Abstract
A negative association between polymorphism Leu-214 and type-1 thymidine analogue mutations (TAM1) and a positive association with a clinically favorable virological response to thymidine analogue-based combination antiretroviral therapy have been described. In this study, the impact of Leu-214 on replication capacity and resistance to zidovudine (ZDV) of viruses containing TAM1 or TAM2 was determined. Leu-214 decreased the growth rate of viruses bearing Tyr-215, as well as their resistance to ZDV. This observation was confirmed by structural and molecular modeling data, suggesting a regulatory role for Leu-214 in the emergence and phenotypic resistance of TAM1.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is responsible for the conversion of the viral single-stranded RNA genome into double-stranded DNA, prior to host-genome integration in target cells. RT has been widely considered a key target for combination antiretroviral therapy. Drugs targeting this viral enzyme include nucleoside analogues (NRTIs) and nonnucleoside RT inhibitors (NNRTIs). One of the mechanisms contributing to decreased HIV susceptibility to NRTIs promotes the removal of the nucleoside analogue from the terminated DNA chain (22). The mutations responsible for this effect, thymidine analogue mutations (TAMs), emerge after long-term therapy with zidovudine (ZDV) and/or stavudine (d4T) and confer resistance to almost all clinically approved RT inhibitors. Two different TAM patterns have been defined: TAM1, including Leu-41, Trp-210, and Tyr-215; and TAM2, including Asn-67, Arg-70, Phe-215, and Gln/Glu-219. TAM1 is more prevalent and confers a higher degree of resistance to thymidine analogues (6, 8, 10, 12, 19, 32-34). To date, the factors driving one mutational pattern or another remain unclear, although the genomic background of the treatment-naive viral population, host factors such as HLA genotype (16), and stochastic effects could be involved.
Leu-214 is a natural polymorphism in the RT coding region, and it is present in ca. 10 to 20% of antiretroviral treatment (ART)-naive and ART-experienced patients carrying any of the major HIV-1 subtypes A, B, or C (3, 4; http://www.hiv.lanl.gov/). A negative association between Leu-214 and the TAM1 pattern and a positive association with the TAM2 pattern have been observed (29). Moreover, it has recently been demonstrated the association of the Leu-214 with a favorable virological response to thymidine analogue-containing ART (3). These data suggest that Leu-214 may regulate divergent resistance pathways by affecting viral fitness and/or drug susceptibility. However, experimental evidence supporting these observations has not yet been reported. The aim of the present study was to compare the in vitro growth rate and relative viral fitness of HIV-1 recombinant mutants containing the Leu-214 polymorphism in RT containing TAM1 or TAM2 patterns, in both the absence and the presence of ZDV. A preliminary structural analysis was also performed in order to explain the putative role of this polymorphism in the RT replicative capacity at the molecular level.
MATERIALS AND METHODS
Site-directed mutagenesis.
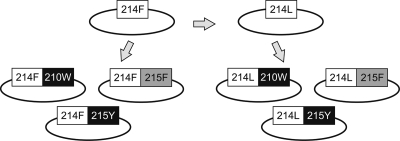
Phe-214 is found in the hemiplasmid p83-2 (9) containing the 5′ half of the genome of the proviral clone pNL4-3 (1). The Leu-214 polymorphic variant was generated by PCR-based site-directed mutagenesis on nucleotide position 3189 (17) by using a QuikChange II site-directed mutagenesis kit (Stratagene). The TAMs Trp-210 (position 3178) and Phe-215 and Tyr-215 (positions 3192 to 3193) were introduced into wild-type variants containing either Phe-214 or Leu-214 by using the same procedure (Fig. 1).
FIG. 1.
Diagram of the viral variants generated by site-directed mutagenesis. Briefly, the Leu-214 (214L) polymorphism was introduced in the pNL4-3 background, which contained the Phe-214 (214F) variant. Trp-210 (210W), Tyr-215 (215Y), or Phe-215 (215F) was then combined with each of these Phe-214 and Leu-214 backgrounds.
Generation of a cloning vector.
The recombinant vector pJM14 (21), reconstructed with an RT-coding region (including its DNA polymerase and RNase H domains) (30), was used to create the new cloning vector pJM16ΔRT (5′-half of the HIV-1 genome without RT). Briefly, the RT-coding region of reconstructed pJM14 was cut with the restriction enzymes SmaI (position 2588) and AgeI (position 3493) (Fermentas), and the resulting fragment was replaced by a polylinker.
