Abstract
Human immunodeficiency virus type 1 (HIV-1) Gag-RNA interactions are required for virus assembly. However, our prior study found that a defect in particle production exhibited by an HIV-1 proviral mutant with a severe deletion in the RNA-binding nucleocapsid (NC) region of Gag, NX, could be reversed by eliminating its protease activity. While our follow-up study indicated that a secondary RNA-binding site in Gag can also provide the required RNA-binding function, how protease activity inhibits NX virion production is still unclear. Therefore, we tested three possible mechanisms: NX virions are unstable and fall apart after budding; NX Gag assembly is slowed, allowing protease processing to start before particle formation; or the protease region within NX Gag-Pol becomes activated prematurely and processes the assembling Gag. We found that NX particles were as stable as wild-type virions. Furthermore, even a modest slowing of protease activity could rescue NX. Pulse-chase analysis revealed that the initial particle production by NC-deleted Gag was delayed compared to that of wild type Gag, but once started, the rate of production was similar, revealing a defect in the initiation of assembly. Wild-type Gag particle production was not eliminated or decreased in the presence of excess NX Gag-Pol, inconsistent with a premature activation of protease. Overall, these results indicate that the particle formation defect of NX is due to delayed initiation of assembly caused by the absence of NC in Gag, making it vulnerable to protease processing before budding can occur. Therefore, NC plays an important initiating role in Gag assembly.
The Gag polyprotein is the primary structural protein in retroviruses and is the only protein strictly required for particle formation (the basics of assembly are reviewed in references 13a, 16, and 53). For human immunodeficiency virus type 1 (HIV-1), Gag is composed of regions that are cleaved by the HIV-1 protease into six protein products in the mature infectious virus: p17MA, matrix (MA); p24CA, capsid (CA); spacer peptide 1 (SP1, also known as p2Gag); p7NC, nucleocapsid (NC); spacer peptide 1 (SP2, also known as p1Gag); and p6Gag. The viral enzymes, protease (PR), reverse transcriptase (RT), and integrase (IN), are expressed as a Pol polyprotein which is produced as a Gag-Pol fusion by a −1 frameshift that occurs immediately after the NC region in approximately 5% of Gag translations. The production of this Gag-Pol fusion protein ensures that the enzymes essential for infection are incorporated into the assembling virion via their Gag portions (47, 52). Once completely liberated by protease processing in the newly budded virion, the mature Gag and Gag-Pol proteins rearrange into the core structure, thereby producing an infectious virion (13, 16, 53).
Protease processing induces a shift in Gag and Gag-Pol function from polyproteins that drive virion assembly by forming tight associations to a structured group of individual proteins poised to rapidly disassociate and establish a provirus during infection. Therefore, because the individual mature proteins do not assemble into virions by themselves in the cell, protease activation needs to be coordinated with assembly and budding to prevent a destructive premature digestion of Gag. It is not known how protease activity is temporally activated during retroviral assembly. For HIV-1, proteolytic processing starts during assembly and is completed after budding (26, 27). Thus, the coordination of protease activation and viral assembly is critical. The sensitivity of assembly to mistimed protease activation is seen in the overexpression of protease in the cell or in mutations that alter the relative timing of assembly, budding, and processing, both of which destroy HIV-1 assembly (3-6, 28-31, 33).
HIV-1 Gag assembly is driven by at least three important types of Gag interactions (reviewed in reference 2): cooperative membrane binding, RNA binding, and intermolecular Gag-Gag binding. NC with its strong RNA binding appears to promote assembly by tethering Gag to either a genomic or cellular RNA, thereby encouraging close-range Gag-Gag interactions. In this way, RNA likely provides a bridge or scaffold upon which Gag-Gag interactions are initiated, thereby stabilizing and promoting virion assembly (7-12, 14, 20, 23, 25, 34, 35, 39, 40, 45, 51, 61). While these three Gag-binding activities likely occur in some sort of order in the cell, this kinetic aspect of assembly is not well understood. Existing biochemical data favors an initial stage during which Gag multimerizes into complexes that rapidly bind to the plasma membrane, leading to the formation of higher-order Gag structures that form particles (54). Recent kinetic imaging in live cells supports this model (24). It is important to note that one prediction of the RNA scaffolding model is that RNA binding by Gag would be an early and potentially initiating event.
Previously, we found that the NX HIV-1 proviral construct (formerly named DelNC) which contains an almost complete deletion of NC (only four N-terminal and three C-terminal NC residues remain) fails to efficiently form particles (43), a result previously observed by others (12, 15, 60). This defect was not due to a decrease in NX Gag expression or its overt stability in the cell, consistent with a severe assembly defect (43, 45). Particles produced by NX are less dense than wild-type virions and exhibit a very aberrant morphology, mostly immature-like forms with some misshapen cores (43). This assembly/budding phenotype could be rescued by eliminating protease activity, either by genetically inactivating/deleting protease or by using an HIV-1 protease inhibitor, restoring efficient particle production of NC-deleted virions with normal virion density and an immature morphology. Therefore, protease activity, rather than the loss of NC-mediated RNA binding per se, is somehow responsible for this NX assembly defect. While this result presented an apparent contradiction to the RNA scaffold concept, our subsequent studies revealed that in the absence of NC, a smaller RNA-binding basic region in the N terminus of HIV-1 MA was able to promote particle formation (45). Similar results were obtained by in vitro Gag binding studies reported by Burniston et al. that implicate both NC and MA sequences in stabilizing Gag-Gag interactions (7). Thus, HIV-1 Gag requires an RNA-binding site, and this assembly function can be fulfilled by either NC or a basic region of MA.
