Abstract
Viruses in the genus Orthobunyavirus, family Bunyaviridae, have a genome comprising three segments (called L, M, and S) of negative-sense RNA. Serological studies have classified the >170 named virus isolates into 18 serogroups, with a few additional as yet ungrouped viruses. Until now, molecular studies and full-length S-segment nucleotide sequences were available for representatives of eight serogroups; in all cases, the S segment encodes two proteins, N (nucleocapsid) and NSs (nonstructural), in overlapping open reading frames (ORFs) that are translated from the same mRNA. The NSs proteins of Bunyamwera virus (BUNV) and California serogroup viruses have been shown to play a role in inhibiting host cell mRNA and protein synthesis, thereby preventing induction of interferon (IFN). We have determined full-length sequences of the S segments of representative viruses in the Anopheles A, Anopheles B, and Tete serogroups, and we report here that these viruses do not show evidence of having an NSs ORF. In addition, these viruses have rather longer N proteins than those in the other serogroups. Most of the naturally occurring viruses that lack the NSs protein behaved like a recombinant BUNV with the NSs gene deleted in that they failed to prevent induction of IFN-β mRNA. However, Tacaiuma virus (TCMV) in the Anopheles A serogroup inhibited IFN induction in a manner similar to that of wild-type BUNV, suggesting that TCMV has evolved an alternative mechanism, not involving a typical NSs protein, to antagonize the host innate immune response.
The family Bunyaviridae is one of the largest taxonomic groupings of RNA viruses, containing well over 300 named viruses (8, 25). Among the unifying characteristics of these viruses is possession of a trisegmented single-stranded RNA genome of negative-sense or ambisense polarity that encodes four structural proteins. The three genome segments (called L [large], M [medium], and S [small]) are encapsidated by the nucleocapsid (N) protein and are associated with the viral RNA-dependent RNA polymerase, the L protein, to form ribonucleoprotein complexes termed nucleocapsids. The three ribonucleoprotein complexes are contained within a lipid envelope into which are embedded two viral glycoproteins, Gn and Gc. Virus replication occurs in the cytoplasm of infected cells, and viruses mature primarily by budding at Golgi membranes (for reviews, see references 14, 16, and 30).
The Bunyaviridae (collectively referred to as bunyaviruses) are classified into five genera—Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus—on the basis of serological and molecular characteristics (25). Within a genus, viruses show similar patterns in the sizes of their genome segments and structural proteins and whether or not nonstructural proteins are encoded. In addition, the three genomic RNA segments characteristically have conserved, complementary terminal sequences that are similar for the three segments of viruses within a genus but differ between genera. Viruses that impinge on human health, either directly by causing disease in humans or indirectly by causing economic losses of domestic animals or crop plants, are found in each of the five genera and are the cause of many examples of emerging diseases (13, 24). Orthobunyaviruses, nairoviruses, and phleboviruses are transmitted by blood-sucking arthropods to vertebrate hosts while tospoviruses are transmitted by various thrips species to plants. The hantaviruses do not have an arthropod vector but are maintained in nature as persistent infections of rodents, and humans become infected by inhaling aerosolized rodent excreta (16, 28, 29).
The largest genus of bunyaviruses is the Orthbobunyavirus genus that contains more than 170 named viruses (8, 25); the prototype of the genus is Bunyamwera virus (BUNV). Among important orthobunyavirus pathogens of humans are La Crosse virus (LACV) that causes pediatric encephalitis, Oropouche virus (OROV) that causes a debilitating febrile illness, and Ngari virus that causes hemorrhagic fever (1) while Aino, Akabane, and Cache Valley viruses are examples of viruses causing disease in domestic animals.
Classification of orthobunyaviruses has proven to be a complex issue. The majority of viruses have been placed in one of 18 serogroups based on serological relatedness of complement-fixing antibodies (mediated by the N protein) and hemagglutinating and neutralizing antibodies (mediated by the glycoproteins) though a number of viruses classified into the Orthobunyavirus genus are currently not assigned to any of these serogroups (8, 25). The 18 serogroups are Anopheles A, Anopheles B, Bakau, Bunyamwera, Bwamba, group C, Capim, California, Gamboa, Guama, Koongol, Minatitlan, Nyando, Olifanstlei, Patois, Simbu, Tete, and Turlock. Serological relatedness varies within a serogroup and is further complicated by the occurrence of natural reassortant viruses, such that viruses may be more related to members of one group or another depending on the assay used (7, 23).
