Abstract
me53 is a highly conserved baculovirus gene found in all lepidopteran baculoviruses that have been fully sequenced to date. The putative ME53 protein contains a zinc finger domain and has been previously described as a major early transcript. We generated an me53-null bacmid (AcΔme53GFP), as well as a repair virus (AcRepME53:HA-GFP) carrying me53 with a C-terminal hemagglutinin (HA) tag, under the control of its native early and late promoter elements. Sf9 and BTI-Tn-5b1 cells transfected with AcΔme53GFP resulted in a 3-log reduction in budded-virus (BV) production compared to both the parental Autographa californica multiple nucleopolyhedrosis virus and the repair bacmids, demonstrating that although me53 is not essential for replication, replication is compromised in its absence. Our data also suggest that me53 does not affect DNA replication. Cell fractionation showed that ME53 is found in both the nucleus and the cytoplasm as early as 6 h postinfection. Deletion of the early transcriptional start site resulted in a 10- to 360-fold reduction of BV yield; however, deletion of the late promoter (ATAAG) resulted in a 160- to 1,000-fold reduction, suggesting that, in the context of BV production, ME53 is required both early and late in the infection cycle. Additional Western blot analysis of purified virions from the repair virus revealed that ME53:HA is associated with both BV and occlusion-derived virions. Together, these results indicate that me53, although not essential for viral replication, is required for efficient BV production.
Autographa californica multiple nucleopolyhedrosis virus (AcMNPV) is the type species for the genus Alphabaculovirus in the family Baculoviridae. Members of the Baculoviridae are characterized by having enveloped, rod-shaped virions with large circular double-stranded DNA genomes (13). Infection is restricted to arthropods, with the vast majority of known permissive species falling within the class Insecta. A hallmark of baculovirus infection is the production of two morphologically distinct virion phenotypes. At late times of infection, nucleocapsids bud from infected cells to form the budded-virus (BV) phenotype, which is responsible for the transmission of infection from cell-to-cell within a host. However, at very late times in infection, nucleocapsids are enveloped and occlusion-derived virions (ODVs) are packaged into intranuclear occlusion bodies (OBs) (3). OBs generally range from 1 to 5 μm in diameter and are responsible for horizontal transmission of the virus from insect to insect (13).
Baculovirus infection is characterized by the temporal cascade of expression consisting of immediate-early, early, late, and very late gene expression, with the onset of DNA replication separating the early and late phases. Early genes are transcribed by host RNA polymerase II, and some early genes are transcribed from promoters containing a TATA-N(24-28)-CAKT at the transcriptional start site (12). Late genes are transcribed by an α-amanitin-resistant viral RNA polymerase, from ATAAG promoter sequence motifs (2, 12).
The AcMNPV me53 open reading frame (ORF) encodes a protein product of 449 amino acids (52.6 kDa) with a predicted 21-amino-acid C-4 zinc-finger domain. Transcriptionally, me53 was initially characterized as a major early transcript (17), with promoter activity similar to that of the other immediate-early genes ie-0, ie-1, ie-2, and pe38. ie-0, ie-1, ie-2, and pe38 regulate essential mechanisms such as transcriptional transactivation (ie-1, ie-0, ie-2, and pe38), DNA replication (ie-1, ie-2, and pe38), cell cycle progression (ie-2), and BV production (ie-1, pe38, and ie-2) (7, 12, 16, 22, 29, 32). However, unlike ie-0, ie-1, ie-2, and pe38, me53 is also transcribed at late times postinfection (p.i.) initiating from a conserved late promoter motif (18). The me53 promoter structure is therefore very similar to that of the gp64 envelope fusion protein, which is regulated by two functional late promoters and a single early promoter (14). gp64 is the major envelope fusion protein in the group I alphabaculoviruses and is essential for the production of infectious BV (23, 25).
A study by Wang et al. (33) revealed that me53 from Trichoplusia ni single nucleocapsid nucleopolyhedrosis virus could be successfully expressed from its native promoters in a recombinant AcMNPV in the Sf9 and BTI-Tn-5b1 (Hi5) insect cell lines. Their study confirmed the transcriptional profile of AcMNPV me53 and indicated that the T. ni single nucleocapsid nucleopolyhedrosis virus me53 was similarly transcribed from unique sites at both early and late times p.i. Recently, Xi et al. (36) reported that me53 was essential for DNA replication, nucleocapsid generation, and BV production.
In the present study, we investigated the role of me53 in AcMNPV infection of Sf9 and BTI-Tn-5b1 cells using an me53-null virus and a virus expressing hemagglutinin (HA)-tagged ME53. The me53-null virus had a significant decrease in BV production and plaque size in both cell lines. In contrast to the results of Xi et al. (36), we found that me53 is not essential for viral replication and had no negative effect on viral DNA replication during a single virus replication cycle. HA-tagged ME53 protein was detected in both the nucleus and the cytoplasm of infected cells as early as 6 h p.i. We further demonstrated that, although transcribed heavily from the early promoter, the late promoter element appears to be crucial for the expression of ME53 in the context of a successful viral infection. Additionally, we demonstrated that the ME53 protein is associated with both purified BVs and purified ODV.
MATERIALS AND METHODS
Cells.
Sf9 (derived from Spodoptera frugiperda) and BTI-Tn-5b1 (derived from Trichoplusia ni) cells were maintained in Grace's insect medium (Invitrogen) supplemented with 3.33 g of lactalbumin hydrolysate/liter, 3.33 g of Yeastolate/liter, 10% fetal bovine serum, 25 U of penicillin/ml, and 25 ng of streptomycin/ml.
AcΔme53 bacmid construction.