Despite these studies, how protease causes the defect in NX particle formation is still unclear. To address the mechanism for this observation and what it might reveal about the role of NC, we have investigated three hypotheses: normal protease processing of Gag in the absence of NC-RNA structural interactions leads to particle instability and virion disintegration after budding (56, 57); protease is activated prematurely and processes Gag before it can form particles; or in the absence of NC, assembly is slowed, thereby allowing Gag to be digested by protease before effective particle formation. The experiments presented here reveal that NC-deleted Gag initiates assembly after a delay in the start of Gag assembly.
MATERIALS AND METHODS
DNA mutagenesis.
The pNL4-3 infectious molecular clone of HIV-1 (1) (GenBank accession number AF324493) was used for these studies and altered by site-directed mutagenesis using PCR-based methods, either by direct amplification with a mutagenic primer or by two rounds of amplification using the overlap extension procedure (21). Proviral NL4-3 mutants used from previous studies were NX (formerly named DelNC) containing a deletion of amino acids 5 to 52 in the NC, leaving seven NC amino acids (43); PX (formerly named PRR57G) containing a protease-inactivating arginine-to-glycine flap mutation at position 57 (44); and NX/PX (formerly named DelNC/PRR57G), which has the NX deletion combined with the PX protease mutation (43). The pNL4-3-based clones constructed for this study were NCK3A, pNL4-3 with nucleotides (nt) 1927 and 1928 changed from AA to GC, resulting in a lysine-to-alanine substitution at NC position 3; NXK3A, NX combined with the K3A mutation, PRT26S, pNL4-3 with nt 2328 changed from A to T, resulting in a threonine-to-serine substitution at protease position 26; NX/PRT26S, NX combined with the PRT26S mutation; PRA28S, pNL4-3 with nt 2334 changed from G to T, resulting in an alanine-to-serine substitution at protease position 28; and NX/PRA28S, NX combined with the PRA28S mutation. The PTAP− mutant (22), carrying a PTAP-to-LIRL change in p6Gag (amino acids 7 to 10), was provided by Eric Freed (Drug Resistance Program, NCI-Frederick, Frederick, MD). The pcDNA3-based Gag expression construct, pGag, was described previously (50). The pDelNC expression construct was produced by overlap PCR of pGag that completely deleted all of the NC sequences, fusing the methionine at the C terminus of SP1 to the N-terminal phenylalanine of SP2.
Cell culture and virion preparations.
293T human embryonic kidney cells (HEK293T cells) and HeLa cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cell culture products were obtained from Invitrogen (Carlsbad, CA). Transient transfections of 293T cells were carried out using calcium phosphate (19)- or Transit293T reagent (Mirus Bio Corp., Madison, WI)-based methods and harvested or radiolabeled 48 h posttransfection. To maintain equal amounts of total DNA in the cotransfection experiments, single DNA construct plasmid controls were combined with amounts of a negative control virus preparation produced from a transfection with sheared salmon sperm DNA (sssDNA) to provide for similar DNA inputs. Saquinavir was obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, NIH. Virions were prepared from clarified (>1,000 × g for 10 min) supernatants by centrifugation through a pad of 20% (wt/vol) sucrose in phosphate-buffered saline (PBS) at >120,000 × g for 1 h at 4°C. For immunoblot samples, free Gag was isolated by centrifuging the material that did not pellet through the density pad by mixing both the previous centrifuged supernatant and the pad, followed by centrifugation at >120,000 × g for 1 h at 4°C to bring down soluble protein.
Metabolic labeling and immunoprecipitation.
Labeling of transfected cells with [35S]Met-Cys and subsequent immunoprecipitation were carried out as previously described (50). Briefly, ∼107 293T cells transfected by calcium phosphate precipitation 48 h earlier were released from the flask, washed in PBS, and then placed in medium lacking Met and Cys (Met−/Cys− medium) for a 1-h starvation period. The cells were then placed in 2 ml of labeling medium containing a mixture of 150 μCi of [35S]Met and 60 μCi of [35S]Cys (ExpreSS labeling mix; Perkin-Elmer Life Sciences, Boston, MA) for 6 h. For pulse-chase labeling, 293T cells were transfected 48 h prior to labeling. As described above, cells (∼107 cells per time point) were released from the flask, washed in PBS, and placed in Met−/Cys− medium (2 ml per time point) for 30 min and then placed in an equal volume of medium containing 200 μCi of [35S]Met and 80 μCi of [35S]Cys for a 1-h pulse labeling. The cells were then centrifuged from the labeling medium and transferred into complete medium to start the chase period. Virions were isolated by density centrifugation through 20% (wt/vol) sucrose in PBS. Gag was immunoprecipitated from detergent-treated sample lysates (virion, cellular, or supernatant) with goat antiserum to p24CA (goat 81; AIDS and Cancer Virus Program, NCI-Frederick) and Protein-G beads (Thermo Scientific, Rockford, IL). Precipitates were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen) and analyzed with a PMI phosphorimager and Quantity One software (Bio-Rad Laboratories, Hercules, CA). The signals measured were adjusted for the relative amount of Met and Cys amino acids in the observed Gag products.