The latest report of the International Committee for the Taxonomy of Viruses delineates the orthobunyaviruses into 44 species (25) though such delineation must be regarded as fluid due to the paucity of molecular characterization. Comprehensive molecular genetic studies have involved viruses in only four serogroups, namely, Bunyamwera, group C, California, and Simbu (5, 12, 26, 27), but recently S-segment nucleotide sequences have been obtained for one or two representatives of the Bakau, Bwamba, Nyando, and Turlock serogroups (20, 35). In all cases the S genome segment encodes two proteins, N and, in an overlapping open reading frame (ORF), a small nonstructural protein termed NSs. N and NSs proteins are translated from the same viral mRNA species as the result of alternate AUG-initiation codon selection (3, 17). The NSs proteins of BUNV and LACV have been shown to play an important role in counteracting the host antiviral defense and in viral pathogenesis (4, 6, 34). This is achieved by inhibition of host cell mRNA and protein synthesis, thereby suppressing production of interferon (IFN) through perturbation of phosphorylation of the C-terminal domain of RNA polymerase II (32). Recent evidence indicates that for BUNV, interaction of NSs with the Mediator protein MED8 is critical in this process (21).
In order to learn more about orthobunyavirus diversity and in particular the role of NSs protein, we have undertaken nucleotide sequence analysis of the S genome segments of representatives of the unstudied serogroups. Here, we report analyses of viruses in the Anopheles A, Anopheles B, and Tete serogroups. The surprising finding is that these viruses do not encode a recognizable NSs overlapping ORF. Most of these viruses behaved like a genetically engineered BUNV isolate, BUNdelNSs, in which the NSs gene was deleted, in inducing IFN-β mRNA in infected cells, whereas wild-type BUNV (wtBUNV) efficiently blocked this induction (6, 34). However, Tacaiuma virus (TCMV) in the Anopheles A serogroup, despite the lack of an NSs protein, was also able to block IFN-β mRNA production.
MATERIALS AND METHODS
Viruses.
The viruses used in this study were graciously provided by the late R. Shope (University of Texas Medical Branch, Galveston, TX) and are listed in Table 1. All viruses except Tete virus (TETEV) were supplied as lyophilized suckling mouse brain extracts; TETEV had been lyophilized after a single passage in Vero cells. After reconstitution in phosphate-buffered saline-2% bovine serum albumen, the viruses were propagated and plaque purified in Vero cells.
TABLE 1.
History of the viruses used in this study
| Serogroup | Virus | Straina | Passage history (animal or cell type and no. of passages)a | Date stock was freeze-dried (mo/day/yr)a | Geographic region of originb | Vertebrate host associationb | Illness in humansb | Vectorb | Isolation yearb |
|---|---|---|---|---|---|---|---|---|---|
| Anopheles A | ANAV | NAc | Mouse, 9 | 12/23/1984 | South America | NA | NA | Mosquito | 1940 |
| TCMV | BeAn73 | NA | 7/21/2001 | South America | Human, bat, primate, bird, horse | Yes | Mosquito | 1955 | |
| Anopheles B | ANBV | NA | Mouse, 179 | 2/15/1968 | South America | NA | NA | Mosquito | 1940 |
| BORV | SPAr395 | Mouse, 4 | 6/21/1965 | South America | NA | NA | Mosquito | 1962 | |
| Tete | TETEV | SA An3518 | Mouse, 7; Vero, 1; mouse, 1 | 7/12/1984 | Africa | Bird | NA | NA | 1959 |
| BMAV | DAK AnB1292 | Mouse, 8 | 12/15/1984 | Africa | Bird | NA | Tick | 1970 |
RNA extraction.
Subconfluent monolayers of Vero cells grown in 175 cm2 flasks were infected at a multiplicity of infection of 0.5 and incubated at 33°C. When 75% of the cells showed cytopathic effect (CPE), the supernatants were collected and clarified by centrifugation at 3,000 rpm for 5 min at 4°C, and virus particles were concentrated by centrifugation at 26,000 rpm for 2 h in an SW41 rotor. The viral pellet was resuspended in 1 ml of Trizol reagent (Invitrogen), and the RNA was extracted according to the manufacturer's protocol. The RNA was dissolved in 30 μl of water.
RT-PCR, cloning, and sequence determination.
A one-step reverse transcription-PCR (RT-PCR) procedure (Access Quick RT-PCR; Promega) was used to synthesize full-length S-segment cDNA according to the manufacturer's instructions. Each 50-μl reaction mixture contained 1 to 5 ng of viral RNA, 20 pmol of forward primer, and 20 pmol of reverse primer. Reaction mixtures were incubated for 60 min at 45°C, followed by 35 PCR cycles, each consisting of 94°C for 45 s, 54°C for 45 s, and 68°C for 90 s. Initially, primers BUNS+ and BUNS− (12) were used for RT-PCR, but these failed to give full-length products; therefore, additional primers were designed based on the partial sequences obtained, and full-length RT-PCR products were obtained using the following primer pairs: S25F (GTCACAGTAGTGTACTCCACWDNAA, where D is A/G/T, N is A/C/G/T, and W is A/T) and S13− (CTGACAGTAGTGTGTGCYCC) for Anopheles A virus (ANAV) and TCMV; S13+ (GTCACAGTAAGTGTACYCC) and ORO2S (27) for Anopheles B virus (ANBV) and Boraceia virus (BORV); and BUNS+ and S13− for TETEV and Batama virus (BMAV). The amplified DNA products were electrophoresed on a 1% agarose gel and purified by using a PCR gel extraction kit from Invitrogen. The purified PCR products were cloned into pGEM-T Easy vector (Promega). Three independent plasmid clones were selected for automated nucleotide sequence determination (Dundee University Sequencing Service), using M13 forward and reverse primers. The terminal sequences of all segments were confirmed by 5′/3′ rapid amplification of cDNA ends, using a kit from Roche, and specific primers based on the previously determined sequence.
