Abstract
Bone marrow stromal cell antigen 2 (BST-2, also known as tetherin) restricts the production of a number of enveloped viruses by blocking virus release from the cell surface. This antiviral activity is counteracted by such viral factors as Vpu of human immunodeficiency virus type 1 (HIV-1). Here, we report that Vpu antagonizes human BST-2 but not BST-2 derived from African green monkeys. The determinants of susceptibility to Vpu map to the transmembrane domain of BST-2. In accordance with this, expression of human BST-2 containing a modified transmembrane domain effectively blocks the replication of wild-type Vpu-expressing HIV-1 in CD4+ T cells. Furthermore, these BST-2 variants, as opposed to wild-type human BST-2, are refractory to Vpu-mediated down-regulation as a result of an attenuated interaction with Vpu. In view of the work by others pointing to a key role of the transmembrane domain of Vpu in promoting virus release, our data suggest that a direct interaction through the transmembrane domain of each of these two proteins is a prerequisite for Vpu to down-modulate BST-2.
Human immunodeficiency virus type 1 (HIV-1) encodes four accessory proteins, Vif, Vpr, Vpu, and Nef. Although they are dispensable for HIV-1 replication in certain transformed cell lines, these accessory proteins play important roles in HIV-1 pathogenesis by modulating host immunity and overcoming antagonism by cellular factors (10). For example, Vif counteracts APOBEC3G by recruiting the cullin 5-elongin B/C ubiquitin ligase complex and sending polyubiquitinated APOBEC3G to proteasomes for degradation (29). In the absence of Vif, newly synthesized APOBEC3G is incorporated into virus particles and hampers the production of infectious proviral DNA in the new round of infection (4, 10, 23). In addition to its role in down-modulating the cell surface expression of CD4 in infected T cells (11), Vpu stimulates HIV-1 production in cells such as HeLa cells (26). The mechanism behind this latter activity of Vpu was unknown until it was recently discovered that bone marrow stromal cell antigen 2 (BST-2, also known as tetherin, CD317, or HM1.24) blocks the release of HIV-1 and that this inhibitory effect is antagonized by viral Vpu (16, 25).
BST-2 harbors an N-terminal transmembrane domain and a C-terminal glycosyl-phosphatidylinositol anchor that together create an unusual topology with both termini of BST-2 inserted into the plasma membrane (8, 18). This unique topology of BST-2 may underlie the mechanism for the retention of progeny virus particles at the cell surface (16). An indirect mechanism behind this tethering effect has not been ruled out, especially in view of the difficulty of detecting BST-2 protein in purified HIV-1 particles (14). In addition to HIV-1, a number of enveloped viruses are subject to inhibition by BST-2, including simian immunodeficiency virus, feline immunodeficiency virus, equine infectious anemia virus, Mason-Pfizer monkey virus, and Lassa virus, as well as Ebola and Marburg viruses (5, 6, 16, 19, 25). This suggests that BST-2 has a broad antiviral effect spectrum.
The bst-2 gene has in its promoter the IRF-1/2 and ISGF3 response elements and thus belongs to the interferon-stimulated gene family (17). In line with its ability to impair the release of enveloped viruses, BST-2 has been demonstrated to be the effector in human embryonic kidney (HEK293T) cells that leads to the interferon-induced block of Vpu deletion-containing HIV-1 production (15). However, the African green monkey kidney cell line COS-7 responds to interferon treatment with a different outcome in that the production of both Vpu deletion-containing and Vpu-expressing HIV-1 is inhibited (15). This indicates that interferon induces a block to HIV-1 in COS-7 cells that cannot be overcome by Vpu. A conceivable candidate that creates this block is BST-2 in COS-7 cells (hereafter named agmBST-2). In this study, we provide evidence that depletion of endogenous BST-2 in COS-7 cells greatly alleviates interferon-induced inhibition of HIV-1 production. The refractoriness of agmBST-2 to Vpu results from a weak association of these two proteins and a resistance of agmBST-2 to Vpu-mediated down-regulation.
MATERIALS AND METHODS
Plasmid DNA and antibodies.
The agmBST-2 cDNA was amplified by reverse transcription (RT)-PCR from RNA samples that were extracted from the African green monkey kidney cell line COS-7 that had been pretreated with alpha 2b interferon (1,000 U/ml; Invitrogen), followed by insertion into the pcDNA3.1 expression vector at the BamHI and NotI sites (Invitrogen). The RT-PCR primers (5′-GCATGGATCCTTCAGCTAGAGGGGAGATCTGG-3′/5′-GCATGCGGCCGCTGAGATCCCAGGAAGCTGGCA-3′) were designed on the basis of the predicted BST-2 mRNA sequence of rhesus monkeys, binding to the 5′ untranslated region and the 3′ untranslated region, respectively. The human BST-2 (hBST-2) cDNA clone was purchased from ATCC (catalog number MGC-45144) and subcloned into pcDNA3.1. The mutated BST-2 DNA clones were created by PCR and verified by sequencing. The BST-2 proteins were all attached to a Flag tag at their N termini. BH10 is an infectious HIV-1 proviral DNA clone. BH10(Vpu−) has the first ATG of Vpu changed to ACG. GPV-CTEx4 DNA carries HIV-1 Gag-Pol coding sequence and is able to generate virus-like particles in a Rev-independent manner (28). Anti-Flag antibody was purchased from Sigma, anti-HIV-1 p24 antibody was from ID Lab Inc., and HIV-1(NL4-3) Vpu and hBST-2 antiserum were from the NIH AIDS Research and Reference Reagent Program (9, 14).