Immunoblot analysis.
Semidry immunoblot analysis was performed as previously described (46). Primary goat antisera against p24CA (goat 81), p17MA (goat 83), or p7NC (goat 77) were obtained from the AIDS and Cancer Virus Program, NCI-Frederick, Frederick, MD. Mouse monoclonal p51/66RT antibody was obtained from Perkin Elmer Life Sciences, Inc. Proteins were detected by developing blots with horseradish peroxidase-conjugated anti-goat (Biochain Institute, Hayward, CA) or anti-mouse (Millipore Corp., Telemuca, CA) secondary antibody and an Immun-star horseradish peroxidase substrate kit (Bio-Rad, Hercules, CA) on LumiFilm (Roche Applied Science, Indianapolis, IN).
Transmission electron microscopy.
Micrographs of positive-stained virions were obtained as previously described (17).
RESULTS
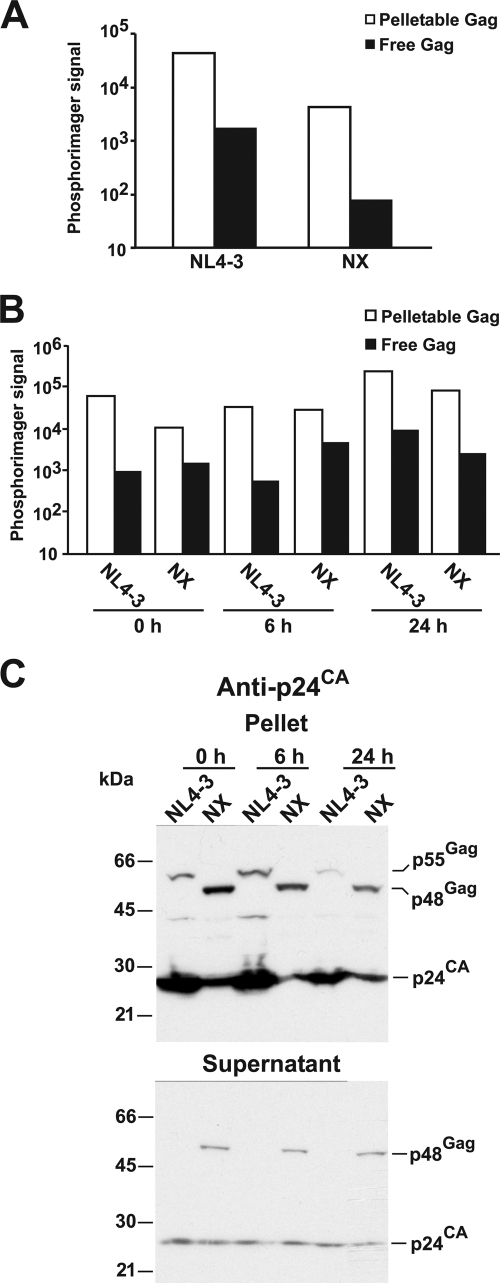
To examine which of the three possibilities (protease-induced virion instability, premature protease activation during assembly, or delayed budding) could be the mechanism for the restoration of NX particle production by eliminating protease activity, we first investigated whether NX particles are less stable due to protease action. This mechanism was proposed in prior work from Wang et al. who studied an HIV-1 mutant with 10 of 13 basic amino acids in the NC of the HXB2 proviral clone changed to alanines and concluded that these newly budded mutant virions rapidly fall apart in the absence of NC-mediated RNA binding due to protease processing of Gag (56, 57). To examine this possibility, both wild-type NL4-3 and NX virions were produced from transfected HEK293T cells in the presence of [35S]methionine-cysteine. The amounts of Gag, both uncleaved Gag and p24CA isolated as virions from the culture supernatants by density centrifugation and as free Gag remaining in the postcentrifugation supernatant, were measured by immunoprecipitation, separated by SDS-PAGE, and measured by a phosphorimager. The results for the wild-type NL4-3 samples revealed that the free Gag signal was only a small fraction (∼7%, on average) of the virion Gag signal (representative data are presented in Fig. 1A). The amount of Gag signal measured for NX virions, both uncleaved NX (Gag Pr48Gag [Pr55Gag minus p7NC]) and p24CA, was lower than that detected in wild-type virions (Fig. 1A and B), consistent with our previous findings (43, 45). Nonetheless, the amount of total free NX Gag in the supernatant was correspondingly lower, at approximately 7% of the virion signal in typical experiments Gag (Fig. 1A and B), similar to the free-to-virion proportion of Gag in the wild-type sample. Thus, NX particles appear to be as stable as the wild type immediately after budding.
FIG. 1.
Stability of wild-type and NX virions. (A and B) Graphs of representative results from 35S-labeled radioimmunoprecipitation analyses of virions and free Gag produced by transfected cells are presented. (C) p24CA immunoblots of wild-type and NX virion and free Gag samples produced by transfected cells is presented. Samples are labeled above each respective lane. For panels B and C, postcollection supernatant incubation times at 37°C are indicated below or above the respective sample labels. The locations of molecular mass markers are presented on the left of the blot, and pertinent Gag bands are identified on the right. Phosphorimager signal is presented as arbitrary pixel density counts.