Sequence analysis.
Nucleotide sequences were analyzed using Sequence Navigator and DNAMAN and were aligned using the Clustal W program. Accession numbers for S-segment sequences used for comparisons are the following: NC_001927 (BUNV), EU564827 (Bwamba virus), ABA54073 (Itaqui virus), EF485033 (LACV), AM711133 (M'Poko virus), AM711134 (Nola virus), AM709781 (Nyando virus), and AAU29359 (OROV).
In vitro transcription and translation.
The viral S-segment cDNAs were subcloned into pBluescript II KS+ plasmid (Stratagene) under the control of the bacteriophage T7 RNA polymerase promoter. Briefly, the inserts in pGEM-T Easy were amplified by PCR using forward primer 5′Xba (5′-TCTAGAAGTAGTGTGCTCCAC) and reverse primer 5′Kpn (5′-GGTACCAGTAGTGTGCTCCAC). The PCR products were digested with KpnI and XbaI, gel purified, and ligated with similarly digested pBluescript II KS+ DNA. The resulting constructs were linearized by digestion with KpnI and used to program coupled in vitro transcription and translation reactions in either rabbit reticulocyte or wheat germ lysates (TnT systems; Promega), according to the manufacturer's directions. Each 25-μl reaction mixture was incubated at 30°C for 90 min, and the radiolabeled products were analyzed by electrophoresis on NuPAGE 6 to 12% gradient gels (Invitrogen).
IFN-β mRNA-specific RT-PCR.
Total RNA was extracted from infected (5 PFU/cell) or mock-infected A549 cells at 24 h postinfection by using Trizol reagent (Invitrogen). For the reverse transcriptase reaction, 1 μg of total RNA was first treated with 2 U of DNase 1 (Promega) and was then reverse transcribed using 100 ng of random hexanucleotide primers and 5 U of avian myeloblastosis virus reverse transcriptase enzyme (Promega) in a 20-μl reaction volume containing 50 mM Tris-HCl, 75 mM KCl, 10 mM MgCl2, 20 U of RNasin RNase inhibitor, and a 10 mM concentration of the deoxynucleoside triphosphates. The mixture was incubated at 42°C for 1 h. The cDNA was amplified by PCR in a final volume of 50 μl containing 10 mM Tris-HCl, 50 mM KCl, and 0.1% Triton X, 1.5 mM MgCl2, 2U of Taq DNA polymerase, a 10 mM concentration of the deoxynucleoside triphosphates, and 20 pmol of each forward and reverse primer. PCR conditions were 30 cycles of 94°C for 30 s, 58°C for 60 s, and 72°C for 60 s, with a final step at 72°C for 10 min. The primers used to amplify mRNA sequences for IFN-β and human γ-actin were as described previously (31).
Reporter assay for IFN-β promoter induction.
Subconfluent monolayers of A549 cells in 35-mm-diameter dishes were transfected with 1 μg of the IFN-β promoter-containing plasmid pIFΔ(−125)lucter, as described previously (6). After 5 h the transfection mixture was removed, and the cells were infected with 1 PFU/cell of the different viruses or mock infected. The cells were harvested after a further 16-h incubation at 37°C, and luciferase activity was measured as described previously (6).
Biological assay for IFN production.
A549 cells were infected with 1 PFU/cell of the different viruses or mock infected and incubated at 37°C for 48 h. The medium was clarified by centrifugation (3,000 × g for 5 min), and any released virus was inactivated by UV light irradiation. Twofold dilutions of the medium were made and applied to A549 cells in 96-well plates, and the cells were incubated for 24 h at 37°C. The cells were then infected with encephalomyocarditis virus (EMCV), which is sensitive to IFN, and incubated for 48 h at 37°C. The cells were fixed with formaldehyde and stained with Giemsa stain to monitor development of CPE.
Nucleotide sequence accession numbers.
Consensus nucleotide sequences of the S RNA segments have been deposited in GenBank under the following accession numbers: FJ660415 (ANAV), FJ660416 (TCMV), FJ660417 (ANBV), FJ660418 (BORV), FJ660419 (TETEV), and FJ660420 (BMAV).