Generating BST-2-inducible SupT1 cell lines.
The hBST-2, CTh, and hTMa1 cDNAs were inserted into the Pur-GOI retroviral vector (Clontech) to create Pur-hBST-2, Pur-CTh, and Pur-hTMa1 DNA clones. The CTh mutant has the cytoplasmic domain of agmBST-2 replaced with the same domain of hBST-2. hTMa1 is a mutant form of hBST-2 that has the first nine amino acids of the transmembrane domain replaced with the corresponding residues in agmBST-2 (see Fig. 3A). Virus stocks were prepared by transfecting each of these DNA clones into GP2-293 packaging cells together with vesicular stomatitis virus (VSV) G DNA, followed by infection of SupT1 cells together with viruses that express the rtTA activator. Stable SupT1 cell lines were selected with G418 (1 mg/ml) and puromycin (2 μg/ml). Expression of hBST-2, CTh, or hTMa1 was induced with doxycycline and examined in Western blot assays with anti-Flag antibody.
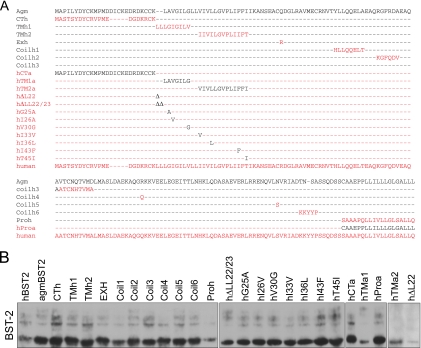
FIG. 3.
Deletion of 22-LL-23 from hBST-2 causes resistance to HIV-1 Vpu. (A) Illustration of the BST-2 mutations studied. HBST-2 and agmBST-2 sequences are shown in red and black letters, respectively. (B) The BST-2 DNA constructs (200 ng) were transfected into HEK293T cells. Expression of BST-2 proteins was assessed by Western blotting with anti-Flag antibody. (C, D) Effects of BST-2 proteins on the production of wild-type HIV-1. HEK293T cells were transfected with various amounts of BST-2 DNA (0, 10, 20, 50, 100, and 200 ng) together with 100 ng of BH10 DNA. Levels of infectious viruses were determined by infecting TZM-bl indicator cells and then measuring luciferase activity (C) or by measuring viral reverse transcriptase activity (D). (E) All BST-2 DNA constructs were tested in one transfection experiment with 100 ng of BST-2 DNA together with 100 ng of BH10 DNA. Levels of infectious HIV-1 particles in the culture supernatants were determined in a single-cycle infection assay by infecting TZM-bl indicator cells. (F) Effects of BST-2 proteins on the production of virus-like particles from GPV-CTEx4 DNA in the absence of Vpu. HEK293T cells were transfected with 50 ng of each BST-2 DNA together with 200 ng of GPV-CTEx4 DNA. Amounts of viruses in culture supernatants were determined by measuring viral reverse transcriptase activity. Expression of viral Gag in the cells was assessed by Western blotting. CPM, counts per minute; RLU, relative luciferase units.
siRNA knockdown.
Short interfering RNA (siRNA) oligonucleotides named siBST-2-1 (5′-AGAAAGTGGAGGAGCTTGA-3′) and siBST-2 (5′-TCACCTATCTCCTGCAACA-3′) were designed to target agmBST-2 mRNA. Together with a control siRNA (catalog no. 4611), they were purchased from Ambion. COS-7 cells were transfected with siRNA (40 nM) by using Lipofectamine 2000 (Invitrogen) before being infected with HIV-1 pseudotyped with VSV G protein at a multiplicity of infection (MOI) of 0.5. Alpha 2b interferon (1,000 U/ml) was added 16 h after infection. Cells and culture supernatants were harvested after 40 h. agmBST-2 knockdown efficiency was assessed by measuring levels of agmBST-2 mRNA in Northern blot assays with α-32P-labeled agmBST-2 cDNA as the probe. Cellular glyceraldehyde-3-phosphate dehydrogenase mRNA was also detected; its level served as an RNA loading control.
Measuring virus production.