There is a possibility that NX particles become less stable over time. To address this question, radiolabeled virions were produced for 6 h and then incubated at 37°C for an additional 6 h or 24 h. The stabilities of the virions were similar for newly budded NL4-3 and NX virions and the incubated viruses (Fig. 1B). Immunoblotting of a parallel nonradiolabeled experiment produced similar results (Fig. 1C). Together, these results show that the decreased NX particle production is not due to instability of the particles.
Reduced protease activity can rescue NX budding.
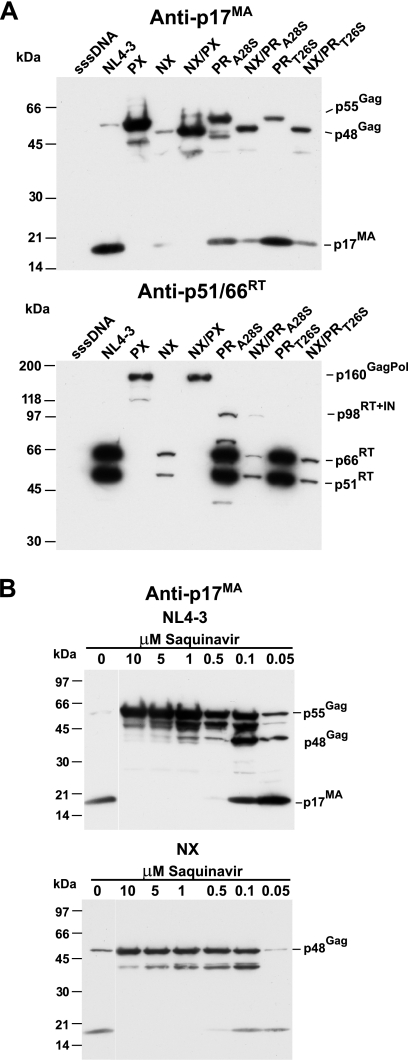
If mistimed protease processing during assembly, either due to slowed assembly or accelerated protease activation, is the root cause of the NX Gag particle production defect, then slowing but not eliminating protease activity might also rescue budding. To test this hypothesis, we produced two versions of NX, one containing an A28S mutation in protease that decreases activity 50-fold and another harboring a T26S protease mutation that provides a more modest 4-fold activity reduction (3, 49). Virion preparations of these mutants produced by HEK293T cell transfection were analyzed by immunoblotting with p17MA antiserum (Fig. 2A). As with our prior results (43), the intensities of the Gag protein signals detected in the NX sample were considerably less than those in the wild-type sample. Also, combining the NX mutation with a PRR57G protease-inactivating mutation (in the NX/PX construct) genetically rescued the particle production defect, as previously observed (43), producing an unprocessed Pr48Gag band with an intensity that was similar to that for the Pr55Gag present in the sample from the NL4-3-based PX mutant, which contains only the protease mutation (Fig. 2A). The NX/PRA28S sample contained a readily detectable Pr48Gag form with the minority of the signal as fully processed MA, consistent with a severe yet not absolute reduction in protease processing. Similarly, the corresponding NL4-3 mutant with only the protease mutation PRA28S also exhibited considerably less processing than the wild type. A comparison of the total intensities of the Gag signals in the NX, NX/PX, and NX/PRA28S samples revealed that NX/PRA28S produced a clearly stronger combined signal than that of the NX sample, though a somewhat weaker signal than that of the NX/PX sample. Thus, the severe decrease in protease activity in NX/PRA28S reverses the defect caused by NC deletion.
FIG. 2.
Genetic and inhibitor-mediated reduction of protease activity. (A) p17MA and p51/66RT immunoblots of virion preparations of NX mutants carrying protease mutations are presented. (B) p17MA immunoblots of wild-type and NX virion preparations produced with transfected HEK293T cells in the presence of decreasing amounts of the protease inhibitor saquinavir are presented. The samples are identified above the respective lanes. The locations of molecular mass markers are presented on the left of the blot, and pertinent Gag bands are identified on the right.
If the NX defect is due to mistiming between protease activation and assembly/budding and not particle stability, then even a more-modest slowing of protease activity should also rescue NX budding by giving Gag more time to form particles yet still allowing for the maintenance of significant levels of processing in the released virions. The sample from the NX construct with the milder PRT26s mutation (NX/PRT26S) contained p17MA and unprocessed Pr48Gag (Fig. 2A). The NL4-3 construct carrying the protease mutation alone, PRT26S, had mostly fully processed p17MA, with a less intense band of Pr55Gag (Fig. 2A), consistent with the fourfold rate decrease seen with the in vitro study (49) The total intensities of the p17MA and Pr48Gag bands in the NX/PRT26S lane were considerably stronger than those in the NX sample, yet weaker than those in the NX/PX sample. Thus, the addition of the PRT26S mutation also rescues the NX virion production defect.