RESULTS AND DISCUSSION
Sequence analyses of S segments.
Two representative viruses from the three serogroups were selected to study: ANAV and TCMV from the Anopheles A serogroup; ANBV and BORV from the Anopheles B serogroup; and BMAV and TETEV from the Tete serogroup. The origins of these viruses are given in Table 1. The viruses were grown in Vero cells, and viral RNA was isolated from released virions. Full-length S-segment cDNAs were obtained by RT-PCR using primers based on the consensus terminal sequences, with modifications as described in Materials and Methods. Three independent cDNA clones were obtained for each virus, and the nucleotide sequences were determined. The characteristics of the S segments are summarized in Table 2 and compared with representatives of the Bunyamwera, Bakau, Bwamba, group C, California, Nyando, Turlock, and Simbu serogroups.
TABLE 2.
Sequence characteristics of Anopheles A, Anopheles B, and Tete serogroup orthobunyavirus S RNA segments compared to previously characterized S segments
| Serogroup | Virus | S-segment characteristic
|
Reference | |||||
|---|---|---|---|---|---|---|---|---|
| Total no. of nucleotides | 5′ NCR length (no. of nucleotides) | 3′ NCR length (no. of nucleotides) | A+U (%) | No. of amino acids ina:
|
||||
| N | NSs | |||||||
| Anopheles A | ANAV | 946 | 52 | 156 | 63 | 245 | — | This study |
| TCMV | 960 | 42 | 181 | 62 | 245 | — | This study | |
| Anopheles B | ANBV | 991 | 35 | 213 | 57 | 247 | — | This study |
| BORV | 982 | 35 | 203 | 56 | 247 | — | This study | |
| Tete | TETEV | 1,022 | 73 | 172 | 58 | 258 | — | This study |
| BMAV | 1,022 | 73 | 172 | 58 | 258 | — | This study | |
| Bakau | NOLAV | 961 | 85 | 174 | 59 | 233 | 101 | 35 |
| Bunyamwera | BUNV | 961 | 85 | 174 | 58 | 233 | 101 | 15 |
| Bwamba | BWAV | 1,096 | 81 | 310 | 65 | 234 | 92 | 20 |
| California | LACV | 978 | 78 | 192 | 58 | 235 | 97 | 5 |
| Group C | ITQV | 920 | 76 | 139 | 54 | 234 | 98 | 26 |
| Nyando | NDOV | 959 | 79 | 178 | 60 | 233 | 101 | 35 |
| Simbu | OROV | 754 | 44 | 14 | 53 | 231 | 91 | 27 |
| Turlock | MPOV | 955 | 85 | 147 | 58 | 240 | 95 | 35 |
The dash indicates that there is no NSs protein.
The N protein ORF is flanked by noncoding regions (NCRs), and like other orthobunyaviruses (except for OROV in the Simbu serogroup), the 5′ NCR is markedly shorter than the 3′ NCR (Table 2). Viruses in the previously analyzed serogroups encode N proteins that range from 213 to 240 amino acids in length (generally, all viruses within a serogroup have the same length of N protein); the newly sequenced S segments encode longer N proteins, with the Tete serogroup viruses displaying the longest N proteins (258 residues) recorded to date. Alignment of the amino acid sequences indicated that the two Tete serogroup virus N proteins have extensions predominantly at the amino terminus compared to the others, with no evidence for extensive internal insertion. In contrast, the Anopheles A and B serogroup virus N proteins have extensions at the carboxy terminus as well as a 2-amino-acid insertion, residues 139/140, relative to other orthobunyavirus N proteins (data not shown).
Comparisons of the amino acid sequences showed that the ANBV and BORV N proteins are 75% identical and that those of TETEV and BMAV are 80% identical, whereas the N proteins of two Anopheles A serogroup viruses, ANAV and TCMV, share only 57% amino acids. This is consistent with serological data that separate ANAV and TCMV into different complexes within the serogroup (9, 10). In addition, the N proteins of the Anopheles B serogroup viruses are 54 to 59% identical to those of the Anopheles A serogroup viruses. Preliminary phylogenetic analysis (data not shown) indicated that these two serogroups comprise two branches of a distinct lineage of orthobunyaviruses. However, we consider more detailed phylogenetic studies premature at this stage until sequence data from other members of the Anopheles A serogroup (which contains 15 viruses) (8) are available.
Absence of an NSs overlapping ORF.