Amounts of viruses in culture supernatants were determined by three methods. First, viral reverse transcriptase activity in culture supernatants was measured by reverse transcriptase assay with poly(rA)-oligo(dT) as the template and [3H]dTTP as the substrate. Incorporation of 3H into poly(rA)-oligo(dT) was scored in a liquid scintillation counter (Perkin-Elmer). Second, amounts of infectious HIV-1 particles in culture supernatants were determined by a single-cycle infection assay. Briefly, TZM-bl indicator cells were seeded into the wells of a 24-well plate 1 day prior to infection with 50 μl of culture supernatants for 40 h, followed by measurement of luciferase activity in cell lysates with a GLOMAX 20/20 luminometer (Promega) with the Luciferase Assay kit (Promega) (27). Third, levels of HIV-1 p24(CA) antigen in culture supernatants were measured by enzyme-linked immunosorbent assay with the Vironostika HIV-1 p24 Microelisa kit (BioMérieux).
Immunoprecipitation.
Transfected 293T cells were lysed on ice for 30 min in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Roche) before clarification at 10,000 × g for 10 min at 4°C. One milligram of cell lysate was incubated with 50 μl of a slurry of agarose beads coated with mouse anti-Flag antibody (Sigma) for 2 h, followed by washing at 4°C three times with a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and protease inhibitor cocktail (Roche). The bound proteins were eluted with 3XFlag peptide (150 μg/μl; Sigma), and protein samples were separated in 0.1% sodium dodecyl sulfate-10% polyacrylamide gels. After being transferred onto polyvinylidene difluoride membranes (Roche), Flag-BST-2 and Vpu were detected in Western blot assays with anti-Flag antibody (1:5,000 dilution) and Vpu antiserum (1:1,000 dilution), respectively.
Flow cytometry.
HEK293T cells were seeded into the wells of 12-well plates and transfected with 100 ng of Flag-hBST-2 or Flag-hΔLL22/23 DNA together with 100 ng of either BH10 or BH10(Vpu−) DNA. Twenty hours after transfection, cells were detached from the plates with 5 mM EDTA (1× phosphate-buffered saline) and washed with ice-cold 1× phosphate-buffered saline containing 2% fetal calf serum, followed by a 60-min incubation on ice with hBST-2 antiserum (1:500 dilution; kindly provided by Klaus Strebel) and a 30-min incubation on ice with Alexa Fluor 488-conjugated anti-rabbit secondary antibody (1:500 dilution; Molecular Probes). Cells were then fixed with 1% paraformaldehyde (in 1× phosphate-buffered saline) and analyzed by flow cytometry.
RESULTS
agmBST-2 inhibits the production of wild-type HIV-1.
To investigate whether agmBST-2 mediates the interferon-induced inhibition of HIV-1 production in COS-7 cells, we first cloned agmBST-2 cDNA from alpha 2b interferon-treated COS-7 cells by RT-PCR. This cloned agmBST-2 protein has 76% homology with hBST-2, with major heterology found in the N-terminal cytoplasmic domain (Fig. 1A). Two siRNA oligonucleotides were accordingly designed, termed siBST-2-1 and siBST-2-2, to target agmBST-2 mRNA. Results of Northern blot assays showed that both siRNA oligonucleotides reduced levels of agmBST-2 mRNA by more than 90% in COS-7 cells that were exposed to alpha 2b interferon (1,000 U/ml) (Fig. 1B). After exposure to HIV-1 particles that had been pseudotyped with VSV glycoprotein, the amounts of progeny viruses produced by COS-7 cells were determined either by measuring viral reverse transcriptase activity or by performing a single-cycle infection assay through infection of TZM-bl indicator cells and then measuring luciferase activity. The results showed that alpha 2b interferon reduced the production of infectious HIV-1 particles from COS-7 cells by more than 50-fold without affecting the expression of viral Gag protein in the cells (Fig. 1C and D), which is in agreement with previous findings (15). Importantly, the siBST-2 RNA oligonucleotides relieved this inhibition by more than 1 order of magnitude (Fig. 1C and D), suggesting that agmBST-2 is a major factor, if not the only one, that leads to the interferon-induced block of HIV-1 production in COS-7 cells.
FIG. 1.
Knockdown of agmBST-2 significantly alleviates the interferon-induced inhibition of HIV-1 production in COS-7 cells. (A) Homology of hBST-2 and agmBST-2 proteins. AGM, African green monkey. (B) Cells were transfected with a siCon or a siBST-2 RNA oligonucleotide prior to infection with VSV G protein-pseudotyped wild-type HIV-1 particles and then treated with alpha 2b interferon (1,000 U/ml). Levels of endogenous agmBST-2 mRNA were measured by Northern blotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was measured as an internal control. (C) Virus production in the culture supernatants was assessed either by measuring viral reverse transcriptase activity or by determining virus infectivity by infecting TZM-bl indicator cells. (D) Western blot assays were performed to assess viral Gag protein expression in the cells. CPM, counts per minute; RLU, relative luciferase units.