Cloning.
To avoid miscarrying mutations that could have occurred during the PCR-based mutagenesis amplification, a 908-bp fragment ranging from positions 2574 to 3482 from the newly generated mutants was amplified by PCR (Platinum Taq High Fidelity; Invitrogen) using primers 2574U29-SmaI (5′-CCA GTA AAA TTA AAG CCC GGG ATG GAT GG-3′) and 3482L31-AgeI (5′-TGG GTC ATA ATA CAC TCC ATG TAC CGG TTC T-3′) and subsequently subcloned into the pJM16ΔRT cloning vector using the SmaI and AgeI restriction enzymes (Fermentas). Sequence integrity from positions 2500 to 3600 was confirmed on the newly generated clones (BigDye Terminator v3.1 cycle sequencing kit; Applied Biosystems).
Generation of viral stocks.
Infectious viral stocks from recombinant proviral clones were obtained by cotransfection with hemiplasmid p83-10 (containing the 3′ half genome of the pNL4-3 [9]) by electroporation of MT-4 cells (NIH AIDS Research and Reference Reagent Program). Seven days after transfection, supernatants were collected and p24Gag content was determined by enzyme-linked immunosorbent assay (HIV-1 p24 ELISA kit; Perkin-Elmer). Chemiluminescence titration assays in HeLa-derived TZM-bl cells (NIH AIDS Research and Reference Reagent Program) were used to determine the mean 50% tissue culture infective dose(s) (TCID50) of each viral stock, as previously described (18).
Replication capacity assays.
Viral replication kinetics of mutant viruses was assayed by inoculation of 5,000 TCID50 of each viral stock onto 5 × 106 peripheral blood mononuclear cells (PBMC) mixed from two healthy donors (multiplicity of infection [MOI] of 0.001), previously stimulated with 3 μg of phytohemagglutinin (Sigma-Aldrich)/ml and 10 U of interleukin-2 (Roche)/ml for 3 days. Replication kinetics assays were performed in parallel, using the same PBMC donor mix in order to guarantee reliable results. After incubation for 2 h at 37°C, the cells were washed twice with phosphate-buffered saline and resuspended in RPMI (Invitrogen) medium supplemented with 20% fetal bovine serum (FBS, Invitrogen) and interleukin-2 (10 U/ml) at a final concentration of 106 cells/ml. When indicated, ZDV (Sigma) was added to the cultures. Viral replication was quantified by measuring HIV-1 p24Gag antigen production in the culture supernatant for 7 days (HIV-1 p24 ELISA kit; Perkin-Elmer). Growth kinetics were analyzed by fitting a linear model to the log-transformed p24Gag data during the exponential growth phase by maximum-likelihood methods using GraphPad Prism 4 software.
Growth competition assays.
Each pair of competing viruses was mixed at ratios of 20:80, 50:50, and 80:20 to infect 106 MT-4 cells in 1 ml of R-10 (RPMI medium supplemented with 10% FBS), with 1,000 TCID50 (MOI of 0.001). After 2 h at 37°C, cells were washed twice with PBS, resuspended in 5 ml of R-10 medium (2 × 105 cells/ml) and cultured in six-well plates. Culture supernatants were collected twice weekly, and 100 μl was used to newly infect 106 of fresh MT-4 cells. Viral RNA was extracted from culture supernatants by using the Qiamp viral RNA minikit (Qiagen). The viral RNA fragment from positions 2212 to 4120 was first amplified by one-step RT-PCR (Superscript High-Fidelity; Invitrogen), followed by nested PCR ranging from positions 2574 to 3482. The relative proportions of the two competing variants were determined at days 7, 14, 21, and 28 based on the relative peak heights of codons 210, 214, and 215 in electropherograms obtained by bidirectional sequencing of the RT coding region (Big Dye Terminator v3.1 cycle sequencing kit and a 3100-Avant genetic analyzer [Applied Biosystems]).
Structural analysis.