To complement the genetic experiments, we exposed transfected HEK293T cells to differing concentrations of the protease inhibitor saquinavir for 24 h and isolated virions from the supernatant in order to observe NX particle production when protease activity is reduced. The p17MA immunoblot results (Fig. 2B) for the wild-type construct preparations showed that Gag was present mostly as Pr55Gag with only a small amount of Gag that was ∼7 kDa smaller (likely Pr55Gag without p6Gag) at the 10 μM saquinavir concentration with increasing processing through the dilutions down to the 0.05 μM-treated sample. The latter was mostly processed, containing only a slight amount of unprocessed and partially processed Gag. As with the wild-type samples, the NX sample with 10 μM saquinavir treatment showed mostly Pr48Gag with only a slight signal from a partially processed form (likely p40MA+CA+SP1) that increased in intensity down to the 0.1 μM-treated sample, which also contained detectable amounts of fully processed p17MA (Fig. 2B). However, unlike the wild-type sample, the 0.05 μM-treated sample contained little Pr48Gag signal and essentially no increase in the p17MA band over that for the 0.1 μM sample band. The similar intensities of the p17MA bands in the 0.1 and 0.05 μM saquinavir samples appear to represent the small fraction of Gag that escapes destructive processing in the cell and produces particles with mature Gag proteins. This result demonstrates that there is a critical level of protease activity above which mistimed Gag processing eliminates NX virion formation. This level appears to be fairly close to full activity, as the Gag in the wild-type sample at this 0.05 μM concentration was mostly processed, with only a small fraction of the p17MA signal in partially processed forms (Fig. 2B). Taken together, our genetic and inhibitor experiments show that even a slight decrease in protease activity can rescue most of the NX particle formation, supporting a mistiming of protease and assembly as the key mechanism for the defect in NX particle production.
Incorporation of Pol proteins is decreased in the presence of protease activity.
The results above show that, as we have noted previously (43, 45), the apparent ratio of Pr48Gag to p24CA in the NX virion preparations is considerably higher than the Pr55Gag-to-p24CA ratio in the wild-type preparations (Fig. 1C and 2A). It is important to note that we routinely observe a small amount of Pr55Gag in wild-type preparations (Fig. 1C and 2A; previous studies). Based on electron microscopic examination and core preparation studies, these proteins are from particles that are composed of mostly, if not entirely, unprocessed Gag (D.E.O. and L.V.C., unpublished observations). Thus, there are always some particles in wild-type virions that either fail to incorporate Gag-Pol or contain inactive protease. If protease dyscoordination with assembly is the basis for the NX effect, then an NX particle population should be enriched with Gag proteins that have escaped protease and be depleted of processed Gag, resulting in a higher ratio of Pr48Gag to p24CA in NX virions compared to that for the wild type. Since Gag-Pol carries protease, we examined the level of this polyprotein in the mutant virion preparations to look for bias against Pol incorporation. The blot in Fig. 2A was stripped of the anti-p17MA reagent and exposed to a high-sensitivity antibody against the RT Pol product, p51/66RT. Intense bands for both the 51- and 66-kDa forms of RT were present in the wild-type sample, and a Pr160GagPol band was present in the PX sample (Fig. 2A), though it was less intense than the two wild-type RT bands together, apparently due to the lower transfer efficiency of higher-molecular-mass proteins. The staining of the two protease rate mutants, PRA28S and PRT26S, was nearly equal to that of the wild-type sample, though the PRA28S sample did exhibit an apparent partially processed Pr98RT/IN form, consistent with its more-severe reduction in processing (Fig. 2A). The NX lane contained a considerably fainter signal than its wild-type counterpart, due in part to fewer particles in the mutant sample (Fig. 2A, p17MA staining). However, the NX/PX Pr160GagPol precursor band was as intense as that in the PX sample; therefore, the absence of NC itself did not have an appreciable effect on the incorporation of Gag-Pol, ruling out a simple NC-mediated packaging defect. However, for both NX protease rate mutants, NX/PRA28S and NX/PRT26S, the amount of RT and hence Pol incorporated into the particles appeared to be quite low by a comparison of the relative intensities of the p51/66RT and p17MA signals (Fig. 2A) and was clearly much lower that that in the wild type and its protease rate mutants. Thus, despite the rescue of particle production by the protease rate mutations, there still is a bias against incorporation of Pol, hence protease, that is not seen in the protease mutant NX/PX. Detection with antiprotease, a less sensitive reagent, produced similar results (data not shown). This bias against the incorporation of Pol is consistent with a selection for Gag molecules that escape processing in particles and explains the increased relative Pr48Gag incorporation in NX virions.
The basic residue in NX NC is dispensable for rescue.