The λ Red recombination system was used to delete the me53 gene from bacmid bMON14272 (Invitrogen) harbored in Escherichia coli DH10 cells as described in Datsenko and Wanner (8). The chloramphenicol acetyltransferase (cat) gene, under the control of a bacterial promoter, with 5′ 50-nucleotide (nt) arms homologous to either the 5′- or 3′-flanking regions of me53 (nt 121154 to 121203 and nt 122555 to 122609 of AcMNPV [NC_001623]) was PCR amplified from plasmid pKD3 by using the primers 5′-ATAACAATACTTTTTATTTAATCTGATTATATTATAACGATACATTTTTAGTGTAGGCTGGAGCTGCTTC-3′ and 5′-TGAACGTGCACAGTATCGTGTTGATTTCTGAGTGCTAACTAACAGTTACACATATGAATATCCTCCTTAGTTC-3′. Amplicons were verified by agarose gel electrophoresis and purified by using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's protocol. Electrocompetent E. coli DH10 cells harboring bMON14272 (AcMNPV bacmid) and pKD46 (λ Red recombinase genes) were transformed with 250 ng of the purified cat amplicon by using the GenePulser Xcell (Bio-Rad). Cells were supplemented with 1 ml of fresh LB medium and incubated for 1 h at 37°C with gentle shaking. Cells were pelleted for 1 min at 10,000 × g, resuspended in 200 μl of LB broth, and plated on LB plates supplemented with kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml). Colonies were subjected to a PCR screen to confirm deletion of me53 and proper genomic insertion of cat.
Repair bacmid construction.
A me53 repair bacmid was constructed by using the Δme53 bacmid and pFACTGFP plasmid, which includes a Tn7 cassette with a gentamicin resistance gene under the control of a bacterial promoter, green fluorescent protein (GFP) under the control of the OpIE1 promoter, and the AcMNPV polyhedrin gene under the control of its native promoter. The entire me53 promoter region, including both the early and the late promoter, and the me53 ORF with an HA tag was PCR amplified by using the primers me53promF (TTACTGAGCTCGTATGTCGGCGTTGTACATG) and me53RHA (TAAGAATTCTTAATAATCAGGAACGTCATAAGGATAGACATTGTTATTTACAATATTAGAATTCTTA; codons for HA tag are underlined, and restriction enzyme sequences are in italics), corresponding to nt 121208 to 122975 of AcMNPV, and cloned into pBluescript by using SacI and EcoRI, creating pBlue53HA. The simian virus 40 poly(A) signal was then amplified from pFastBacHta (Invitrogen) by using the primers polya40F (TAAGAATTCGATCATAATCAGCCATACCAC) and polya40R (TAACTCGAGTCAAGCAGTGATCAGATCC) and cloned into the EcoRI and XhoI sites of pBlue53HA, creating pBlueME53:HApolya. The entire me53 gene fused with the HA tag was then subcloned into pFACTGFP by using SacI and XhoI, creating pFACTGFPme53HA. The Tn7 cassette from pFACTGFPme53HA was transferred to the AcΔ53 bacmid, as described in the Bac-to-Bac expression manual (Invitrogen), creating AcRepME53:HA-GFP.
Transfection.
Bacmid DNA was purified from 500-ml LB cultures using a Qiagen midi-plasmid kit, and concentrations were determined spectrophotometrically by using NanoDrop ND-1000. Sf9 or BTI-Tn-5b1 cells were plated at a density of 9 × 105 cells per well in six-well plates. Cells were transfected with 2 μg of bacmid DNA by using Cellfectin (Invitrogen) according to the manufacturer's protocol for insect cells. Cells were incubated in the presence of the transfection mixture for 5 h, at which time the transfection mixture was removed and replaced with fresh Grace's medium.
Determination of virus titer and viral growth curve.
To monitor the production of BV, Sf9 or BTI-Tn-5b1 monolayers (3 × 106 cells) were seeded in 75-cm2 flasks and transfected with 10 μg of either AcGFP, AcΔme53GFP, or AcRepME53:HA-GFP. At the indicated time points, 200-μl samples of the medium were collected and centrifuged at 1,000 × g for 5 min to pellet the cells. Each supernatant was transferred to a fresh microcentrifuge tube and used to determine the virus titer by endpoint dilution (26), using a range from 100 to 10−7. Plates were scored based on the presence or absence of GFP expression.
Viral plaque assay.
Sf9 or BTI-Tn-5b1 cells were plated at a density of 9 × 105 cells in 35-mm dishes. Cells were transfected with 1 μg of either AcGFP, AcΔme53GFP, or AcRepME53:HA-GFP bacmid using Cellfectin according to the manufacturer's protocol for insect cells. Monolayers were then overlaid with 1% agarose for cell culture (Gibco) in complete Grace's medium. At 5 days posttransfection (p.t.), plaques were photographed and measured. Only well-isolated plaques and plaques greater than about four cells in diameter were included in measurements.
Viral DNA accumulation assay.
A real-time quantitative PCR assay was used to determine the levels of intracellular and total (intracellular plus extracellular, BV) viral DNA for each virus construct. Cell monolayers were seeded into 35-mm dishes at a density of 9 × 105 cells and transfected with 2 μg of either AcGFP, AcΔme53GFP, or AcRepME53:HA-GFP. At 0, 8, 16, and 24 h p.t., DNA was isolated from cells (intracellular DNA) or from both cells and culture medium (total DNA). For intracellular DNA, cell monolayers were harvested into the medium, pelleted at 500 × g for 5 min, resuspended in 200 μl of phosphate-buffered saline (PBS), and stored at −20°C. For total DNA, cell monolayers were harvested directly into the culture medium with a cell scraper and stored together at −20°C. DNA from either preparation was isolated by using a blood minikit (Qiagen) according to the manufacturer's protocol.
The quantitative PCR assay was performed as follows. A PCR master mix included 10 μl of FastStart SYBR green master (Roche), 0.5 μM IE1 forward primer (CACAACACGAACATTTGCACG), 0.5 μM IE1 reverse primer (GTAAACCACATTGCTCACGTAC) and sterile water to a total volume of 18 μl per reaction. A 4-ng portion (in 2 μl) of total intracellular DNA was used as a template for each reaction. The standard curve consisted of eight serial fivefold dilutions of AcMNPV DNA purified from BV ranging from 64 pg to 5 μg. PCR was performed by using a Corbett Rotor-Gene 6200, with the following reaction parameters: 95°C for 15 min and then 30 cycles of 95°C for 15 s, 55°C for 15 s, 72°C for 30 s, and 76°C for 5 s (for data acquisition). This cycling was followed by melting-curve analysis. The data were analyzed by using Rotor-Gene series 6000 software (Corbett Robotics). All products were separated and examined by agarose gel electrophoresis to confirm the expected sizes.
Western blot analysis.