In order to directly assess the effect of agmBST-2 on wild-type HIV-1 production, we transfected COS-7 cells with HIV-1 proviral DNA clone BH10 and agmBST-2 DNA. As opposed to the neutral effect of hBST-2 on HIV-1 production in the presence of Vpu, agmBST-2 reduced the yield of infectious HIV-1 particles by as much as 20-fold without any effect on viral Gag expression in the cells (Fig. 2A). The ability of agmBST-2 to block the production of wild-type HIV-1 may result either from its extremely potent antiviral activity or from its resistance to the countering action of HIV-1 Vpu. The first possibility was tested by measuring the effects of hBST-2 and agmBST-2 on virus production in the absence of Vpu. To this end, we used two DNA constructs in the cotransfection experiments, a Vpu deletion-containing HIV-1 proviral DNA named BH10(Vpu−) and the GPV-CTEx4 DNA that expresses Gag and Gag-Pol proteins (28). The results showed that hBST-2 inhibited virus production 10-fold more potently than agmBST-2 in the absence of Vpu (Fig. 2B and C). Together, these data suggest that agmBST-2 is not antagonized by Vpu and is thus able to inhibit wild-type HIV-1 production.
FIG. 2.
Ectopic expression of agmBST-2, but not hBST-2, inhibits wild-type HIV-1 production. (A) Various amounts of hBST-2 or agmBST-2 DNA (0, 10, 20, 50, 100, and 200 ng) were cotransfected into COS-7 cells with 100 ng BH10 DNA. Virus production was measured as described in Materials and Methods. Levels of viral Gag protein and BST-2 protein in the cells were assessed in Western blot assays. (B) The BH10(Vpu−) proviral DNA does not express Vpu and was used in the cotransfection experiments together with hBST-2 or agmBST-2 DNA. Virus production was examined by infecting TZM-bl indicator cells or by measuring levels of viral reverse transcriptase activity and levels of viral p24(CA) antigen. Levels of Gag protein and BST-2 protein in the cells were measured by Western blotting. (C) HBST-2 more efficiently blocks the production of virus-like particles from GPV-CTEx4 DNA in the absence of Vpu than agmBST-2. Levels of virus-like particles in culture supernatants were determined by measuring viral reverse transcriptase activity. Gag and BST-2 protein expression in the cells was examined by Western blotting. CPM, counts per minute; RLU, relative luciferase units.
We also noted that hBST-2 (in 200 ng DNA) diminished viral reverse transcriptase activity by 10-fold, in contrast to a 1,000-fold reduction of infectious Vpu deletion-containing virus particles, as measured in the single-cycle infection assay (Fig. 2B). The amount of progeny HIV-1 virions in the culture supernatants was further quantified by measuring levels of viral p24(CA) antigen. The enzyme-linked immunosorbent assay results revealed a fivefold reduction of p24(CA) production when 200 ng BST-2 DNA was used for transfection, which supports the relatively moderate decrease in viral reverse transcriptase activity (Fig. 2B). Since the levels of viral reverse transcriptase activity or viral p24(CA) antigen reflect the total number of virus particles (infectious and noninfectious) whereas the luciferase data score the titers of infectious virus particles in the culture supernatants, the results suggest that hBST-2 causes defects in progeny HIV-1 particles in the absence of Vpu in addition to blocking virus release from the cell surface. Results in Fig. 2A and B suggest that agmBST-2 is slightly more efficient at restricting the production of wild-type BH10 viruses than are the Vpu-negative viruses. One possible explanation is that elimination of Vpu translation affects the production of viral envelope protein from the same mRNA, which in turn moderately interferes with the antiviral activity of BST-2.
The transmembrane domain of hBST-2 determines its sensitivity to HIV-1 Vpu.
agmBST-2 differs from hBST-2 at 44 amino acid positions, with 13 of these clustered in the N-terminal cytoplasmic domain (Fig. 1A). We next wished to determine which of these 44 amino acids confer Vpu resistance on agmBST-2. Accordingly, 11 DNA constructs were generated by replacing distinct regions of agmBST-2 with the corresponding sequences of hBST-2 (Fig. 3A). These BST-2 variants were expressed at comparable levels (Fig. 3B). We first transfected different doses of each BST-2 DNA construct into HEK293T cells together with wild-type BH10 DNA. Levels of infectious HIV-1 particles in the culture supernatants were determined by infecting TZM-bl indicator cells (Fig. 3C) or by measuring viral reverse transcriptase activity (Fig. 3D). Expression of Gag protein in the cells was assessed by Western blotting. No major change in Gag expression was observed (data not shown). We also tested all of the BST-2 DNA constructs in one set of transfection experiments with 100 ng of each BST-2 DNA and 100 ng of BH10 DNA. The single-cycle infection assay results are shown in Fig. 3E. Results of both studies showed that the TMh1 and TMh2 proteins exhibited the least inhibitory activity of the 11 mutant agmBST-2 proteins (Fig. 3C to E). The inability of the TMh1 and TMh2 proteins to efficiently block wild-type HIV-1 production is not due to their attenuated antiviral activity in the absence of Vpu, since both mutant proteins inhibited the production of virus-like particles from GPV-CTEx4 DNA as much as wild-type agmBST-2 in the absence of Vpu (Fig. 3F). This indicates that TMh1 and TMh2 are sensitive to HIV-1 Vpu.
