identify the polyadenylation signals of each mRNA. Unfortunately, the 3′ RACE with NS1 gene-specific primers did not yield any PCR product, suggesting that the polyadenylation signal is located far from the 3′ terminus of the ORF. We decided to further investigate the NS1 transcript by RT-PCR using a 5′ primer specific to the NS1 gene and 3′ primers in the NP1 gene or further downstream. This revealed that the complete NP1 gene is situated adjacent to the NS1 gene. Furthermore, RT-PCR with a 3′ primer at position 3605 was negative (data not shown). This suggests that NS1 mRNA is polyadenylated downstream of the NP1 gene.
The 3′ RACE results with NP1 and VP2 gene-containing transcripts were positive (Fig. 3C). The VP2 PCR fragment of approximately 100 bp and the NP1 PCR fragment of approximately 790 bp (Fig. 3C) were properly primed on a polyadenosine tail and revealed that the NP1 containing mRNAs are polyadenylated at two positions proximal to the 3′ terminus of the NP1 gene (position 3219 and 3260; Fig. 3D). Upstream of position 3219, a polyadenylation signal is present (position 3199), there is also a polyadenylation signal situated upstream of position 3260 (position 3233; Fig. 3D), and we conclude that both polyadenylation signals proximal to the NP1 gene are in use. The VP2 gene-containing transcript is polyadenylated at position 5075, downstream of a polyadenylation signal at position 5057 (Fig. 3D).
With the identification of the spliced mRNAs, we provide further evidence that a productive HBoV infection was established. For a final control, we tested whether the spliced mRNA could have originated from the clinical samples themselves, instead of being produced in the infected cells. An RT-PCR on the 5′ terminus of the NP1 gene-containing mRNAs was performed on the input materials (0 hpi; Fig. 5) and the cellular RNA at 95 hpi. All clinical samples, including Bonn-1, were negative for spliced mRNA, whereas at 95 hpi, the spliced NP1 mRNAs are present in the cellular mRNA pool of the Bonn-1 virus-inoculated culture (Fig. 5).
FIG. 5.
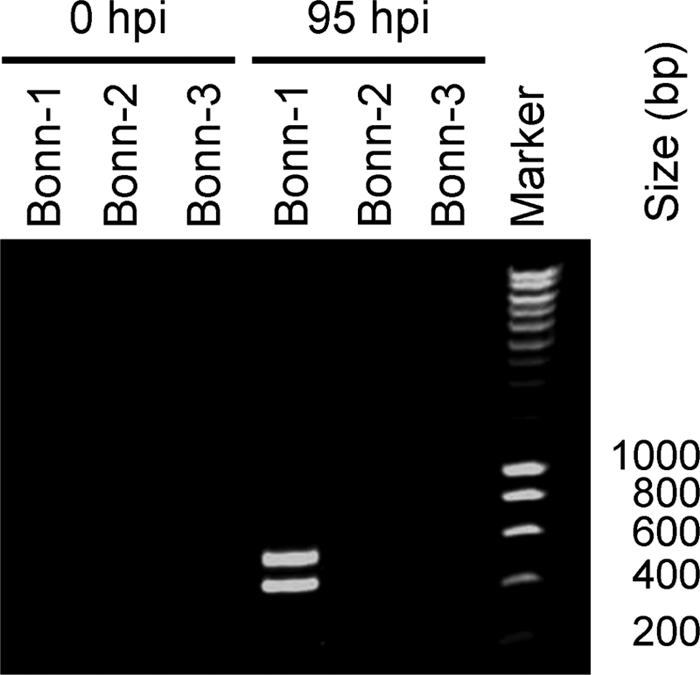
Spliced mRNAs are not present in the inoculum. Agarose gel with ethidium bromide-stained RT-PCR products. Primers spanning the splice junctions in the NP1 mRNA were used for amplification (primers BOCA_5UTR1 and BOCA_R12; Table 1). At 95 hpi, the spliced mRNA RT-PCR is positive (mRNA 2, 445 bp; mRNA 3, 565 bp) for the Bonn-1 HBoV, whereas the inoculum did not contain HBoV mRNA (0 hpi).