Protein samples were fractionated into cytoplasmic and nuclear fractions as described in Fang et al. (11). Briefly, cell monolayers (in 35-mm dishes) were infected with AcRepME53:HA-GFP at a multiplicity of infection (MOI) of 4. The supernatant was removed, and the cells were scraped with a cell scraper into 1 ml of PBS and pelleted at 1,000 × g for 5 min. The PBS was discarded, and the cell pellets were resuspended in NP-40 lysis buffer (10 mM Tris-Cl [pH 7.9], 10 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.5% [vol/vol] NP-40) in the presence of 1 mM phenylmethylsulfonyl fluoride and complete EDTA-free protease inhibitor cocktail (Roche) and incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 1,000 × g for 3 min. The supernatant containing the cytoplasmic fraction was harvested. Nuclei were resuspended in NP-40 lysis buffer. Samples were mixed with 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and heated to 100°C for 10 min prior to loading. Protein was separated on SDS-10% PAGE gels and transferred to polyvinylidene difluoride membranes. Membranes were blocked by drying and then incubated with primary antibody (monoclonal anti-HA antibody from Sigma at 1:1,000), anti-IE-1 at 1:5,000 (30), and anti-gp64 at 1:1,000 (15) diluted in Tris-buffered saline with Tween 20 (TBST; 50 mM Tris, 150 mM NaCl, 0.1% Tween 20) overnight. Membranes were washed three times for 5 min each time in TBST. Secondary antibody (rabbit anti-mouse antibody-horseradish peroxidase conjugate [Sigma]) was diluted 1:20,000 in TBST. Detection was accomplished by using SuperSignal West Pico chemiluminescent substrate (Pierce).
Deletion mutagenesis.
Both the early and/or the late promoters were separately deleted from pFACTGFPme53 by using QuikChange site-directed mutagenesis (Stratagene). Primers EarlyXO1 (CGATGGCTGAACGTGATCGTGTTGATTTC) and EarlyXO2 (GAAATCAACACGATCACGTTCAGCCATCG) were used to delete the early promoter (creating pFACTGFPxoEme53), and primers LateXO1 (CAATCTTATCTCTATAGGTTTCCATATATAAAGC) and lateXO2 (GCTTTATATATGGAAACCTATAGAGATAAGATTG) were used to delete the late promoter (creating pFACTGFPxoLme53). The dual promoter knockout was created using pFACTGFPxoEme53 as a template with lateXO1 and lateXO2 primers. The bacmids AcΔEme53, AcΔLme53, and AcΔEΔLme53 were created by transformation of DH10B cells harboring AcΔme53 bacmid and helper plasmid pMON7124 (Invitrogen) with pFACTGFPxoEme53, pFACTGFPxoLme53, and pFACTxoEΔLme53, respectively. Appropriate deletions were confirmed by sequencing pFACTGFPxoEme53, pFACTGFPxoLme53, and pFACTxoEΔLme53.
Northern analysis.
Cell monolayers (1.5 × 106) were seeded into 60-mm dishes and transfected with 5 μg of either AcRepME53:HA-GFP, ΔEme53, AcΔLme53, or AcΔEΔLme53. Total RNA was purified at 24 and 48 h p.t. with TRIzol (Invitrogen) according to the manufacturer's protocol. For each sample, total RNA (10 μg) was separated by electrophoresis through a 2.2 M formaldehyde-1.5% agarose gel and blotted onto Immobilon-NY+ membrane (Millipore) using the Turboblotter system (Schleicher & Schuell). An me53-specific 32P-labeled single-stranded RNA probe complementary to me53 RNA (nt 121529 to 121849 of the AcMNPV genome) was synthesized by in vitro transcription as described by Sambrook and Russell (31). Blots were hybridized overnight at 68°C in UltraHyb buffer (Ambion), washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 68°C, washed twice in 0.1× SSC-0.1% SDS at 68°C, exposed to a Phosphoscreen (Amersham), and scanned with a personal molecular imager (Bio-Rad).
BV and ODV purification and Western blot analysis.
BV was purified from supernatants as described in O'Reilly et al. (26). Briefly, Sf9 cells were infected with either AcGFP or AcRepME53:HA-GFP at an MOI of 0.1. At 7 days p.i., viral supernatants were collected from two 125-cm2 flasks and centrifuged at 5,000 × g for 10 min in a Beckman JS 5.3 rotor, and the pellet was kept for isolation of ODV. The BV in the supernatant was pelleted through a 30% sucrose cushion by centrifugation at 24,000 rpm (80,000 × g) in a Beckman SW28.1 rotor for 1 h. The pellet was resuspended in 2 ml of PBS and loaded onto a 25 to 60% continuous sucrose gradient in PBS and spun at 26,500 rpm (96,000 × g) in a Beckman SW28.1 rotor for 3 h at 4°C. The virus band was collected, diluted in PBS, and centrifuged at 24,000 rpm (80,000 × g in a Beckman SW28.1 rotor) for 1 h. The virus pellet was resuspended in 500 μl of 0.1 M Tris (pH 7.4).
ODV was purified as described by O'Reilly et al. (26). Briefly, infected cells were pelleted and resuspended in 0.5% SDS, pelleted, and washed in 0.5 M NaCl. To release ODV, the OBs were pelleted and resuspended in 1 M sodium carbonate and 0.1 M sodium thioglycolate and then incubated at room temperature for 1 h. Liberated ODVs were pelleted through a 30% sucrose cushion by centrifugation at 80,000 × g in a Beckman SW28.1 rotor for 1 h. ODV pellets were resuspended in 0.1 M Tris.
Whole BV and purified ODV for AcGFP and AcRepME53:HA-GFP were separated by SDS-PAGE (10%) and transferred to polyvinylidene difluoride membrane. The membrane was dried and incubated with anti-HA in TBST at a 1:5,000 dilution. The membrane was washed three times in TBST and incubated with rabbit anti-mouse antibody-horseradish peroxidase conjugate (Sigma) at a 1:25,000 dilution in TBST plus 1% skim milk.
RESULTS
Virus replication of AcΔme53GFP, AcRepME53:HA-GFP, and AcGFP bacmids.