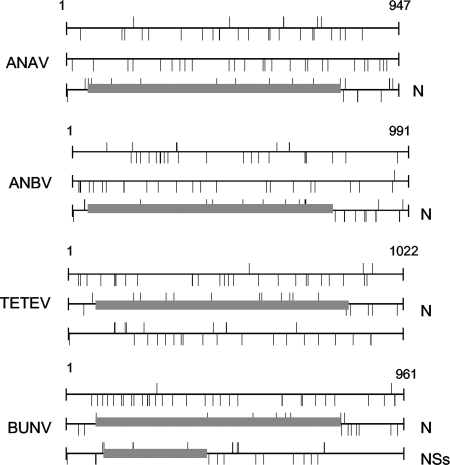
A characteristic of the S segments of viruses in the Bunyamwera, Bakau, Bwamba, group C, California, Nyando, Turlock, and Simbu serogroups is that they encode a second protein, NSs, in an ORF overlapping that of N, that is quite variable in size between different viruses, ranging from 83 to 109 residues (12). However, inspection of the newly acquired sequences did not reveal a similarly placed second overlapping ORF. Analysis of the distribution of start and stop codons in the sequences indicated that there were numerous translational stop codons in the two reading frames other than that of N (Fig. 1), indicating a real loss of the NSs ORF rather than merely mutation of the NSs initiation codon or introduction of a single stop codon into a presumptive NSs ORF. The positions of these stop codons are not conserved between the different viruses. Based on phylogenetic analyses, Bowen et al. (5) suggested that the evolutionary trend of orthobunyaviruses is toward encoding a smaller NSs protein, with truncation occurring at the carboxy end of the protein. Such viruses may have an evolutionary advantage in that fewer codons are constrained by the need to encode both N and NSs proteins in the same sequence. Hence, it will be highly instructive to investigate the phylogeny of Anopheles A serogroup viruses once more sequence data become available.
FIG. 1.
ORF analysis of Orthobunyavirus S segments. Sequences were analyzed using DNAMAN software (Lynnon corporation). Each reading frame is shown on a separate line, with ticks above the line indicating AUG codons and ticks below the line indicating translational stop codons. The boxed areas represent the N and NSs overlapping ORFs as indicated.
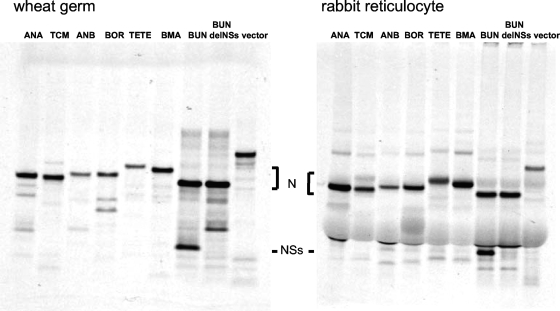
To confirm the absence of a second S-segment gene product, the cDNAs were cloned into an expression plasmid under the control of a T7 RNA polymerase promoter and were used to program protein synthesis in coupled transcription-translation systems. Both wheat germ and rabbit reticulocyte lysates were used in this study, with S-segment cDNAs from wtBUNV and BUNdelNSs viruses included as controls. The translation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2). For wtBUNV two major products were identified in both types of lysates, corresponding to N and NSs proteins, whereas the NSs band was not seen in translation products from BUNdelNSs cDNA. For the lysates programmed with cDNA from the other viral S segments, major bands corresponding to N protein were seen, which varied in mobility according to their predicted molecular weights. No major bands smaller than the N band were seen; minor, fast-migrating products probably represent internal initiation products on the N ORF, as analogous bands are seen in both wtBUNV and BUNdelNSs cDNA-programmed reactions.
FIG. 2.
Cell-free translation of proteins expressed from S-segment cDNA clones. S-segment cDNAs were cloned under the control of the T7 promoter and used to program either wheat germ or rabbit reticulocyte-coupled transcription/translation systems as directed by the manufacturer (TnT; Promega). Empty plasmid vector was included as a control. Radiolabeled proteins were separated on NuPAGE 6 to 12% gradient gels. Positions of viral N and NSs proteins are indicated.
Induction of IFN-β mRNA.