The structural analysis was performed by using the X-ray crystallographic coordinates of HIV-1 RT deposited in the Protein Data Bank (http://www.rcsb.org/PDB) with code 1N6Q (27). This structure corresponds to a HIV-1 RT complex bound to a DNA-DNA template primer, where the primer is blocked at its 3′ terminus with ZDV. In this Protein Data Bank entry, the ZDV-monophosphate (ZDVMP) moiety is located at the nucleotide binding site, in the so-called “pretranslocation” complex. The starting geometries of the mutants containing Tyr-215 in combination with Leu-214 and Phe-214 were generated by making single-residue substitutions in the crystallographic structure. In order to analyze the potential interactions occurring in the absence of the drug, the ZDVMP residue was replaced by deoxythymidine-monophosphate (dTMP). The obtained structures were energy minimized, using the MMFF force field (11), a dielectric constant equal to 1, and a convergence criterion fixed at 0.5 kJ/mol·Å. RT residues within 30 Å from the catalytic Asp residues were free to move, while the rest of the enzyme was kept frozen. Subsequently, the MOLINE method (2) was adopted to evaluate the interaction energies of the four complexes derived from the combination of both RT variants and both DNA primer templates with 3′-terminus ZDVMP or dTMP. All simulations were performed with MacroModel version 7.2 (24). The LigPlot program was used to identify the most relevant interactions between the nucleotide and p66 subunit of the RT (31).
Statistical analysis.
Statistical data were estimated by using GraphPad Prism v.4 software. A Student t test was applied to compare data means.
RESULTS
Variations in the replication capacity.
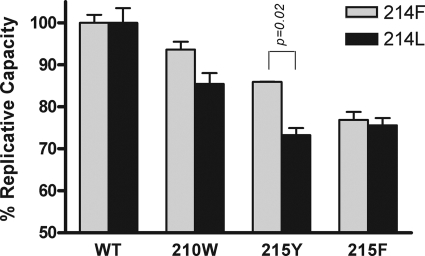
Viral replication kinetics of mutant viruses was first assayed in the absence of drug. As expected, all HIV-1 variants Phe-214 or Leu-214 in combination with Trp-210, Tyr-215, or Phe-215 showed decreased replication capacity compared to the wild-type virus. The loss of fitness was more evident in viruses with mutations at codon 215 (Fig. 2). In fact, the role of Trp-210 in replication capacity is controversial, with some previous studies reporting a significant effect on viral fitness (13, 14) and others showing limited change (5, 20).
FIG. 2.
Replication kinetics assay. The slope of p24 antigen production of each TAM-mutated virus after infection of PBMC was compared to the slope of the corresponding wild-type virus harboring the same 214 background (Phe-214 in gray bars and Leu-214 in black bars). Error bars represent the standard deviations of two independent experiments. Statistical analyses were performed by using the Student t test. WT, wild type.
No statistically significant difference on replication capacity was observed between the two viruses containing Phe-214 or Leu-214 within the wild-type background. However, pairwise comparisons of the growth rate of viral variants that contained Phe-214 or Leu-214 in combination with the TAM Trp-210, Tyr-215, or Phe-215 showed that Leu-214 significantly decreases the growth rate of the Tyr-215 mutant, slightly decreases replication of the Trp-210 variant, and has no effect on the Phe-215 mutant, with respect to viral variants that contained Phe-214 (Fig. 2). The data also suggest that Tyr-215, the major mutation in the TAM1 pattern, resulted in a better fitness than the Phe-215 mutation in the Phe-214 background, whereas no clear advantage was observed in the Leu-214 background.
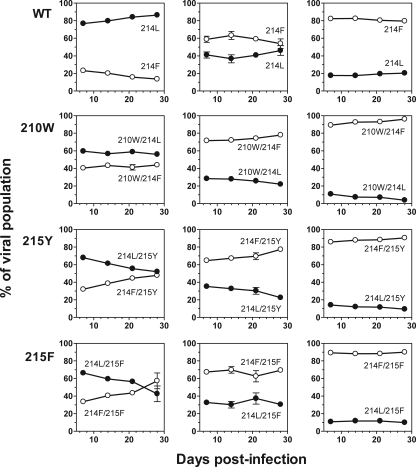
Growth competition assays were performed to assess the previous results. Recombinant viruses with either Phe-214 or Leu-214 in an otherwise isogenic viral backbone were competed. A total of four different competition viral variant pairs were established: Phe-214 versus Leu-214, Trp-210/Phe-214 versus Trp-210/Leu-214, Phe-214/Tyr-215 versus Leu-214/Tyr-215, and Phe-214/Phe-215 versus Leu-214/Phe-215. The data obtained from the growth competition experiments are consistent with those previously obtained in the replication kinetics assays: the clearest evidence of comparative disadvantage was observed between the two viruses containing Tyr-215 (Fig. 3), since Leu-214/Tyr-215 shows a 8.4% ± 0.8% loss of fitness when competed with Phe-214/Tyr-215.