To determine whether me53 was essential for virus replication in tissue culture and what role it might play, we constructed an me53-null virus (AcΔme53GFP) and a repair virus expressing HA-tagged ME53 (AcRepME53:HA-GFP) (Fig. 1). AcΔme53GFP, AcRepME53:HA-GFP, and AcGFP bacmids were transfected into Sf9 or BTI-Tn-5b1 cells and monitored daily for GFP expression indicating initiation of replication and for cell-to-cell spread of infection. Early, from 12 to 24 h p.t., all bacmid-transfected monolayers displayed equivalent numbers of GFP positive cells, indicating relatively equal levels and efficiencies of transfection. By 48 h p.t., for both AcRepME53:HA-GFP and AcGFP there was an increased spread of fluorescence to neighboring cells in both Sf9 and BTI-Tn-5b1 cells. In contrast, only localized fluorescence was seen at 48 h p.t. in the AcΔme53GFP-infected cells (Fig. 2A). By 72 h p.t., GFP fluorescence spread throughout most of the monolayer for AcGFP and AcRepME53:HA-GFP, while for AcΔme53GFP only a limited spread of fluorescence was observed, with GFP-positive cells being found either individually or in small clusters of two to three cells each (data not shown). At 96 h p.t., the fluorescence had spread completely to the entire monolayer for AcGFP and AcRepME53:HA-GFP, while for AcΔme53GFP only a limited spread of fluorescence was observed (Fig. 2A). Similar patterns of fluorescence were seen in both Sf9 and BTI-Tn-5b1 cells. Additionally, the limited spread of fluorescence observed for cells infected with AcΔme53GFP virus was maintained throughout three rounds of serial passage of the BV (data not shown).
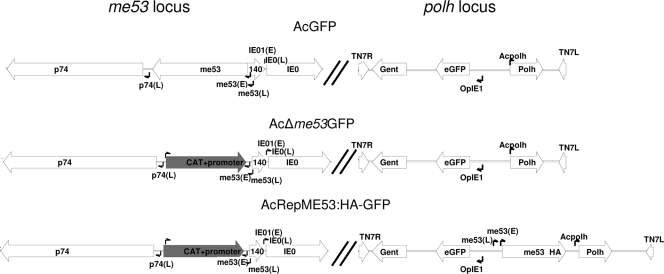
FIG. 1.
Schematic representation of the me53 and TN7 insertion loci for AcGFP, AcΔme53GFP, and AcRepME53:HA-GFP. All GFP viruses contain an eGFP reporter and polyhedrin under the control of its native promoter at the Tn7 site. AcΔme53GFP, me53 was replaced with cat under the control of a prokaryotic promoter. AcRepME53:HA-GFP carries me53 with a C-terminal HA tag under the control of its native early and late promoter at the Tn7 site. Promoter sites for p74, me53, ie0, and Opie1 are indicated by small arrows (E, early; L, late).
FIG. 2.
Cytopathology and GFP fluorescence of AcGFP, AcRepME53:HA-GFP, and AcΔme53GFP in Sf9 and BTI-Tn-5b1 cells. (A) Fluorescence images of monolayers of AcGFP, AcRepME53:HA-GFP, and AcΔme53GFP transfected cells at 48 and 96 h p.t. as indicated. (B) Representative viral plaques from AcGFP, AcRepME53:HA-GFP and AcΔme53GFP transfected Sf9 and BTI-Tn-5b1 monolayers at 120 h p.t. The inset shows the means and standard deviations of plaque sizes from 20 well- isolated plaques from each bacmid.
In order to monitor the effect of the me53 deletion on viral spread, a GFP fluorescent plaque assay was performed at 5 days p.t. on cell monolayers transfected with either AcGFP, AcRepME53:HA-GFP, or AcΔme53GFP. The diameters of only well-isolated plaques were measured (20 for each virus). Both AcGFP and AcRepME53:HA-GFP produced large plaques with mean diameters of 941 ± 104 μm and 938 ± 119 μm, respectively, in the Sf9 cells (Fig. 2B). The largest isolated plaques were 1,138 μm and 1,211 μm for AcGFP and AcRepME53:HA-GFP, respectively. The AcΔme53GFP plaques were significantly smaller in size, with a mean diameter of 387 ± 140 μm for plaques larger than four cells in diameter (Fig. 2B). The plaque sizes for AcΔme53GFP varied greatly from single cells to the largest plaque measuring 624 μm in diameter. A similar pattern was observed in transfected BTI-Tn-5b1 cells. Both AcGFP and AcRepME53:HA-GFP produced plaques of similar sizes (1,245 ± 152 μm and 1,207 ± 167 μm, respectively), while AcΔ53GFP produced significantly smaller plaques (357 ± 131 μm). Since only plaques greater than about four cells were included in the above calculations, and ca. 50% of the plaques from the AcΔ53GFP virus were smaller than four cells in diameter, the actual average plaque sizes for AcΔ53GFP are smaller than indicated in Fig. 2B.
Since smaller plaques for AcΔ53GFP could be due to release of fewer BVs and therefore reduced transmission to neighboring cells, a time course of extracellular BV production was monitored over 96 h after transfection with either 2 μg of AcΔme53GFP, AcRepME53:HA-GFP, or AcGFP bacmid DNA (Fig. 3A and B). The yield and growth kinetics of BV for AcRepME53:HA-GFP were similar to those of AcGFP (data not shown). For AcRepME53:HA-GFP and AcGFP, BV production was detected by 24 h p.t. in both cell lines tested. Infectious BV production for AcΔme53GFP was not detected until 48 h p.t. Throughout the time course, the infectious BV titer of AcΔme53GFP was consistently at least 3 to 4 logs lower than the titers of either AcRepME53:HA-GFP or AcGFP for both the Sf9 and the BTI-Tn-5b1 cell lines.
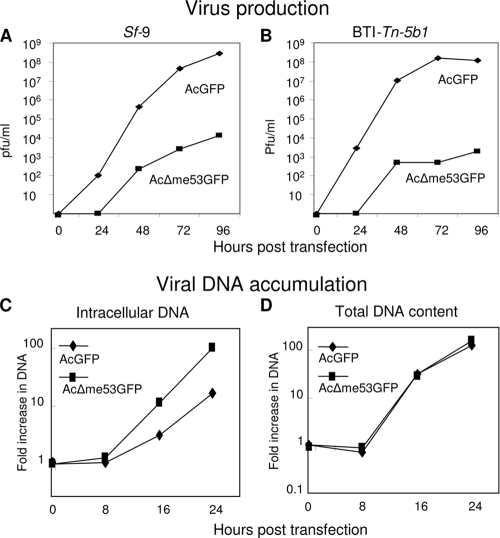
FIG. 3.