Since the TMh1 and TMh2 mutations introduce changes into the transmembrane domain of agmBST2, the data described above suggest a key role for the transmembrane domain in regulating the sensitivity of BST-2 to Vpu. In support of this notion, an hBST-2 variant named hTMa1, which has a portion of its transmembrane domain replaced with the relevant sequence from agmBST-2 (Fig. 3A), drastically blocked the production of infectious HIV-1 particles in the presence of Vpu (Fig. 3C to E).
The hBST-2 and agmBST-2 proteins differ by nine amino acids in their transmembrane domains (Fig. 3A). We next changed each of these nine amino acids in the context of hBST-2 to the corresponding residues in agmBST-2 with the aim of identifying the key amino acids that are associated with Vpu resistance (Fig. 3A). Results of transfection experiments showed that deletion of two leucine residues at positions 22 and 23 (named hΔLL22/23) efficiently blocked wild-type HIV-1 production (Fig. 3C to E). In addition, mutations hV30G and hT45I also conferred resistance to Vpu, albeit to a lesser extent than hΔLL22/23 (Fig. 3C to F). Therefore, the dileucine residues 22-LL-23, which exist in hBST-2 but not in agmBST-2, constitute the key motif in the transmembrane domain that determines the sensitivity of hBST-2 to HIV-1 Vpu.
Deletion of 22-LL-23 diminishes the association of hBST-2 with Vpu.
We next assessed whether hBST-2 associates with Vpu and whether deletion of 22-LL-23 affects this putative interaction. To this end, we transfected HEK293T cells with BH10 DNA together with hBST-2, agmBST-2, or hΔLL22/23 DNA. BST-2 proteins were immunoprecipitated with anti-Flag antibody, and the precipitates were subjected to Western blotting to measure the presence of Vpu. The results showed that Vpu was efficiently coprecipitated with hBST-2, but to a significantly lesser extent (four- to fivefold lower) than with agmBST-2 or hΔLL22/23 when the amount of immunoprecipitated Vpu protein was adjusted for the amount of each precipitated BST-2 protein (Fig. 4). This significantly attenuated association of Vpu with either agmBST-2 or hΔLL22/23 was consistently observed in three independent transfection and immunoprecipitation experiments. We also noted that HIV-1 Gag protein was not coprecipitated with any of these BST-2 proteins (Fig. 4). Therefore, deletion of 22-LL-23 significantly attenuates the association of hBST-2 with Vpu.
FIG. 4.
Association of Vpu with hBST-2, agmBST-2, and hΔLL22/23. HEK293T cells were cotransfected with BH10 DNA and hBST-2, agmBST-2, or hΔLL22/23 DNA. BST-2 proteins were immunoprecipitated (IP) with anti-Flag antibody, and the precipitates were subjected to Western blotting (W.B.) with antibodies to Flag, Vpu, or HIV-1 p24. Also shown are the results of one Western blot assay in which the amounts of immunoprecipitated hBST-2, agmBST-2, and hΔLL22/23 proteins were adjusted to similar levels in order to facilitate comparison of the amounts of coprecipitated Vpu protein. Relative intensities of the BST-2 and Vpu signals in Western blot assays were determined with the ImageJ program (NIH). The Vpu signal in the hBST-2 sample was arbitrarily set at 100. The results shown are representative of three independent transfection and immunoprecipitation experiments. Standard deviations were also calculated and are shown in parentheses.
Levels of hBST-2 but not agmBST-2 are substantially down-modulated by Vpu.
It has been reported that Vpu significantly down-modulates hBST-2 expression, especially from the cell surface (1, 2, 14, 25). We therefore asked whether, in contrast to hBST-2, the agmBST-2 or hΔLL22/23 protein escapes from down-regulation by Vpu as a result of its weak association with this viral protein. To answer this question, we transfected HEK293T cells with BH10 or BH10(Vpu−) DNA together with hBST-2, agmBST-2, or hΔLL22/23 DNA. Results of Western blot assays showed that levels of hBST-2 protein were substantially lower in cells that were cotransfected with BH10 DNA than in cells that were cotransfected with BH10(Vpu−) DNA (Fig. 5A, lanes 2 and 3). In contrast, the expression of the agmBST-2 and hΔLL22/23 proteins was not affected, regardless whether Vpu was present or not (Fig. 5A, lanes 5 and 6 and lanes 8 and 9).
FIG. 5.