FIG. 3.
Growth competition assays between Phe-214 (214F) and Leu-214 (214L) in wild-type (WT), Trp-210 (210W), Tyr-215 (215Y) or Phe-215 (215F)-mutated HIV-1 variants; the Leu-214 variant is represented with solid symbols and Phe-214 with open symbols. Each pair of viruses was mixed at initial ratios of 20:80 (first column), 50:50 (second column) and 80:20 (third column), and the proportion of each viral variant was determined at days 7, 14, 21, and 28. The error bars represent the standard error of two independent experiments.
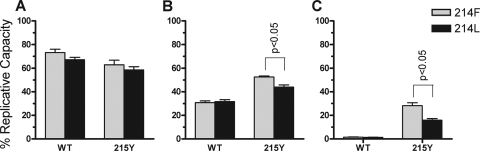
Susceptibility to ZDV.
The viral variants containing Phe-214 and Leu-214 in combination with the wild-type Thr-215 or the thymidine analogue-associated mutation Tyr-215 were selected to perform replication kinetics assays in the presence of 0.02, 0.2, and 1 μM ZDV. When cultured at 0.02 μM ZDV, the wild-type Thr-215 variants remained more replicative than the Tyr-215 mutants (Fig. 4A), as expected, because the ZDV IC50 in wild-type-susceptible virus is 0.033 μM (25). In contrast, both Tyr-215 mutants—whether containing Phe-214 or Leu-214—replicated better at 0.2 μM ZDV (Fig. 4B). In these conditions, Phe-214/Tyr-215 replicated better than Leu-214/Tyr-215. When 1 μM ZDV was used in the cell culture, the replication advantage of Phe-214/Tyr-215 was even greater (Fig. 4C). Our findings demonstrate that the Tyr-215 mutation in a Phe-214 background results in better replication kinetics and greater fitness than with Leu-214 in the absence or presence of ZDV.
FIG. 4.
Replication kinetics of wild-type (WT) and Tyr-215 (215Y) HIV-1 variants in the presence of 0.02 μM (A), 0.2 μM (B), and 1 μM (C) ZDV. The slope of p24 antigen production in each experiment was compared to the slope of the corresponding nonmutated wild-type virus harboring the same 214 background in the absence of drug (Phe-214 in gray bars and Leu-214 in black bars). Error bars represent the standard deviations of two independent experiments. Statistical analyses were performed by using the Student t test.
Molecular modeling studies.
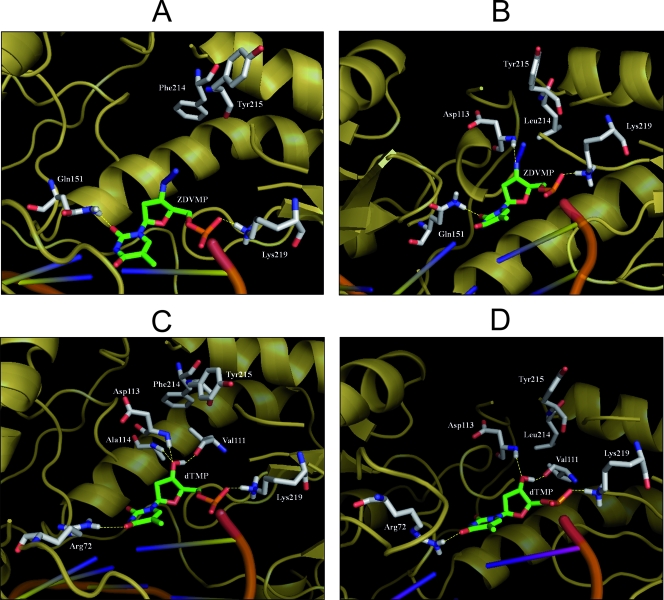
In order to test whether the structural proximity of the side chains at positions 214 and 215 could explain the different levels of viral replication and susceptibility to ZDV, we built molecular models of the RT variants containing the Tyr-215 mutation in complex with a template primer containing ZDV- or dTMP-terminated primers and evaluated the interaction energy (ΔE) for the four RT complexes. According to the theoretical models, the calculated ΔE of ZDVMP primer template with the RT containing Phe-214 and Tyr-215 was higher than that observed with the enzyme containing Leu-214 and Tyr-215 (ΔE in kcal/mol, −4,876.42 versus −4,938.92). In addition, we observed an increased number of hydrogen bonds for ZDVMP-oligonucleotide in the optimized RT complex containing Leu-214/Tyr-215 compared to the enzyme containing Phe-214/Tyr-215 (Fig. 5A and B). In agreement with the results of the replication kinetics assays carried out in the absence of drugs, the Phe-214/Tyr-215 variant bound to the dTMP-terminated primer showed a favorable energetic profile, since the interaction energy for the dTMP moiety was slightly lower than for the Leu-214/Tyr-215 combination (ΔE in kcal/mol, −4,836.35 versus −4,830.22). Such an observation was further supported by the higher stabilization of the dTMP due to the specific hydrogen bond established between the nucleotide and the p66 Ala-114 backbone only in the RT harboring the Phe-214/Tyr-215 combination (Fig. 5C and D).