Virus growth curves of AcGFP and AcΔme53GFP in Sf9 (A) or BTI-Tn-5b1 cells (B) and viral DNA accumulation (C and D). Monolayers were transfected with either AcGFP or AcΔme53GFP bacmid, and supernatant samples were collected at the indicated time points. Note that time zero is when the transfection supernatant is removed and replaced with fresh medium. Sf9 monolayers were transfected with 2 μg of either AcGFP or AcΔme53GFP, and the fold increase in intracellular DNA (C) or total DNA (intracellular and budded virus) (D) was determined at the times noted. For panels C and D, the fold increase was calculated by dividing by viral DNA copy number at time zero. The data represent the averages of three independent experiments.
DNA replication analysis of AcΔme53GFP, AcRepME53:HA-GFP, and AcGFP bacmids.
To determine whether the decreased level of AcΔme53GFP BV production was due to decreased viral DNA replication, a quantitative PCR assay was utilized to monitor the accumulation of just intracellular and of total DNA (intracellular plus extracellular BV DNA) in Sf9 monolayers transfected with either AcΔme53GFP or AcGFP bacmids. This assay was restricted to a single replication cycle over the first 24 h p.t. to avoid the inclusion of viral DNA resulting from secondary infection of nontransfected cells. There was no increase in viral DNA content between 0 and 8 h p.t. for both viruses. The level of intracellular DNA started to accumulate at 16 h p.t., and by 24 h p.t. the level of DNA for AcΔme53GFP increased 102-fold, while that for AcGFP increased only 16.3-fold (Fig. 3C). The lower level of intracellular viral DNA for AcGFP compared to AcΔme53GFP might have been due to the loss of viral DNA from the cell in the form of extracellular BV. Therefore, we also monitored the level of total viral DNA (intracellular and extracellular) at different time points. By 16 h p.t. there were 30- and 33-fold increases in the total viral DNA over time zero for AcGFP and AcΔme53GFP, respectively (Fig. 3D). By 24 h p.t., with AcGFP and AcΔme53GFP, there were 120- and 156-fold increases, respectively, in viral DNA accumulation relative to time zero (Fig. 3D). Thus, the total DNA produced for AcΔme53GFP was equivalent to that of AcGFP, except that most of the DNA for AcΔme53GFP-infected cells was intracellular, while that for AcGFP was extracellular in BV. This demonstrated that deletion of me53 did not affect overall viral DNA replication kinetics.
Time course of ME53:HA protein production.
AcRepME53:HA-GFP demonstrated identical growth kinetics to AcGFP and allowed for the utilization of HA specific antibodies to follow the expression of ME53 during the infection cycle. Cell monolayers were infected with AcRepME53:HA-GFP at an MOI of 4, collected at various times p.i. and fractionated into nuclear and cytoplasmic fractions. The fractions were analyzed by SDS-PAGE and Western blot analysis with either anti-HA, anti-IE1, or anti-gp64 monoclonal antibodies (Fig. 4A). ME53:HA bands were detected predominantly in the nuclear fraction of both Sf9 and BTI-Tn-5b1 cells, from as early as 6 h p.i., and the intensity increased through to 48 h p.i. ME53:HA could also be detected in the cytoplasmic fractions of both cell lines beginning at 6 h p.i., although at lower levels than those seen in the nuclear fractions. The nuclear proteins IE-0 and IE-1 (24) and the cytoplasmic GP64 (27) were detected only in the nuclear and cytoplasmic fractions, respectively, confirming the effectiveness of cellular fractionation (Fig. 4A).
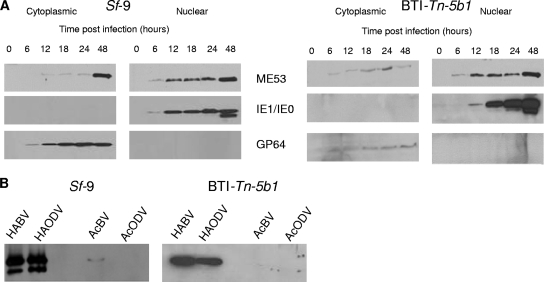
FIG. 4.
Western immunoblot of infected cells or virus particles. Sf9 and BTI-Tn-5b1 cells were infected with AcRepME53:HA-GFP. (A) Cells were collected and fractionated into cytoplasmic and nuclear fractions at various times p.i. as indicated. Blots were probed with anti-HA, anti-IE1, or anti-gp64 antibodies. (B) AcRepME53:HA-GFP BVs (HABV) and ODVs (HAODV) were purified and probed with anti-HA to detect HA-tagged ME53. Parental AcMNPV virus (AcBV and AcODV) was included as a control to ensure there was no cross-reactivity with the antibodies. Electron microscopy was used to confirm the isolation of intact BVs.
Western blot analysis of BV and ODV.
Since AcGFPΔme53 demonstrated a reduced ability to produce BV and was found at late times by Western blot analysis, we hypothesized that ME53 is important for virion morphogenesis and might be physically associated with BVs. BVs were purified from the extracellular medium of AcRepME53:HA-GFP-infected monolayers of both Sf9 and BTI-Tn-5b1cells. The HA-tagged ME53 was detected in purified BV samples from the AcRepME53:HA-GFP monolayers (HABV, Fig. 4B). To ensure that the detected protein was not a result of cross-reactivity with either the primary or secondary antibodies, Western analysis, using anti-HA, was also conducted on purified AcGFP BV and ODV (Fig. 4B, lanes AcBV and AcODV). Slight cross-reactivity was detected in the AcBV sample with a protein similar in size to ME53:HA; however, this was at much lower levels compared to the signal for the HA-tagged samples. ODVs were also isolated from OBs in infected monolayers. ME53:HA was associated with ODV (HAODV), as well as the BVs derived from either Sf9 or BTI-Tn-5b1-infected cells (Fig. 4B). BV and ODV samples from Sf9 cells displayed two anti-HA specific bands, while samples from BTI-Tn-5b1 did not. This may be the result of protein processing or posttranslational modification in Sf9 cells.