Effect of Vpu on BST-2 expression. (A) hBST-2, agmBST-2, or hΔLL22/23 DNA (100 ng) was cotransfected into HEK293T cells either with the empty expression vector pcDNA3.1 (100 ng) or with BH10 (100 ng) or BH10(Vpu−) DNA (100 ng). Western blot assays were performed with anti-Flag antibody, anti-tubulin antibody, anti-HIV-1 p24 antibody, or Vpu antiserum. (B) The number of cells that expressed BST-2 proteins on their surfaces was scored by flow cytometry after immunostaining with hBST-2 antiserum. The relatively low percentage (15%) of agmBST-2-positive cells is likely due to the poor recognition of agmBST-2 by hBST-2 antiserum. Results shown are the average of two independent transfection experiments.
BST-2 exerts its tethering activity at the cell surface (16). HIV-1 Vpu has been shown to diminish the hBST-2 level at this cellular site (25). We therefore further measured the cell surface levels of agmBST-2 and hΔLL22/23 in the presence or absence of Vpu by fluorescence-activated cell sorting. Results in Fig. 5B show that, in contrast to a fourfold reduction of hBST-2 at the cell surface, Vpu did not affect agmBST-2 expression and caused a 30% decrease in the level of hΔLL22/23. These data together suggest that changing the transmembrane domain of hBST-2 disrupts its association with Vpu and leads to its escape from Vpu-mediated down-modulation.
The Vpu-resistant BST-2 variants potently inhibit the replication of wild-type HIV-1 in CD4+ T cells.
We next assessed the extent to which BST-2 affects HIV-1 replication in human CD4+ T cells. To this end, we generated SupT1 cell lines that expressed the hBST-2, hTMa1, or CTh protein with induction by doxycycline. The latter two BST-2 variants were chosen because of their extremely potent inhibition activity against wild-type HIV-1 production in HEK293T cells (Fig. 3). Of note, SupT1 cells do not express endogenous hBST-2 at a level that can be detected by Western blotting (data not shown). We first measured the effect of Vpu on the expression of these BST-2 proteins in SupT1 cells. After exposure to either wild-type (BH10) or Vpu deletion-containing [BH10(Vpu−)] HIV-1 for 40 h in the presence of doxycycline (50 ng/ml), the SupT1 cell lines were assessed for the expression of different BST-2 proteins in Western blot assays. Results in Fig. 6A show that hBST-2 expression was virtually depleted in cells that were infected with BH10 virus (MOI = 1). In contrast, the hTMa1 and CTh proteins were present at similar levels in cells that had been infected with either the BH10 or the BH10(Vpu−) virus. These data demonstrate the susceptibility of hBST-2 to down-regulation by Vpu in T cells.
FIG. 6.
The hTMa1 and CTh proteins profoundly inhibit the replication of wild-type HIV-1 in SupT1 cells. (A) The hBST-2, hTMa1, and CTh SupT1 cell lines were exposed to doxycycline (50 ng/ml) prior to infection with either wild-type HIV-1 (BH10) or Vpu deletion-containing HIV-1 [BH10(Vpu−)]. Virus stocks were prepared by transfecting HEK293T cells with BH10 or BH10(Vpu−) DNA together with VSV G DNA. Three doses of viruses with MOIs of 0.5, 1, and 2 were used to infect hBST-2 SupT1 cells. hTMa1 and CTh cells were exposed to viruses with a MOI of 1. Levels of BST-2 proteins in each cell line were assessed 40 h after infection in Western blot assays with anti-Flag antibody. Expression of the viral Gag and Vpu proteins was also examined by Western blotting. Control represents uninfected SupT1 cells. (B) Growth of the BH10 and BH10(Vpu−) viruses in SupT1 cell lines. The control cell line was generated with the control retroviral vector Pur-GOI, which does not contain an inserted gene within its expression cassette. SupT1 cells were infected with the BH10 or BH10(Vpu−) virus (MOI of 0.5) in the presence of different doses of doxycycline (0, 50, and 500 ng/ml). Levels of viral reverse transcriptase activity were measured and plotted over days of infection. Results shown are the average from two independent infections.
We next used the BH10 or BH10(Vpu−) virus to challenge hBST-2, hTMa1, and CTh cells, as well as a control cell line that was created with the empty retroviral vector Pur-GOI. Cells were exposed to different doses of doxycycline (0, 50, and 500 ng/ml) in order to induce the expression of BST-2 proteins at different levels. Virus growth was monitored by measuring viral reverse transcriptase activity in culture supernatants. The results in Fig. 6B showed that expression of hBST-2 effectively inhibited the growth of BH10(Vpu−) but not the wild-type BH10 virus, whereas the hTMa1 and CTh proteins blocked the replication of both the BH10 and BH10(Vpu−) viruses by more than 1 order of magnitude when 500 ng/ml doxycycline was administered. These data demonstrate that in T cells, Vpu is able to deplete hBST-2, but not hTMa1 and CTh, which effectively impair the replication of wild-type HIV-1.