In silico analysis of HBoV transcripts.
The VP2 protein is one of the capsid proteins of HBoV. Since humans develop antibodies to this protein after infection, it is clear that expression of the protein indeed occurs (14). As we found no VP2-specific mRNA, it is most likely that mRNA 4 and/or mRNA 5 is the template for VP2 translation. By in silico analysis, we searched for indications of the mechanism of translation of VP2. We searched for IRESs upstream of the VP2 gene with the RegRNA version 1.0 software but did not find any at this location. Next, we examined whether a VP1/VP2 polyprotein might be a target for proteases, in case VP2 is expressed as part of a larger polyprotein. However, no known protease cleavage site is located at the amino-terminal region of the VP2 protein. Finally, we investigated mRNA 4 and 5 in detail to examine whether one of these two transcripts might be better for VP2 expression, in comparison to VP1 expression, but detected no clear signals that link mRNA 4 and mRNA 5 to expression of VP1 and VP2, respectively, or vice versa. However, there is one interesting feature in mRNA 5. This mRNA contains the genome segment from positions 2044 to 2164. Strikingly, this segment contains an AUG codon (position 2116). An AUG codon upstream of the VP1 gene is an indication that translation of VP1/VP2 is inefficient from mRNA 5, although it must be mentioned that the Kozak sequence is poor (Fig. 4B), so deficient scanning of the ribosome complex is not unlikely. Still, in the case that the 2116 AUG in mRNA 5 is used to start translation, a small protein (UP2) is generated, as a stop codon is encountered after 81 codons (TAA at position 3067).
Also in mRNA 3, the genome segment from positions 2044 to 2164 is included. Inspection of mRNA 3 reveals that in the case that the position 2116 AUG is used, a protein (UP1) of 161 amino acids is generated. The first 17 amino acids of UP1 and UP2 are identical to amino acids 622 to 638 of the NS1 protein.
DISCUSSION
This is the first study that presents a culture system for HBoV. Pseudostratified human airway epithelium cell culture can be used as a model for HBoV replication. We observed virus replication after inoculation with a nasopharyngeal washing from an HBoV-infected patient. Apical secretion of HBoV was observed at 72 hpi, but there were no obvious morphological changes to the cells as seen for respiratory syncytial virus (34).
The difficulty of isolating respiratory viruses from clinical material on conventional cell lines has been described in the mid-1960s, leading to ex vivo culturing of human embryo respiratory tract explants (29, 30). Nowadays, the usage of human embryo respiratory tract explants raises ethical issues, but efforts to isolate HBoV from clinical respiratory material on conventional cell lines, like LLC-MK2, HEp-2, Vero, and MRC-5 cells were not successful (10, 19, 35). It is likely that these cell lines are not susceptible to certain respiratory viruses, e.g., because they are deficient in receptor expression or no longer exhibit their cell-specific phenotypes, such as basal, secretory, and ciliated cells. We show that pseudostratified human airway epithelium cell culture is a convenient tool for the isolation and characterization of newly identified respiratory viruses that cannot be cultured on cell lines. This culture system morphologically and functionally resembles the human airways in vivo, and the susceptibility toward viruses coincides with the degree of differentiation. The system also allows characterization of the mode of release and infection of a virus, a major advantage over the respiratory tract explants.