FIG. 5.
Molecular modeling analysis of ZDVMP and dTMP interactions in the optimized complex of RT Phe-214/Tyr-215 (A and C) and RT Leu-214/Tyr-215 (B and D) bound to template primers blocked at their 3′ ends. The enzyme and DNA are shown, respectively, as yellow-orange and orange diagrams; the 3′-terminal nucleotides and the RT residues involved in hydrogen bonding are represented, respectively, as green and gray carbon sticks. Hydrogen bonds are indicated with yellow dashed lines.
DISCUSSION
In the absence of drug, Tyr-215 and Trp-210, both included in the TAM1 pattern, confer higher replication capacities when in combination with Phe-214, whereas the Phe-215 mutation, considered a TAM2, did not show any significant advantage. The impact of the Leu-214 polymorphism on viral replication when associated with other changes in the RT structure may explain the strong negative association observed in NRTI-failing patients between TAM1 and Leu-214, probably by affecting the stability of the dTMP-enzyme complex by interactions between aromatic side chains at close positions (Fig. 5).
The proportion of patients who experienced virological failure at 48 months after starting thymidine analogue-based combination ART was 16% in patients with Leu-214 and 36% in those with Phe-214, if previously ART-naive patients, or 47% in patients with Leu-214 and 72% in those with Phe-214, if ART-experienced patients (3). Our results provide additional insights into the favorable impact of Leu-214 on the virological response to thymidine analogue-based treatments. This effect could be due to the fact that Leu-214 somehow inhibits the development of TAM1. It has been reported that this mutational profile confers higher phenotypic resistance to NRTIs than TAM2, thus providing a favorable effect on virological response (6, 7, 15, 26). Moreover, our results suggest that Leu-214 not only influences viral fitness but could also play a direct role in drug susceptibility. Thus, viruses containing Tyr-215 in their RT were more susceptible to ZDV in the presence of Leu-214 than in the presence of Phe-214. Our molecular modeling studies highlighted a favorable interaction energy of ZDVMP primer template for the variant containing Leu-214/Tyr-215. These data would be consistent with a higher genetic barrier for excision of the inhibitor in the presence of a pyrophosphate donor. However, geometrical constraints arising from the different conformation of the ZDVMP moiety in the binary complex could also affect the removal of the inhibitor. Our results also support previous reports showing that Tyr-208, Lys-211 and Phe-214 substitutions, despite lacking a direct effect on drug resistance per se, could increase ZDV resistance and cross-resistance when they appear in combination with amino acid substitutions of the TAM1 pathway (7, 23, 28).
The present study evaluates the in vitro effect of Leu-214 on the single TAM1 Trp-210 and Tyr-215, since they showed a stronger negative association with Leu-214 in ART-experienced patients (3, 29). In a similar way, the effect of Leu-214 on the single TAM2 Phe-215 was also assessed. In the latter case, the absence of a significant fitness effect when Leu-214 was combined with Phe-215 is not surprising since this mutation did not showed a positive per se association with Leu-214 in ART-experienced patients (3). Since in vivo TAM patterns are highly complex, involving a larger number of mutations clustering together, other TAMs, including Leu-41, Asp-44, Asn-67, Arg-70, and Gln-219, have yet to be evaluated in vitro. More complex approaches with diverse combinations of these TAMs could also improve our understanding of this association mechanism. Further modeling studies by means of molecular dynamics simulations could provide interesting insights into each mutation effect on the enzyme conformational properties and additional correlations with resistance profiles.