DISCUSSION
Our results demonstrate that BST-2 from COS-7 African green monkey kidney cells is resistant to Vpu and effectively blocks HIV-1 production. Reciprocally, hBST-2 restricts the production of SIVagm (5). In addition to Trim5alpha and APOBEC3G, which exhibit species-specific restriction activity and thus limit cross-species transmission of infectious viruses (20, 21, 24), BST-2 represents another cellular factor with similar properties. This notion is further supported by the finding that BST-2 from monkeys and mice also inhibits HIV-1 production in the presence of Vpu (2, 3, 12).
The Vpu resistance property of agmBST-2 allowed us to map within hBST-2 the key residues that determine its sensitivity to HIV-1 Vpu. The amino acids thus identified, including 22-LL-23, V30, and T45, are all located in the transmembrane domain. Deletion of 22-LL-23 creates resistance to Vpu at a much higher level than the V30G or T45I mutations. While our manuscript was in preparation, Paul Bieniasz's group reported similar findings (12). In addition to V30 and T45, they observed that deletion of 25-GI-26, as well as the I33V and I36L substitutions, also diminished the sensitivity of hBST-2 to Vpu to variable extents. Maximal resistance to Vpu was observed when delGI was recombined with T45I (12). The role of 22-LLL-24 in this regard was, however, not reported in this study, likely because McNatt and colleagues created and tested the LL23,24VI mutation, which involves changing leucine to valine and isoleucine, all of which harbor similar hydrophobic side chains (12). For similar reasons, we did not observe the Vpu resistance phenotype associated with delGI because these two amino acids were mutated individually to alanine and valine in our study. Given the essential role of the transmembrane domain of Vpu in promoting virus release (22), we propose that Vpu interacts with hBST-2 via the transmembrane domains of both proteins. In support of this notion, mutating the transmembrane domain sequence of hBST-2, e.g., deletion of 22-LL-23, substantially reduces the association of hBST-2 with Vpu. In contrast with the aforementioned findings with agmBST-2, Vpu-resistant determinants appear to be dispersed in rodent BST-2 proteins (2), which indicates that rodent BST-2 proteins escape from Vpu-mediated down-modulation by a different mechanism.
The molecular basis of the antagonism of Vpu against hBST-2 has not been fully elucidated. Studies by John Guatelli's group indicate that Vpu is able to down-modulate the cell surface level of hBST-2 (25). Vpu is also able to down-regulate the total level of hBST-2 (1, 2), which is further confirmed by studies performed with macrophages (14). Our results further demonstrate that levels of hBST-2 are substantially reduced in SupT1 cells by wild-type HIV-1 infection. Interestingly, the agmBST-2 and hΔLL22/23 proteins, which potently inhibit wild-type HIV-1 production, are resistant to down-modulation by Vpu. This observation further strengthens the concept that down-regulation is the major mechanism whereby Vpu counteracts BST-2.
It remains unknown whether the loss of cell surface expression of hBST-2 in the presence of Vpu is attributable to depletion of cellular pools of hBST-2 or is a result of interference with the trafficking of hBST-2 to the cell surface. The cellular pathway that Vpu hijacks to antagonize hBST-2 remains a mystery, although a recent study suggests a likely involvement of the endosomal/lysosomal pathway in the down-modulation of BST-2 (13). We were unable to detect the hemagglutinin-ubiquitin signal associated with hBST-2 when hemagglutinin-ubiquitin DNA was cotransfected with hBST-2 and BH10 DNAs (data not shown), which casts a cloud on the notion of direct ubiquitination of hBST-2. It appears that Vpu does not function in the same way in different cell lines in terms of hBST-2 depletion. For example, very little effect of Vpu on hBST-2 expression was observed in CEMx174 cells (14). We speculate that CEMx174 cells do not possess the cellular factors that Vpu needs to down-modulate hBST-2.
Our data suggest that hBST-2 is more potent than agmBST-2 in blocking the release of HIV-1 virus-like particles that were produced from the GPV-CTEx4 DNA in the absence of Vpu expression (Fig. 3F). Exchange of the cytoplasmic domains between hBST-2 and agmBST-2 created two mutant proteins named CTh and hCTa that preserved the antiviral activity associated with the origin of the cytoplasmic domain (Fig. 3F). We thus speculate that the cytoplasmic domain regulates the potency of BST-2 in restricting virus production.