The transcription profile that we present for HBoV displays features that are similar to the transcription map of the closely related BPV and MVC, which belong to the same Bocavirus genus (22, 27). The NS1 mRNAs of BPV and MVC are not spliced, similar what we show for HBoV. Downstream of the NS1 gene, the complete NP1 gene is present on the NS1 mRNA of HBoV. The same has been found in MVC (mRNA R1 and mRNA R2) and in BPV (mRNA R1b), yet in BPV, one of the NS1 mRNAs (R1a) is polyadenylated upstream of the stop codon of NP1 (22, 27). It is, however, unlikely that the NP1 protein of the bocaviruses is expressed from the nonspliced NS1 gene-containing mRNA. In HBoV, the pre-mRNA is alternatively spliced, and the spliced mRNAs are probably used for translation of NP1, VP1, and VP2. In BPV and MVC, two NP1 mRNAs which differ in the site of polyadenylation are found (22, 27). We detected two HBoV NP1 mRNAs (mRNA 2 and mRNA 3) that differ due to alternative splicing. Alternative splicing is also operational for the HBoV transcripts containing the VP1/VP2 ORF. There are two forms, one which is double spliced (mRNA 4) and one which is triple spliced (mRNA 5). Also, in MVC- and BPV-infected cells, two mRNAs that encode VP1/VP2 are generated. One mRNA is spliced once, and the other is spliced three times. The implications of the two alternatively spliced VP1/VP2 mRNAs have not been reported for either BPV or MVC (22, 27). The most remarkable HBoV transcript feature we found was the creation of an ORF by splicing in mRNA 3 and mRNA 5. Due to the retaining of a small part of the NS1 gene, these mRNAs have the potential to encode two previously unknown viral proteins: UP1 and UP2 (18.1 and 8.6 kDa, encoded by mRNA 3 and mRNA 5, respectively). The proteins display no clear homology with other known viral or cellular proteins, and future research is needed to determine whether expression occurs during infection.
The transcript map that we propose was determined for an ST2 type HBoV. Inspection of the ST1 complete genome sequences in GenBank revealed that ST1 will probably generate the same five mRNAs, because the promoter and all splice donor and acceptor sites are conserved for ST1 and ST2 strains. In the case that splicing in ST1 occurs in an manner identical to that in ST2 also, the putative UP1 and UP2 proteins can be produced, as the start and stop codons of UP1 and UP2 are present in the ST1 genomes. Even the ORFx gene is present in ST1 and ST2 strains, although not in all isolates. The reference sequence of ST1 (DQ000495) contains a stop codon 175 nt downstream of the AUG codon, and the ST2 strains from Taiwan (EU984241, EU984240, EU984236, EU984239, EU984242, EU984231, and EU984245) contain an AUG which is located 24 nt upstream of the ST2 ORFx AUG codon. Whether the ORFx protein of ST1 and ST2 is produced during infection is a question that will be subject to future research. We analyzed whether ORFx could be alternatively expressed via an IRES and identified a putative IRES of 98 nt in length located directly upstream of the putative ORFx gene on mRNA transcripts 4 and 5 (data not shown). Translation of ORFs located within the coding region of the VP2 mRNA is not an unusual feature of the parvovirus family (32). Still, whether the IRES is functional remains to be determined.
The capacity to culture HBoV broadens the scope of research possibilities, among which are the identification of the cellular receptor and tropism. Furthermore, and perhaps most important, it provides the possibility to analyze antiviral compounds for their capacity to inhibit viral replication.
Acknowledgments
R.D. and O.S. are supported by the sixth framework grant LSHM-CT-2006-037276 from the European Union. L.V.D.H. is supported by VIDI grant 016.066.318 from The Netherlands Organization for Scientific Research (NWO).
We thank Eric Claas for providing HBoV primer and probe sequences, Marta Canuti for useful discussions, and Ben Berkhout for critical reading of the manuscript.
Footnotes
Published ahead of print on 27 May 2009.