In conclusion, the effect of Leu-214 on the RT structure of HIV-1 directly affects the growth rate and drug susceptibility of drug-resistant variants containing TAM1. These results help to explain the putative role of Leu-214 in regulating the emergence of these viral variants and in enhancing the virological response to thymidine analogue-containing ART.
Acknowledgments
This study was supported by the Fundación para la Investigación y Prevención del Sida en España through grant 36771/08, by the the Spanish AIDS network Red Temática Cooperativa de Investigación en SIDA (RD06/0006), and by grants from the Italian National Institute of Health and the Italian Ministry of University and Scientific Research. We acknowledge support from CHAIN, Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project no. 223131, funded by the European Commission Framework 7 Program.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaro, S., F. Gasparrini, O. Incani, S. Mecucci, D. Misiti, M. Pierini, and C. Villani. 2000. A quasi-flexible automatic docking processing for studying stereoselective recognition mechanisms. Part I. Protocol Validation. J. Comp. Chem. 21515-530. [DOI] [PubMed] [Google Scholar]
- 3.Ceccherini-Silberstein, F., A. Cozzi-Lepri, L. Ruiz, A. Mocroft, A. N. Phillips, C. H. Olsen, J. M. Gatell, H. F. Gunthard, P. Reiss, C. F. Perno, B. Clotet, and J. D. Lundgren. 2007. Impact of HIV-1 reverse transcriptase polymorphism F214L on virological response to thymidine analogue-based regimens in antiretroviral therapy (ART)-naive and ART-experienced patients. J. Infect. Dis. 1961180-1190. [DOI] [PubMed] [Google Scholar]
- 4.Ceccherini-Silberstein, F., F. Gago, M. Santoro, C. Gori, V. Svicher, F. Rodriguez-Barrios, R. d'Arrigo, M. Ciccozzi, A. Bertoli, A. d'Arminio monforte, J. Balzarini, A. Antinori, and C. F. Perno. 2005. High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J. Virol. 7910718-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong, M. E., W. Heneine, and J. G. Garcia-Lerma. 2007. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 813037-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cozzi-Lepri, A., L. Ruiz, C. Loveday, A. N. Phillips, B. Clotet, P. Reiss, B. Ledergerber, C. Holkmann, S. Staszewski, and J. D. Lundgren. 2005. Thymidine analogue mutation profiles: factors associated with acquiring specific profiles and their impact on the virological response to therapy. Antivir. Ther. 10791-802. [PubMed] [Google Scholar]
- 7.De Luca, A., S. Di Giambenedetto, L. Romano, A. Gonnelli, P. Corsi, M. Baldari, M. Di Pietro, S. Menzo, D. Francisci, P. Almi, and M. Zazzi. 2006. Frequency and treatment-related predictors of thymidine-analogue mutation patterns in HIV-1 isolates after unsuccessful antiretroviral therapy. J. Infect. Dis. 1931219-1222. [DOI] [PubMed] [Google Scholar]
- 8.Flandre, P., D. Descamps, V. Joly, V. Meiffredy, C. Tamalet, J. Izopet, and F. Brun-Vezinet. 2004. A survival method to estimate the time to occurrence of mutations: an application to thymidine analogue mutations in HIV-1-infected patients. J. Infect. Dis. 189862-870. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10343-350. [DOI] [PubMed] [Google Scholar]
- 10.Gonzales, M. J., T. D. Wu, J. Taylor, I. Belitskaya, R. Kantor, D. Israelski, S. Chou, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS 17791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halgren, T. A. 1996. Merck molecular force field. I, II, III, IV, and V. J. Comput. Chem. 17490-641. [Google Scholar]
- 12.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. J. Brown, A. V. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181904-911. [DOI] [PubMed] [Google Scholar]
- 13.Harrigan, P. R., S. Bloor, and B. A. Larder. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 723773-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrigan, P. R., I. Kinghorn, S. Bloor, S. D. Kemp, I. Najera, A. Kohli, and B. A. Larder. 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 705930-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Japour, A. J., S. Welles, R. T. D'Aquila, V. A. Johnson, D. D. Richman, R. W. Coombs, P. S. Reichelderfer, J. O. Kahn, C. S. Crumpacker, D. R. Kuritzkes, et al. 1995. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients after long-term zidovudine treatment. J. Infect. Dis. 1711172-1179. [DOI] [PubMed] [Google Scholar]
- 16.John, M., C. B. Moore, I. R. James, and S. A. Mallal. 2005. Interactive selective pressures of HLA-restricted immune responses and antiretroviral drugs on HIV-1. Antivir. Ther. 10551-555. [PubMed] [Google Scholar]
- 17.Korber, B. T., B. T. Foley, K. L. Kuiken, S. K. Pillai, and J. G. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG, p. 102-111. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 1998. Theoretical Biology and Biophysics Group, Los Alamos, NM.