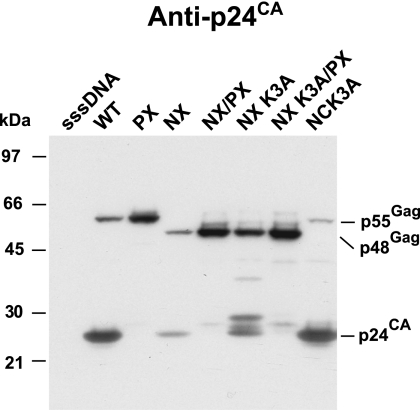
In constructing NX, we had retained a basic residue, lysine at NC position 3, to maintain protease processing of the SP1-NC site. While one lysine is unlikely to provide RNA-binding activity to NX, a previous study found that two arginines (positions 3 and 7) in NC in the HXB2 proviral clone were necessary for an interaction (I) domain function (51). The I domain is defined as sequences that allow for close Gag-Gag packing and dense particles (59). Because of its potential to contribute to assembly, we mutated lysine 3 of NC in the NX clone to alanine to produce the NXK3A proviral mutant. Examination of NX and NXK3A virion preparations from transfection supernatants by p24CA immunoblotting revealed that most of the Gag in the NXK3A mutant sample was in the unprocessed Pr48Gag form, with several partial cleavage products, including p25CA+SP1 and a band that is likely a p28CA+SP1+delNC+SP2 product (Fig. 3). As seen above, only weak signals for p24CA and Pr48Gag were present in the NX sample. The combined bands in the NXK3A sample constituted considerably more total signal than that in the NX sample, comparable to that of the NX/PX sample (Fig. 3). Therefore, instead of inhibiting NX Gag particle production, the loss of this basic residue reduces the extent of protease processing, thereby rescuing the NX defect, similar to the results of the genetic and inhibitor studies presented above. A construct containing both the NX and PRR57G protease mutations, NXK3A/PX, produced a Pr48Gag band of an intensity similar to that of the corresponding NX/PX band (Fig. 3), confirming the dispensability of this lysine for NX particle formation even in the absence of protease activity. NL4-3 containing only the NCK3A mutation did not show any overt signs of a processing defect, demonstrating that the effect of this mutation on processing is unique to the NX background. Taken together, these results show that lysine 3 in NX was not responsible for particle production. Instead, it was the partial decrease of protease processing in the NXK3A mutant that rescued budding, consistent with the mistiming-of-protease model.
FIG. 3.
Analysis of NC lysine 3 mutants. A p24CA immunoblot of NX mutant virion preparations produced by transfection is presented. The samples are identified above the respective lanes. The locations of molecular mass markers are presented on the left of the blot, and pertinent Gag bands are identified on the right.
NX/PX particle production is not cell line dependent.
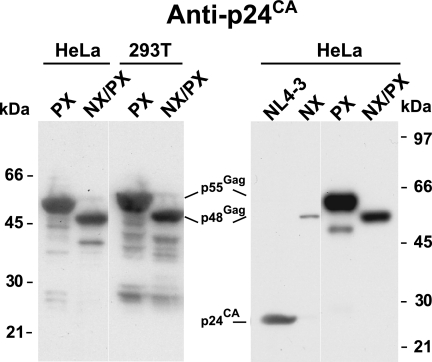
We have studied the NX and NX/PX constructs exclusively with the HEK239T cell line, which produces relatively high levels of virions. However, HeLa cells are a commonly used alternative model system for studying HIV-1 assembly, though they typically express less protein upon transfection, because not only are fewer cells transfected but also the expression levels in the cells are lower (D.E.O., unpublished results), likely reflecting reduced DNA uptake. It is conceivable that the higher relative levels of NX/PX expression in HEK293T cells are responsible for the rescue of the NC deletion by protease inactivation. To explore this question, we transfected both HeLa and HEK293T cells with either the PX or the NX/PX mutant. The overall intensities produced in a p24CA immunoblot analysis of the virion samples from the HeLa transfections were somewhat lower that those from the HEK293T cell preparations, as expected (Fig. 4). However, the relative intensities of the PX and NX/PX samples were similar for the two cell lines, with that of PX being somewhat more intense than that of NX/PX, consistent with our prior data (43, 45). An immunoblot of NL4-3, NX, PX, and NX/PX samples (Fig. 4) produced from transfected HeLa cells produced data similar to those obtained from HEK293T cells (Fig. 2 and 3). Thus, the rescue of the NX mutation by protease inactivation is not dependent on production in HEK293T cells or its somewhat higher expression levels.
FIG. 4.
Virion production of NX/PX in HeLa cells. p24CA immunoblots of NX/PX virion preparations produced by transfection are presented. The virion samples and the cell lines used to produce them are identified above the respective lanes. The locations of molecular mass markers are presented on the outer margins of the blots, and pertinent Gag bands are identified between the blots.
NX Gag-Pol does not interfere with wild-type Gag expression.