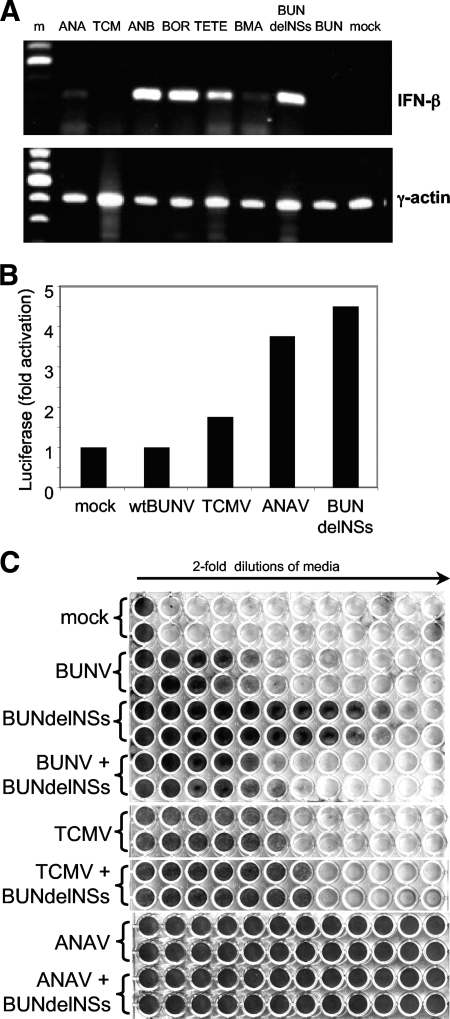
The BUNV and LACV NSs proteins have been shown to inhibit the synthesis of IFN by blocking the transcription of IFN genes. However, induction of the IFN-β mRNA was observed in cells infected with NSs-deleted versions of these viruses by using a specific RT-PCR approach (4, 33). Therefore, total RNA was extracted from A549 cells infected with the different viruses, with RNA from wtBUNV- or BUNdelNSs-infected cells included as controls. The RNA was used in RT-PCRs to detect IFN-β or actin mRNA as previously described (4, 31, 33). No IFN-β-specific product was seen in RNA from wtBUNV-infected cells, whereas a strong product was seen in RNA from BUNdelNSs-infected cells, as observed previously (Fig. 3A). All viruses except TCMV behaved similarly to BUNdelNSs in inducing the production of IFN-β mRNA, to a greater or lesser extent. However, no product was seen with RNA from TCMV-infected cells. In all cases RT-PCR specific for actin mRNA gave a strong signal, indicating that the RNA preparations were of similar quality and similar concentrations. In fact the TCMV sample gave the strongest actin signal, indicating that the lack of IFN-β signal was not due to poorer quality RNA, and this result has been repeated using an independently prepared RNA sample.
FIG. 3.
IFN production in infected cells. (A) Induction of IFN-β mRNA. Total RNA was extracted from mock- or virus-infected cells as indicated and reverse transcribed with random hexamer primers, and then cDNA was PCR amplified using primers specific for human IFN-β mRNA (upper panel) or γ-actin mRNA (lower panel). Amplified DNA products were electrophoresed in an agarose gel and stained with ethidium bromide. (B) Induction of IFN-β promoter. A549 cells were transfected with the IFN-β promoter containing reporter plasmid and 5 h later infected with the different viruses or mock infected. Cells were lysed 16 h later, and luciferase activity was determined. The activity in mock-infected cells was taken as 1, and the activity in virus-infected cells was expressed relative to this. (C) Biological activity for IFN production. Twofold dilutions of medium from A549 cells infected with different viruses, either singly or in combination as indicated, or mock infected were used to treat fresh A549 cells in 96-well plates for 24 h. The cells were then infected with EMCV, and the development of CPE was monitored 48 h later by staining with Giemsa. m, molecular ladder.
To confirm that TCMV was capable of suppressing IFN-β induction, two further assays were used. A549 cells were transfected with a plasmid containing the luciferase gene under the control of the IFN-β promoter and subsequently infected with various viruses or mock infected. As shown in Fig. 3B, luciferase activity in wtBUNV-infected cells was scarcely above background, whereas there was a fivefold increase in cells infected with BUNdelNSs. In TCMV-infected cells a slight increase in luciferase activity was measured (1.75-fold), but activity in ANAV-infected cells showed a nearly fourfold increase.
Second, a biological assay to measure the presence of secreted IFN in the medium of virus-infected cells was employed. Twofold dilutions of medium from virus-infected cells were used to treat fresh A549 cells in order to induce an antiviral state, and the cells were subsequently infected with the IFN-sensitive EMCV. As seen in Fig. 3C, cells treated with medium from uninfected cells were completely susceptible to infection with EMCV, whereas cells treated with medium from BUNdelNSs- or ANAV-infected cells were resistant to EMCV infection over the dilution range used. In contrast, medium from wtBUNV- or TCMV-infected cells induced protection at low dilutions only, indicating that much less IFN had been secreted. Coinfection experiments were also performed in which A549 cells were infected with BUNdelNSs together with either wtBUNV, TCMV, or ANAV, and the medium was assayed for IFN production. The medium from cells coinfected with BUNdelNSs and ANAV showed the same level of protection as that from the single ANAV infection (Fig. 3C). However, medium from cells coinfected with BUNdelNSs and wtBUNV or with BUNdelNSs and TCMV showed a lower level of protection than the protection provided by medium from cells infected with BUNdelNSs alone. These data thus confirm that TCMV behaved like wtBUNV in its ability to block IFN production in infected cells, whereas ANAV behaved like BUNdelNSs virus. Furthermore, in the dually infected cells, wtBUNV and TCMV were able to block a significant proportion of the IFN induced by BUNdelNSs infection.
Conclusions.