- 18.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcelin, A. G., C. Delaugerre, M. Wirden, P. Viegas, A. Simon, C. Katlama, and V. Calvez. 2004. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J. Med. Virol. 72162-165. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Picado, J., and M. A. Martinez. 2008. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 134104-123. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Picado, J., T. Wrin, S. D. Frost, B. Clotet, L. Ruiz, A. J. Brown, C. J. Petropoulos, and N. T. Parkin. 2005. Phenotypic hypersusceptibility to multiple protease inhibitors and low replicative capacity in patients who are chronically infected with human immunodeficiency virus type 1. J. Virol. 795907-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 9513471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, M. D., N. Margot, B. Lu, L. Zhong, S. S. Chen, A. Cheng, and M. Wulfsohn. 2004. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J. Infect. Dis. 189837-846. [DOI] [PubMed] [Google Scholar]
- 24.Mohamadi, F., N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson, and W. C. Still. 1990. MacroModel: an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 11440-467. [Google Scholar]
- 25.Ntemgwa, M., M. A. Wainberg, M. Oliveira, D. Moisi, R. Lalonde, V. Micheli, and B. G. Brenner. 2007. Variations in reverse transcriptase and RNase H domain mutations in human immunodeficiency virus type 1 clinical isolates are associated with divergent phenotypic resistance to zidovudine. Antimicrob. Agents Chemother. 513861-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee, S. Y., T. Liu, J. Ravela, M. J. Gonzales, and R. W. Shafer. 2004. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob. Agents Chemother. 483122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarafianos, S. G., A. D. Clark, Jr., K. Das, S. Tuske, J. J. Birktoft, P. Ilankumaran, A. R. Ramesha, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2002. Structures of HIV-1 reverse transcriptase with pre- and posttranslocation AZTMP-terminated DNA. EMBO J. 216614-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturmer, M., S. Staszewski, H. W. Doerr, B. Larder, S. Bloor, and K. Hertogs. 2003. Correlation of phenotypic zidovudine resistance with mutational patterns in the reverse transcriptase of human immunodeficiency virus type 1: interpretation of established mutations and characterization of new polymorphisms at codons 208, 211, and 214. Antimicrob. Agents Chemother. 4754-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svicher, V., T. Sing, M. M. Santoro, F. Forbici, F. Rodriguez-Barrios, A. Bertoli, N. Beerenwinkel, M. C. Bellocchi, F. Gago, A. d'Arminio Monforte, A. Antinori, T. Lengauer, F. Ceccherini-Silberstein, and C. F. Perno. 2006. Involvement of novel human immunodeficiency virus type 1 reverse transcriptase mutations in the regulation of resistance to nucleoside inhibitors. J. Virol. 807186-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villena, C., J. G. Prado, M. C. Puertas, M. A. Martinez, B. Clotet, L. Ruiz, N. T. Parkin, L. Menendez-Arias, and J. Martinez-Picado. 2007. Relative fitness and replication capacity of a multinucleoside analogue-resistant clinical human immunodeficiency virus type 1 isolate with a deletion of codon 69 in the reverse transcriptase coding region. J. Virol. 814713-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8127-134. [DOI] [PubMed] [Google Scholar]
- 32.Wolf, K., H. Walter, N. Beerenwinkel, W. Keulen, R. Kaiser, D. Hoffmann, T. Lengauer, J. Selbig, A. M. Vandamme, K. Korn, and B. Schmidt. 2003. Tenofovir resistance and resensitization. Antimicrob. Agents Chemother. 473478-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, F. Ariasi, F. Volot, J. A. Gastaut, H. Gallais, J. Moreau, and J. Fantini. 1999. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J. Clin. Microbiol. 374099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, and J. Fantini. 2000. Mutation L210W of HIV-1 reverse transcriptase in patients receiving combination therapy. Incidence, association with other mutations, and effects on the structure of mutated reverse transcriptase. J. Biomed. Sci. 7507-513. [DOI] [PubMed] [Google Scholar]