The bst-2 gene has within its promoter the IRF-1/-2 and ISGF3 response elements (17), allowing BST-2 expression to be enhanced by interferon. Blocking the release of various enveloped viruses suggests that BST-2 acts as an important element in interferon-mediated antiviral pathways through a unique mechanism (5, 6, 12, 19). Yet, the physiological relevance of the anti-HIV activity of BST-2 has not yet been demonstrated by experiments with primary T cells and ultimately in vivo. One possibility is that HIV-1 particles that are tethered to the cell surface may facilitate the formation of viral synapses and thus promote cell-to-cell virus transmission. This may be particularly true in infected solid organs such as the lymph nodes and gut. This issue may partially be addressed by our data showing that hBST-2 not only blocks the release of HIV-1 particles in the absence of Vpu but, more importantly, abrogates the infectivity of HIV-1 virions. In this light, hBST-2 is expected to pose a severe restriction on HIV-1 infection if Vpu is absent. This implication awaits further support from study of the impact of BST-2 on HIV-1 infectivity in primary CD4+ T cells and macrophages. We noted that another group reported an approximately 10-fold decrease in p24 production in the culture supernatants from NL4-3 proviral DNA with ectopic expression of BST-2, albeit that they observed a similar 10-fold decrease in the yield of infectious HIV-1 particles (2). In addition, the Vpu-negative HIV-1 progeny exhibited kinetics of replication in A3.01 cells similar to that of wild-type HIV-1 (7). However, the endogenous level of BST-2 in A3.01 cells has not been examined. The discrepancy between our observation and the results of other groups in regard to the effect of BST-2 expression on the infectivity of HIV-1 virions likely results from the different experimental systems utilized, such as cell lines and virus strains.
In summary, our study further supports the species-specific sensitivity of BST-2 to Vpu. Importantly, results of detailed mutagenesis and biochemical analysis suggest that Vpu and BST-2 interact via their transmembrane domains. Elucidation of the molecular mechanism behind the antagonism by Vpu may lead to the development of new strategies for HIV intervention.
Acknowledgments
We thank Klaus Strebel for providing Vpu and hBST-2 antiserum through the NIH AIDS Research and Reference Reagent Program and Rongtuan Lin for helpful discussions.
This research was supported by funding from the Canadian Institutes of Health Research and the Canadian Foundation for AIDS Research.
Footnotes
Published ahead of print on 27 May 2009.
REFERENCES
- 1.Bartee, E., A. McCormack, and K. Fruh. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goffinet, C., I. Allespach, S. Homann, H. M. Tervo, A. Habermann, D. Rupp, L. Oberbremer, C. Kern, N. Tibroni, S. Welsch, J. Krijnse-Locker, G. Banting, H. G. Krausslich, O. T. Fackler, and O. T. Keppler. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5285-297. [DOI] [PubMed] [Google Scholar]
- 3.Gupta, R. K., S. Hue, T. Schaller, E. Verschoor, D. Pillay, and G. J. Towers. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113803-809. [DOI] [PubMed] [Google Scholar]
- 5.Jouvenet, N., S. J. Neil, M. Zhadina, T. Zang, Z. Kratovac, Y. Lee, M. McNatt, T. Hatziioannou, and P. D. Bieniasz. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 831837-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaletsky, R. L., J. R. Francica, C. Agrawal-Gamse, and P. Bates. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. USA 1062886-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klimkait, T., K. Strebel, M. D. Hoggan, M. A. Martin, and J. M. Orenstein. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kupzig, S., V. Korolchuk, R. Rollason, A. Sugden, A. Wilde, and G. Banting. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4694-709. [DOI] [PubMed] [Google Scholar]
- 9.Maldarelli, F., M. Y. Chen, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 675056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malim, M. H., and M. Emerman. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3388-398. [DOI] [PubMed] [Google Scholar]
- 11.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1565-574. [DOI] [PubMed] [Google Scholar]
- 12.McNatt, M. W., T. Zang, T. Hatziioannou, M. Bartlett, I. B. Fofana, W. E. Johnson, S. J. Neil, and P. D. Bieniasz. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell, R. S., C. Katsura, M. A. Skasko, K. Fitzpatrick, D. Lau, A. Ruiz, E. B. Stephens, F. Margottin-Goguet, R. Benarous, and J. C. Guatelli. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyagi, E., A. J. Andrew, S. Kao, and K. Strebel. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. USA 1062868-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neil, S. J., V. Sandrin, W. I. Sundquist, and P. D. Bieniasz. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451425-430. [DOI] [PubMed] [Google Scholar]
- 17.Ohtomo, T., Y. Sugamata, Y. Ozaki, K. Ono, Y. Yoshimura, S. Kawai, Y. Koishihara, S. Ozaki, M. Kosaka, T. Hirano, and M. Tsuchiya. 1999. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem. Biophys. Res. Commun. 258583-591. [DOI] [PubMed] [Google Scholar]
- 18.Rollason, R., V. Korolchuk, C. Hamilton, P. Schu, and G. Banting. 2007. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci. 1203850-3858. [DOI] [PubMed] [Google Scholar]
- 19.Sakuma, T., T. Noda, S. Urata, Y. Kawaoka, and J. Yasuda. 2009. Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 832382-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 1022832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert, U., S. Bour, A. V. Ferrer-Montiel, M. Montal, F. Maldarell, and K. Strebel. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 24.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 796111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varthakavi, V., R. M. Smith, S. P. Bour, K. Strebel, and P. Spearman. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 10015154-15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodrich, H., A. Schambach, and H. G. Krausslich. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]