Our data indicate that the six viruses representing the Anopheles A, Anopheles B, and Tete serogroups contain S RNA segments that, in contrast to previously characterized orthobunyaviruses, do not encode an NSs gene. From knowledge of the role of other orthobunyavirus NSs proteins (34), it was thus not unexpected that five of these viruses behaved like BUNV or LACV lacking the NSs overlapping ORF in that they induced IFN-β mRNA and IFN in IFN-competent A549 cells. However, the result with TCMV was surprising in that this virus behaved like wtBUNV or wtLACV in suppressing IFN-β induction and IFN production. Furthermore, the results are consistent with protein labeling experiments (data not shown) where only TCMV and wtBUNV proteins were detected in infected A549 cells; for the other viruses and BUNdelNSs, no viral proteins could be detected in whole-cell lysates.
Previous results using BUNV and LACV engineered to delete the NSs overlapping ORF showed that NSs was not required for replication of either virus in tissue culture or in mice, but the NSs-expressing viruses were attenuated in IFN-competent animals. This was interpreted as indicating that NSs protein is a nonessential virulence factor (34). Blakqori et al. (4) noted no difference in the replication of wtLACV and LACV with a deletion of the NSs protein in mosquito cells, suggesting that NSs does not play a role in infection of the vector. Thus, naturally occurring NSs-null orthobunyaviruses would have reduced virulence in mammalian hosts, and theoretically an insect-only cycle involving both horizontal (venereal) and vertical (transovarial) transmission between mosquitoes might be more important for their maintenance in nature. Of the six newly characterized viruses, only TCMV has been associated with human disease, causing a febrile illness (11, 19). The virus was originally isolated from a sentinel capuchin monkey (Cebus apella) and subsequently from Anopheles mosquito, and serological surveys showed antibodies in humans, horses, cattle, pigs, bats, rodents, and birds in Brazil and other South American countries (2, 11, 18). Livonesi et al. (22) investigated the effect of IFN-α on TCMV replication in vitro and in vivo. Like other tested orthobunyaviruses, TCMV replication was completely inhibited if IFN was applied to cells 24 h before or 2 h after infection. However, the virus was more sensitive to IFN when IFN was added to infected cells up to 48 h postinfection, whereas OROV, which expresses NSs protein, was resistant to inhibitory effects if IFN was added to cells more than 2 h after infection. On the other hand, prophylactic administration of IFN failed to prevent death of mice infected with TCMV, and IFN-treated mice were protected from OROV-mediated encephalitis. Taken together, the data suggest that TCMV does have the ability to overcome the host innate immune response, but this may function differently from the described effects of the orthobunyavirus NSs protein. Attempts to determine the viral product responsible for surmounting innate defenses are in progress.
Acknowledgments
We thank Alain Kohl for fruitful discussions, Elina Koudriakova for technical assistance, and the late Bob Shope, a true gentleman of science, for providing viruses used in this study.
M.M. was supported by a studentship from the Malaysian Ministry of Science, and work in R.M.E.'s group was funded by grants from the Wellcome Trust.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Barrett, A. D. T., and R. E. Shope. 2005. Bunyaviridae, p. 1025-1058. In B. W. J. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, 10th ed., vol. 2. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 2.Beaty, B. J., and C. H. Calisher. 1991. Bunyaviridae—natural history. Curr. Top. Microbiol. Immunol. 16927-78. [PubMed] [Google Scholar]
- 3.Bishop, D. H. L., K. G. Gould, H. Akashi, and C. M. Clerx-Van Haaster. 1982. The complete sequence and coding content of snowshoe hare bunyavirus (S) viral RNA species. Nucleic Acids Res. 103703-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakqori, G., S. Delhaye, M. Habjan, C. D. Blair, I. Sanchez-Vargas, K. E. Olson, G. Attarzadeh-Yazdi, R. Fragkoudis, A. Kohl, U. Kalinke, S. Weiss, T. Michiels, P. Staeheli, and F. Weber. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 814991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen, M. D., A. O. Jackson, T. D. Bruns, D. L. Hacker, and J. L. Hardy. 1995. Determination and comparative analysis of the small RNA genomic sequences of California encephalitis, Jamestown Canyon, Jerry Slough, Melao, Keystone and Trivittatus viruses (Bunyaviridae, genus Bunyavirus, California serogroup). J. Gen. Virol. 76559-572. [DOI] [PubMed] [Google Scholar]
- 6.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calisher, C. H. 1988. Evolutionary significance of the taxonomic data regarding bunyaviruses of the family Bunyaviridae. Intervirology 29268-276. [DOI] [PubMed] [Google Scholar]
- 8.Calisher, C. H. 1996. History, classification and taxonomy of viruses in the family Bunyaviridae, p. 1-17. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, NY.