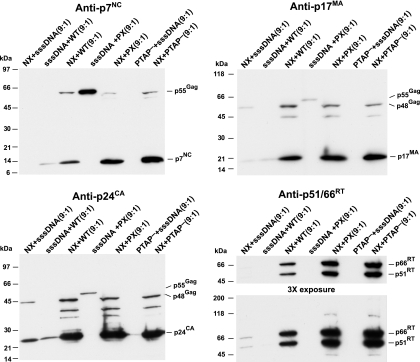
The deletion of NC in NX Gag-Pol might somehow cause premature protease activation and Gag processing. If so, then the presence of an excess of NX Gag-Pol would be able to digest wild-type Gag before it assembles. Overexpressing Gag-Pol and protease has been found to eliminate particle formation by Gag (3-6, 28-31, 33). We have previously found that NX Gag-Pol can restore processing to the protease-deficient PX construct and that PX can rescue NX budding in trans (45), demonstrating that all four different Gag and Gag-Pol molecules can interact and be packaged together to form mixed particles. To examine whether the protease in NX Gag-Pol is activated inappropriately, we cotransfected NX with NL4-3, PX, or the PTAP− L domain mutant constructs at a 9-to-1 ratio. Under these conditions, if the protease in the NX construct becomes activated prematurely, then the gross excess of NX Gag-Pol would interact with the wild-type Gag and digest it before it can bud. In contrast, if the defect in NX is reflected by slower Gag assembly and budding, then particle production by the wild-type Gag should be at least normal, and the slowed budding of the PTAP− mutant should be rescued by the presence of NX Gag, because both L-domain and NX mutants can be rescued in trans with small amounts of wild-type Gag (36-38, 41, 45, 58). The relative contribution of each cotransfection partner can be measured by immunoblotting: p17MA or p24CA antisera detect both NX and wild-type Gag while p7NC antiserum reacts with only that of the wild type. Examining the cotransfection samples for Pr55Gag by a p7NC immunoblot analysis revealed that the NC signal was markedly more intense in the 9 NX-to-1 NL4-3 cotransfection sample than the 9 sssDNA-to-1 NL4-3 control (Fig. 5). This indicates that excess NX increases the efficiency of wild-type Gag particle production rather than inhibiting it. Exposing the blot to p17MA and p24CA antisera showed that the 9 NX-to-1 sssDNA sample contained a weak signal, similar to that of the 9 sssDNA-to-1 NL4-3 control, reflecting its defect in particle production. However, the two constructs together (9 NX-to-1 NL4-3) produced a strong signal, indicating that each of these constructs efficiently interact, increasing the assembly and budding of the other. Thus, virion production is increased for both viruses.
FIG. 5.
Coexpression of NX and wild-type Gag-Pol. P7NC, p24CA, and p17MA immunoblots of virion preparations produced by cotransfection are presented. The samples are identified above the respective lanes, with the ratio of the cotransfected DNAs above. The locations of molecular mass markers are presented on the left of the blot, and pertinent Gag bands are identified on the right.
The ability of these two constructs to interact was further demonstrated by a cotransfection of NX with the protease mutant PX and the L-domain mutant PTAP−. The latter mutant contains a PTAP-to-LIRL mutation in the N-terminal L-domain motif of p6Gag and results in a reduction of virion production due to an inability of the particles to detach from the plasma membrane (22, 46). The blots showed that production of virions from either of these constructs was not decreased by the presence of NX. The NX protease was able to process the wild-type Gag of the PX construct (Fig. 5, anti-p7NC blot), further showing that the protease in NX Gag-Pol is active and can assemble with the PX proteins and rescue their processing defect. Similarly, the excess of NX was clearly able to rescue the PTAP− construct: little or no p7NC signal was detectable for the 9 sssDNA-to-1 PTAP− control, while there was a strong NC signal in the 9 NX-to-1 PTAP− sample (Fig. 5, anti-p7NC blot). A p51/66RT blot of the samples revealed that RT was clearly present in the 9-to-1 cotransfections of NX with the other proviral constructs and either barely present or undetectable in the cotransfections with sssDNA. Thus, unlike the bias against Gag-Pol observed with the NX protease rate mutants (Fig. 2A), the presence of even small amounts of Gag that contains NC rescues both NX particle production and efficient Pol protein incorporation. In turn, NX Gag increases the particle formation of wild-type Gag and restores function to two different mutants, demonstrating the ability of the two types of Gag proteins to complement each other. Because the presence of excess NX Gag-Pol did not result in decreased Gag particle production, it is unlikely that a premature activation of protease in NX Gag-Pol is the mechanism for the poor particle formation by NX.
Particle production is slowed in the absence of NC.