REFERENCES
- 1.Allander, T., T. Jartti, S. Gupta, H. G. Niesters, P. Lehtinen, R. Osterback, T. Vuorinen, M. Waris, A. Bjerkner, A. Tiveljung-Lindell, B. G. van den Hoogen, T. Hyypia, and O. Ruuskanen. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns, K., and C. R. Parrish. 2007. Parvoviridae, p. 2437-2477. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 4.Berns, K. I. 1990. Parvovirus replication. Microbiol. Rev. 54316-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blom, N., J. Hansen, D. Blaas, and S. Brunak. 1996. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 52203-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. Van der Noordaa. 1990. A rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6271-281. [Google Scholar]
- 8.Duckert, P., S. Brunak, and N. Blom. 2004. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17107-112. [DOI] [PubMed] [Google Scholar]
- 9.Fauquet, C. M., and D. Fargette. 2005. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol. J. 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulongne, V., Y. Olejnik, V. Perez, S. Elaerts, M. Rodiere, and M. Segondy. 2006. Human bocavirus in French children. Emerg. Infect. Dis. 121251-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulcher, M. L., S. Gabriel, K. A. Burns, J. R. Yankaskas, and S. H. Randell. 2005. Well-differentiated human airway epithelial cell cultures, p. 183-206. In J. Picot (ed.), Human cell culture protocols. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 12.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 13.Huang, H. Y., C. H. Chien, K. H. Jen, and H. D. Huang. 2006. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 34W429-W434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn, J. S., D. Kesebir, S. F. Cotmore, A. D'Abramo, Jr., C. Cosby, C. Weibel, and P. Tattersall. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J. Infect. Dis. 19841-50. [DOI] [PubMed] [Google Scholar]
- 15.Kapoor, A., E. Slikas, P. Simmonds, T. Chieochansin, A. Naeem, S. Shaukat, M. M. Alam, S. Sharif, M. Angez, S. Zaidi, and E. Delwart. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kesebir, D., M. Vazquez, C. Weibel, E. D. Shapiro, D. Ferguson, M. L. Landry, and J. S. Kahn. 2006. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J. Infect. Dis. 1941276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiemer, L., O. Lund, S. Brunak, and N. Blom. 2004. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kupfer, B., J. Vehreschild, O. Cornely, R. Kaiser, G. Plum, S. Viazov, C. Franzen, R. L. Tillmann, A. Simon, A. Muller, and O. Schildgen. 2006. Severe pneumonia and human bocavirus in adult. Emerg. Infect. Dis. 121614-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, X., R. Endo, N. Ishiguro, T. Ebihara, H. Ishiko, T. Ariga, and H. Kikuta. 2006. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J. Clin. Microbiol. 441132-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning, A., V. Russell, K. Eastick, G. H. Leadbetter, N. Hallam, K. Templeton, and P. Simmonds. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J. Infect. Dis. 1941283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 726014-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu, J., F. Cheng, F. B. Johnson, and D. Pintel. 2007. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 8112080-12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2651-56. [DOI] [PubMed] [Google Scholar]
- 24.Sayers, E. W., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, I. Mizrachi, J. Ostell, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, E. Yaschenko, and J. Ye. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37D5-D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schildgen, O., T. Glatzel, T. Geikowski, B. Scheibner, B. Matz, L. Bindl, M. Born, S. Viazov, A. Wilkesmann, G. Knopfle, M. Roggendorf, and A. Simon. 2005. Human metapneumovirus RNA in encephalitis patient. Emerg. Infect. Dis. 11467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 7915511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, Y., A. Y. Chen, F. Cheng, W. Guan, F. B. Johnson, and J. Qiu. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 833956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, C. I., W. S. Barclay, M. C. Zambon, and R. J. Pickles. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J. Virol. 808060-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyrrell, D. A., M. L. Bynoe, and B. Hoorn. 1968. Cultivation of “difficult” viruses from patients with common colds. Br. Med. J. 1606-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyrrell, D. A. J., and M. L. Bynoe. 1965. Cultivation of novel type of common-cold virus in organ cultures. Br. Med. J. 11467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicente, D., G. Cilla, M. Montes, E. G. Perez-Yarza, and E. Perez-Trallero. 2007. Human bocavirus, a respiratory and enteric virus. Emerg. Infect. Dis. 13636-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zádori, Z., J. Szelei, and P. Tijssen. 2005. SAT: a late NS protein of porcine parvovirus. J. Virol. 7913129-13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 791113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 765654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, L. L., L. Y. Tang, Z. D. Xie, X. J. Tan, C. S. Li, A. L. Cui, Y. X. Ji, S. T. Xu, N. Y. Mao, W. B. Xu, and K. L. Shen. 2008. Human bocavirus in children suffering from acute lower respiratory tract infection in Beijing Children's Hospital. Chin. Med. J. 1211607-1610. [PubMed] [Google Scholar]