Analysis of me53 early and late promoters.
Given the strong expression of ME53:HA at late times, we decided to further characterize the relative importance of the early and late me53 promoter elements in virus replication. Site-directed mutagenesis was used to delete either the CACAGT (−40) early promoter element transcriptional start site (AcΔEme53), the ATAAG late promoter and transcriptional start site (−82) (AcΔLme53) (1), or both promoter elements (AcΔEΔLme53) from the pFACTme53 repair plasmid (Fig. 5A). The deletions in the resulting plasmids were confirmed by sequencing and these plasmids were used to repair AcΔme53 bacmid at the Tn7 transposition sites. The amount of infectious BV was determined for each bacmid at 5 days p.t. with equal amounts of bacmid DNA. The BV titer for Sf9 monolayers transfected with AcΔEme53 was 360-fold lower than with AcRepME53:HA-GFP, while titers of the AcΔLme53 and AcΔEΔLme53 transfected monolayers were even further compromised with more than 1,000- and 5,000-fold reductions, respectively, compared to AcRepME53:HA-GFP levels (Fig. 5B). Promoter deletion had less of an effect on titers from transfected BTI-Tn-5b1; however, a similar reduction in BV production appeared, with AcΔEme53 demonstrating a 10-fold reduction and AcΔLme53 and AcΔEΔLme53 showing 160- and 200- fold reductions, respectively, compared to AcRepME53:HA-GFP (Fig. 5B). Northern analysis was used to ensure that promoter deletion resulted in a reduction of me53 transcription (Fig. 5C). Deletion of the promoter elements resulted in a significant reduction in me53 expression at both 24 and 48 h p.t. compared to the repair construct.
FIG. 5.
(A) Nucleotide sequence of the me53 upstream region for AcGFP, AcΔEΔLme53, AcΔLme53, and AcΔEme53. The early promoter is italicized and underlined, the late promoter is boldface and underlined, and the TATA element is underlined. Crossed-out nucleotides indicate those deleted in the constructs. (B) Titers of AcΔEme53, AcΔLme53, AcΔEΔLme53, or AcGFP bacmid transfected monolayers at 5 days p.t. of Sf9 and BTI-Tn-5b1 cells. The data represent the averages of two independent experiments. (C) Northern blot analysis of me53 transcripts in cells transfected with AcRepME53:HA-GFP, AcΔEΔLme53, AcΔLme53, and AcΔEme53 at 24 and 48 h p.t. Ribosomal bands are shown below to indicate relative RNA loading.
DISCUSSION
AcMNPV me53, a zinc finger protein gene found in all lepidopteran baculoviruses sequenced to date, is strongly transcribed by 1 h p.i. as a major immediate-early transcript (17, 18). However, the function and localization of ME53 in baculovirus infection has remained mainly uncharacterized. Here, we describe the functional characterization of a me53 deletion virus (AcΔme53GFP). In addition, we repaired the deletion virus with a HA-tagged version of ME53, which behaved similarly to the native ME53. Our results demonstrated that deletion of me53 resulted in greatly reduced BV titers leading to reduced levels of cell-to-cell virus transmission and reduced plaque size. Despite the reduced ability to produce BV, cells initially transfected with AcΔme53GFP demonstrated typical CPE and OB production, as determined by light microscopy. Furthermore, these observations were consistent in cell lines from two different lepidopteran species, demonstrating the phenotype observed is likely not species specific.
Xi et al. (36) concluded that no BV was produced for a me53-null bacmid and suggested that me53 was essential for DNA replication. However, we detected infectious BV in the supernatant of AcΔme53GFP transfected monolayers, albeit at 3 to 4 orders of magnitude lower than AcGFP. Since Dai et al. (7) demonstrated that DNA replication from secondary infection of adjacent cells in a transfected monolayer likely begins around 28 to 30 h p.t., we restricted our DNA replication analysis to the first 24 h p.t. to eliminate detecting viral DNA derived from secondary infection. Our analysis of DNA replication over the first 24 h p.t., which is thus more representative of a single virus replication cycle, revealed that deletion of me53 did not affect the accumulation of viral DNA in the initially transfected cells compared to both parental and repair bacmids. Although the AcΔme53GFP virus showed greater accumulation of intracellular viral DNA than AcGFP, there was no significant difference in total viral DNA (i.e., intracellular and BV) levels between AcΔme53GFP and AcGFP. This observation is consistent with the delay in BV production seen from AcΔme53GFP-transfected monolayers (Fig. 3A and B), since viral DNA is mostly retained within the cell, while for AcGFP most of the viral DNA is found in the BVs, but the same total amount of viral DNA is made by both viruses. Our data clearly indicate that the defect in BV production is not due to lack of DNA replication, as suggested by Xi et al. (36), but must lie beyond DNA replication, possibly in the virus morphogenesis pathway or another later part of the viral replication cycle. In the study by Xi et al. (36), their first DNA replication time point was 24 h p.t. at which point they observed no significant differences between the knockout and wild-type viruses in agreement with the results of the present study. It is possible that these authors did not detect viral DNA replication because the first 24 h p.t. were not examined in detail. In addition, they reported that for parental bacmid, by 72 h p.t. all cells are completely filled with OBs, which represents the end of the viral replication cycle. In contradiction to this, however, they did not detect significant viral DNA replication levels for their wild-type control virus until 96 to 120 h p.t.
Western blot analysis showed that HA-tagged ME53 was present in both the nucleus and the cytoplasm as early as 6 h p.i. in both cell lines. This expression profile is consistent with the findings of Knebel-Morsdorf et al. (17), who detected me53 specific transcripts by 1 h p.i. A subsequent study by Knebel-Morsdorf et al. (18) using primer extension demonstrated a switch from an early to a late promoter at around 12 h p.i. Transcription from a late promoter is also consistent with the expression pattern seen in our Western blot analysis since ME53:HA was readily detectable at late (24 h) and very late (48 h) times p.i. Although found in both the nucleus and cytoplasm, the predominant localization of ME53:HA was to the nucleus, in keeping with its possible function as a transcription factor. Nevertheless, at late times the level in the cytoplasm remained constant, suggesting that a proportion remains sequestered there, similar to the situation for the herpesvirus transcription factor VP16 (35).