- 9.Calisher, C. H., J. S. Lazuick, D. J. Muth, O. de Souza Lopes, G. T. Crane, R. E. Elbel, and R. E. Shope. 1980. Antigenic relationships among Tacaiuma complex viruses of the Anopheles A serogroup (Bunyaviridae). Bull. Pan Am. Health Organ. 14386-391. [PubMed] [Google Scholar]
- 10.Calisher, C. H., D. R. Sasso, K. S. Maness, V. N. Gheorghiu, and R. E. Shope. 1973. Relationships of anopheles A group arboviruses. Proc. Soc Exp. Biol. Med. 143465-468. [DOI] [PubMed] [Google Scholar]
- 11.Causey, O. R., C. E. Causey, O. M. Maroja, and D. G. Macedo. 1961. The isolation of arthropod-borne viruses, including members of two hitherto undescribed serological groups, in the Amazon region of Brazil. Am. J. Trop. Med. Hyg. 10227-249. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, E. F., D. C. Pritlove, and R. M. Elliott. 1994. The S RNA genome segments of Batai, Cache Valley, Guaroa, Kairi, Lumbo, Main Drain and Northway bunyaviruses: sequence determination and analysis. J. Gen. Virol. 75597-608. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, R. M. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3572-577. [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, R. M. 1990. Molecular biology of the Bunyaviridae. J. Gen. Virol. 71501-522. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, R. M. 1989. Nucleotide sequence analysis of the small (S) RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. J. Gen. Virol. 701281-1285. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, R. M. (ed.). 1996. The Bunyaviridae. Plenum Press, New York, NY.
- 17.Elliott, R. M., and A. McGregor. 1989. Nucleotide sequence and expression of the small (S) RNA segment of Maguari bunyavirus. Virology 171516-524. [DOI] [PubMed] [Google Scholar]
- 18.Iversson, L. B., R. A. Silva, A. P. da Rosa, and V. L. Barros. 1993. Circulation of eastern equine encephalitis, western equine encephalitis, Ilhéus, Maguari and Tacaiuma viruses in equines of the Brazilian Pantanal, South America. Rev. Inst. Med. Trop. São Paulo 35355-359. [DOI] [PubMed] [Google Scholar]
- 19.Karabatsos, N. (ed.). 1985. International catalogue of arboviruses including certain other viruses of vertebrates, 3rd ed. American Society of Tropical Medicine and Hygiene, San Antonio, TX.
- 20.Lambert, A. J., and R. S. Lanciotti. 2008. Molecular characterization of medically important viruses of the genus Orthobunyavirus. J. Gen. Virol. 892580-2585. [DOI] [PubMed] [Google Scholar]
- 21.Leonard, V. H. J., A. Kohl, T. J. Hart, and R. M. Elliott. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with Mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 809667-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livonesi, M. C., R. L. de Sousa, S. J. Badra, and L. T. Figueiredo. 2007. In vitro and in vivo studies of the interferon-alpha action on distinct Orthobunyavirus. Antivir. Res. 75121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 24.Nichol, S. T., J. Arikawa, and Y. Kawaoka. 2000. Emerging viral diseases. Proc. Natl. Acad. Sci. USA 9712411-12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichol, S. T., B. J. Beaty, R. M. Elliott, R. Goldbach, A. Plyusnin, C. S. Schmaljohn, and R. B. Tesh. 2005. Family Bunyaviridae, p. 695-716. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 26.Nunes, M. R., A. P. Travassos da Rosa, S. C. Weaver, R. B. Tesh, and P. F. Vasconcelos. 2005. Molecular epidemiology of group C viruses (Bunyaviridae, Orthobunyavirus) isolated in the Americas. J. Virol. 7910561-10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeed, M. F., H. Wang, M. R. Nunes, P. F. Vasconcelos, S. C. Weaver, R. E. Shope, D. M. Watts, R. B. Tesh, and A. D. T. Barrett. 2000. Nucleotide sequences and phylogeny of the nucleocapsid gene of Oropouche virus. J. Gen. Virol. 81743-748. [DOI] [PubMed] [Google Scholar]
- 28.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 395-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmaljohn, C. S., and S. T. Nichol (ed.). 2001. Hantaviruses. Springer-Verlag, Berlin, Germany.
- 30.Schmaljohn, C. S., and J. W. Hooper. 2001. The Bunyaviridae, p. 1581-1602. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 31.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 792079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 27931471-31477. [DOI] [PubMed] [Google Scholar]
- 33.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 767949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, F., and R. M. Elliott. 2009. Bunyaviruses and innate immunity, p. 287-299. In A. Brasier, A. Garcia-Sastre, and S. Lemon (ed.), Cellular signaling and innate immune responses to RNA virus infections. ASM Press, Washington, DC.
- 35.Yandoko, E. N., S. Gribaldo, C. Finance, A. Le Faou, and B. H. Rihn. 2007. Molecular characterization of African orthobunyaviruses. J. Gen. Virol. 881761-1766. [DOI] [PubMed] [Google Scholar]