Regulation of me53 by both early and late promoters is similar to that of AcMNPV gp64, which codes for the BV-specific GP64 fusion protein (4, 14, 18). To determine the importance of the me53 early and late promoters in viral replication, we deleted either the early (CACAGT), the late (ATAAG), or both promoters from the upstream region of the pFACTGFPme53HA repair plasmid. BV titers after transfection of Sf9 cells with bacmids lacking the me53 early promoter, but still containing the late promoter, were ∼300-fold lower than the AcRepME53:HA-GFP control, indicating that early expression of me53 is needed for efficient BV production. However, deletion of the late promoter, while leaving the early promoter intact, resulted in an even greater, >1,000-fold, reduction in the BV titer. Deletion of both early and late promoters, resulted in the greatest (>5,000-fold) reduction compared to AcRepME53:HA-GFP, which is similar to the reduction seen in AcΔme53GFP. Titers obtained from transfection of BTI-Tn-5b1 cells displayed a lower AcRepME53:HA-GFP yield but a pattern similar to that of AcΔEme53, AcΔLme53, and AcΔEΔLme53 with 10-, 160-, and 200- fold reductions, respectively. Although the CACAGT early promoter element was deleted, this deletion is likely not sufficient to completely abolish early me53 transcription. Blissard et al. (5) demonstrated that baculovirus early gene transcription can initiate in the presence of a TATA element and in the absence of a CAGT transcription initiation site, but at reduced levels. In addition, common sequence elements such as GATA and CACGTG, present in the upstream region of me53 (−202 and −296, respectively [data not shown]), can contribute to TATA-dependent transcription (19). In consideration of these findings, it is difficult to determine the relative importance of early versus late expression of me53 in the context of BV production based on the present study. Nevertheless, our results indicate that the late promoter is a crucial element in terms of the regulation of me53 expression and a concomitantly high level of BV production. These findings suggest that ME53 plays a vital role in viral replication, especially during the late phase of the infection cycle, when BV and ODV production occurs.
Since deletion of the late promoter resulted in a 1,000-fold reduction in BV and ME53:HA was detected late in the infection cycle, at the time of virion morphogenesis, ME53 might act as a scaffolding protein or be a component of BVs or ODVs. Western blot analysis of BVs and ODVs from recombinant viruses demonstrated that ME53 was associated with both the BVs and ODVs. Interestingly, mass spectrometry (MS) analysis of AcMNPV ODV by Braunagel et al. (6) did not detect ME53 as a component of the ODVs. A more recent study by Deng et al. (9) on Helicoverpa armigera SNPV (HearNPV) ODVs also did not conclusively identify ME53, but MS peptide footprints that matched ME53 with low MOWSE scores were reported. There are other examples of baculoviral proteins that have been identified as components of virions but were not found in any of the MS analyses to date. For example, both HA122 and PIF-2 from HearNPV were identified as components of HearNPV ODVs by Western blotting but were not detected by MS (9, 10, 20). In addition, Fang et al. (11) identified EXON-0 (Ac141) as a component of both AcMNPV BVs and ODVs; however, this protein was also not detected by MS analysis (6). These results highlight the limitation of MS analysis and do not preclude the possibility that virion proteins might escape detection by MS (6).
There are several possibilities why ME53 would be associated with baculovirus virions. The first is that it may be acting as a scaffolding protein, such as ICP35 from herpes simplex virus (HSV), which coordinates virion assembly within the nuclei of infected cells (28). In the case of HSV, ICP35 is released from the virion during the maturation process. If ME53 is acting in a similar, transient manner, it may explain why MS analysis has failed to detect ME53 in ODVs (6, 9). A second possibility is that ME53 is acting as a transcriptional regulator of viral and/or insect genes and that having it present immediately upon infection of host cells confers an advantage. One classical example of this type of transcription factor is the HSV VP16, a protein found within the tegument of HSV particles that interacts with host factors Oct-1 and HCF-1 to induce immediate-early viral promoters (35). Association of transcription factors with virions has also been noted for AcMNPV, since Braunagel et al. (6) identified the early gene transactivator IE-1 and the very late transactivator VLF-1 as components of AcMNPV ODVs. A third possibility is that ME53 is packaged indiscriminately within virions only as a consequence of its presence within the nucleus of infected cells at late times during the infection cycle when virion morphogenesis and assembly occurs.
Homologues of me53 are found in the genomes of all fully sequenced lepidopteran baculoviruses. The equivalent results obtained from the two cell lines used in the present study, derived from different lepidopterans (S. frugiperda and T. ni), suggests that me53 plays a fundamental role, in a species-independent manner, in successful baculovirus replication. In contrast, deletion of the highly conserved immediate-early gene ie-2 results in different viral phenotypes in Sf21, BTI-Tn-5b1, and Tn-368 cell lines (21, 29). Therefore, me53 may serve a more fundamental role in the baculovirus life cycle than other immediate-early genes such as ie-2.
Deletion of me53 resulted in a phenotype that is very similar to that seen for an AcMNPV exon0 (ac141) knockout virus (7). Deletion of exon0 also results in decreased BV production (>10,000-fold decrease) and, similar to what we observed for AcΔme53GFP, exon0-null AcMNPV, had no negative effect on DNA replication. Much like ME53, the EXON0 protein product is a structural component of BVs and ODVs and is associated with nucleocapsids (11). This same study also indicated that exon0 interacts with other nucleocapsid proteins, FP25 and BV/ODV-C42, and may play a role in the egress of nucleocapsids from the nucleus, rather than in nucleocapsid assembly.
Our data show that ME53 is important for morphogenesis of infectious BV that is not directly related to DNA replication. It is possible that ME53 may act as a transcriptional regulator, as is common of zinc finger proteins (34), regulating genes involved in nucleocapsid or virion morphogenesis. Alternatively, since ME53 appears to be a structural component of both BVs and ODVs, it is possible that ME53 may be acting as a scaffolding or packaging protein. Production of OBs did not appear to be affected by the loss of ME53, but further studies such as electron microscopy analysis are needed to assess any impact on ODV morphogenesis. Since some BV is produced in the absence of ME53, it is possible that another protein assumes the same role if ME53 is lacking, although at a significantly reduced efficiency.
Acknowledgments
This study was funded by an NSERC discovery and strategic grant to P.J.K. and B.M.A.
We acknowledge D. Leishman for his assistance.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Becker, D., and D. Knebel-Mosdorf. 1993. Sequence and temporal appearance of the early transcribed baculovirus gene HE65. J. Virol. 675867-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beniya, H., C. J. Funk, G. F. Rohrmann, and R. F. Weaver. 1996. Purification of a virus-induced RNA polymerase from Autographa californica nuclear polyhedrosis virus. Virology 202586-592. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 2073-93. [DOI] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: modulating early transcription and transactivation by IE1. J. Virol. 655820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blissard, G. W., P. H. Kogan, R. Wei, and G. F. Rohrmann. 1992. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology 190783-793. [DOI] [PubMed] [Google Scholar]
- 6.Braunagel, S. C., W. K. Russell, G. Rosas-Acosta, D. H. Russell, and M. D. Summers. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. USA 1009797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, X., T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 789633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, F., R. Wang, M. Fang, Y. Jiang, X. Xu, H. Wang, X. Chen, B. M. Arif, L. Guo, H. Wang, and Z. Hu. 2007. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. J. Virol. 819377-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, M. G., H. Wang, L. Yuan, X. Chen, J. M. Vlak, and Z. Hu. 2006. Open reading frame 132 of Helicoverpa armigera nucleopolyhedrovirus encodes a functional per os infectivity factor (PIF-2). J. Gen. Virol. 872563-2569. [DOI] [PubMed] [Google Scholar]
- 11.Fang, M. G., X. Dai, and D. A. Theilmann. 2007. Autographa californica multiple nucleopolyhedrovirus EXON0 (ORF141) is required for efficient egress of nucleocapsids from the nucleus. J. Virol. 819859-9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-170. In L. K. Miller (ed.), The baculoviruses. Plenum Press, Inc., New York, NY.
- 13.Funk, C.J., S. C. Braunagel, and G. F. Rohrmann. 1997. Baculovirus structure, p. 7-32. In L. K. Miller (ed.), The baculoviruses. Plenum Press, Inc., New York, NY.
- 14.Garrity, D. B., M.-J. Chang, and G. W. Blissard. 1997. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology 231167-181. [DOI] [PubMed] [Google Scholar]
- 15.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125432-444. [DOI] [PubMed] [Google Scholar]
- 16.Imai, N., N. Matsuda, K. Tanaka, A. Nakano, S. Matsumoto, and W. Kang. 2003. Ubiquitin ligase activities of Bombyx mori nucleopolyhedrovirus RING finger proteins. J. Virol. 77923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knebel-Morsdorf, D., A. Kremer, and F. Jahnel. 1993. Baculovirus gene ME53, which contains a putative zinc finger motif, is one of the major early-transcribed genes. J. Virol. 67753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knebel-Morsdorf, D., J. T. Flipsen, R. Roncarati, F. Jahnel, A. W. Kleefsman, and J. M. Vlak. 1996. Baculovirus infection of Spodoptera exigua larvae: lacZ expression driven by promoters of early genes pe38 and me53 in larval tissue. J. Gen. Virol. 77815-824. [DOI] [PubMed] [Google Scholar]
- 19.Kogan, P. H., X. Chen, and G. W. Blissard. 1995. Overlapping TATA-dependent and TATA-independent early promoter activities in the Baculovirus gp64 envelope fusion protein gene. J. Virol. 691452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, G., X. Chen, D. Peters, J. M. Vlak, and Z. Hu. 2003. Open reading frame 122 of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus encodes a novel structural protein of occlusion-derived virions. J. Gen. Virol. 84115-121. [DOI] [PubMed] [Google Scholar]
- 21.Lu, A., and L. K. Miller. 1995. Differential requirements for baculovirus late expression factor genes in two cell lines. J. Virol. 696265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milks, M. L., J. O. Washburn, L. G. Willis, L. E. Volkman, and D. A. Theilmann. 2003. Deletion of pe38 attenuates AcMNPV genome replication, budded virus production, and virulence in Heliothis virescens. Virology 310224-234. [DOI] [PubMed] [Google Scholar]
- 23.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 704607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254297-314. [DOI] [PubMed] [Google Scholar]
- 26.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1994. Baculovirus expression vectors: a laboratory manual. W. H. Freeman and Company, New York, NY.
- 27.Pearson, M. N., R. L. Q. Russel, and G. F. Rohrmann. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 2922-31. [DOI] [PubMed] [Google Scholar]
- 28.Pelletier, A., F. Dô, J. J. Brisebois, L. Lagace, and M. G. Cordingley. 1997. Self-association of herpes simplex virus type I ICP35 is via coiled-coiled interactions and promotes stable interaction with the major capsid protein. J. Virol. 715197-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prikhod'ko, E. A., A. Lu, J. A. Wilson, and L. K. Miller. 1999. In vivo and in vitro analysis of baculovirus ie-2 mutants. J. Virol. 732460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross, L., and L. A. Guarino. 1997. Cycloheximide inhibition of delayed early gene expression in baculovirus-infected cells. Virology 232105-113. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Theilmann, D. A., and S. Stewart. 1992. Molecular analysis of the transactivating IE-2 gene of Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 18784-96. [DOI] [PubMed] [Google Scholar]
- 33.Wang, W., S. Davison, and P. J. Krell. 2004. Identification and characterization of a major early-transcribed gene of Trichoplusia ni single nucleocapsid nucleopolyhedrovirus using the baculovirus expression system. Virus Genes. 2919-29. [DOI] [PubMed] [Google Scholar]
- 34.Wolfe, S. A., H. A. Greisman, E. I. Ramm, and C. O. Pabo. 1999. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 2851917-1934. [DOI] [PubMed] [Google Scholar]
- 35.Wysocka, J., and W. Herr. 2003. The Herpes Simplex Virus VP16 induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28294-304. [DOI] [PubMed] [Google Scholar]
- 36.Xi, Q., J. Wang, R. Deng, and X. Wang. 2007. Characterization of AcMNPV with a deletion of me53 gene. Virus Genes 34223-232. [DOI] [PubMed] [Google Scholar]