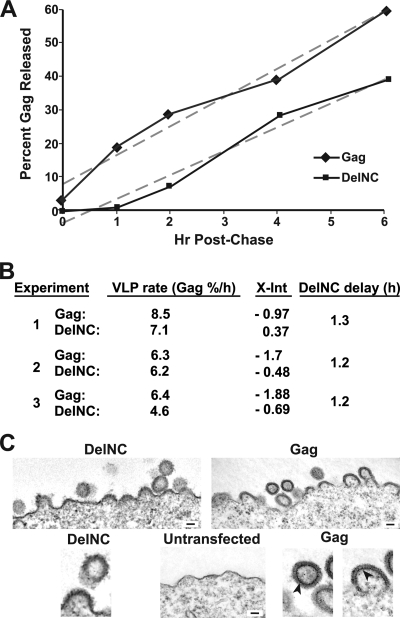
To examine whether Gag without NC assembles and buds more slowly than the wild type, we constructed a cytomegalovirus promoter-driven Gag expression plasmid, pDelNC, by eliminating all of the NC region from a Pr55Gag expression construct, pGag. The kinetics of Pr48Gag and Pr55Gag virus-like particle (VLP) production were measured by pulse-chase radiolabeling experiments with transfected HEK293T cells. DelNC- or Gag-expressing cells were pulsed with [35S]cysteine-methionine for 1 h and chased with complete medium with samples taken at various time points. Immunoprecipitates of Gag from VLP preparations and cell lysates were analyzed by SDS-PAGE and measured by phosphorimager analysis. A graph of VLP Gag as a percentage of total Gag produced during the chase shows that less Pr48Gag was produced at each time point relative to Pr55Gag (Fig. 6A, experiment 1 results; data not shown). These data are consistent with our previous steady-state labeling and immunoblotting data showing that the NX/PX proviral construct was produced somewhat less efficiently (∼80%) than the PX mutant (see Fig. 2 and 3 presented in reference 45). However, a comparison of the production rates of the two VLPs (percent produced per hour, measured as the slope of each least-squares regressed line) revealed no drastic difference, with the DelNC Pr48Gag being produced at a rate between 98% and 71% of the rate of wild-type Pr55Gag (Fig. 6B). Rather, the graph reveals that the onset of DelNC VLP production was delayed relative to that for wild-type Gag (Fig. 6A). The x-axis intercept of each regressed line extrapolates to the time at which particles were first produced into the medium. Since these are pulse-chase experiments, a negative number indicates that particle production began during the pulse-labeling, as is the case for wild-type Gag in experiment 1 (Fig. 6B). A comparison of the difference in the x-axis intercepts from three independent experiments detected a delay of ∼1.3 h in the initial production of the DelNC Gag particles (Fig. 6B and data not shown). An electron microscopic examination of DelNC- and Gag-expressing cells did not reveal any marked difference in the cellular assembling and budding forms (Fig. 6C), except that the wild-type Gag multimers contained a thin dark electron-dense region at the membrane distal end of Gag (Fig. 6C). This is most likely RNA bound to NC that is absent in DelNC Gag, as we have noted before (43). Neither sample contained noticeable arrested budding forms indicative of a failure to release from the cell, effectively ruling out any L-domain-type failure by the assembled DelNC VLPs (18, 22). Thus, based on our kinetic results, this delay in DelNC VLP production appears to be due to a bottleneck in the initiation of assembly. After this stage is completed, later assembly events apparently occur at near-normal rates.
FIG. 6.
Pulse-chase analysis of VLP production. (A) Graph of VLP production during a 35S-labeled metabolic pulse-chase labeling experiment (experiment 1) is presented as percent VLP Gag (i.e., Gag isolated as dense particles) over time postchase for both wild-type and DelNC Gag expression constructs. The linear regression for each construct is presented as a gray dashed line. Hr, hours. (B) A table of the results from three independent experiments is presented. The VLP rate is the production of particles as measured by the slope of the regressed line, equal to the percent of total Gag produced into the medium as pelletable Gag per hour postchase. The x-axis intercept from the regressed line, labeled X-Int, provides the time VLP production was initiated postchase. The DelNC delay is the absolute difference in x-axis intercepts between the wild-type and DelNC linear regressions expressed as hours for each experiment. (C) Thin-section electron micrographs (×10,000) of DelNC- and Gag-expressing HEK293T cells. The inset bar denotes 100 nM. Arrows in the enlarged sections point to the thin bands of electron-dense material. Enlarged portions of electron micrographs are provided below the respective ×10,000 fields. The enlarged DelNC section was taken from a region that was cropped out of the ×10,000 DelNC field.
DISCUSSION
The data presented here clarify several properties of the NX clone (an HIV-1 virus with a nearly complete deletion of NC) and provide a clearer picture of the role of NC in assembly. Based on our experiments, the primary particle production defect in the NX mutant is caused by a delay in the initiation of virion assembly in the cell, which results in a mistiming in the normally coordinated process of assembly and cleavage of Gag by the viral protease, resulting in the processing of Gag before it can form virions. This model is supported by data from our various experiments: there is an observed delay in the initiation of virion production by DelNC Gag (Fig. 6), a modest decrease in protease activity rescues NX particle formation (Fig. 2), a partial reduction in Gag processing induced by the NCK3A mutation rescues particle formation (Fig. 3), and a small amount of wild-type Gag is sufficient to rescue the NX particle production phenotype (Fig. 5). Together, these data clarify the mechanism for the restoration of NX budding by incapacitating protease (43) and point to a role for NC in promoting the initiation of viral assembly.
As observed both here and in our prior publications (43, 45), NX virion preparations contain a considerably higher ratio of unprocessed Gag polyprotein to processed Gag than that found in wild-type preparations. This difference is predicted by our model, because Gag that assembles slowly is more susceptible to protease digestion in the cells. This dyscoordination of processing and assembly results in a marked decrease in particles containing mature proteins. In contrast, the Pr48Gag molecules that do not interact with protease will escape digestion and can form particles. Thus, there is a selection for incorporation of Pr48Gag in NX particles and against that of the mature proteins. This bias against protease is also manifested in the reduced Pol incorporation by the NX constructs with PR rate mutations. The absence of partially cleaved Gag products in NX particles suggests that there is no gross defect in NX protease, indicating that PR is either missing from or inactive in the Pr48Gag-containing virions. The NX construct can complement the protease-deficient PX mutation (45) (Fig. 5, p7NC blot), supporting the activity of the NX protease. Taken together, these observations explain the altered unprocessed Gag-to-processed Gag ratio in NX and support a delay in NX assembly and budding that makes Gag susceptible to mistimed protease digestion.
The other potential mechanisms for the rescue of NX by protease inactivation are not supported by our data. One alternative hypothesis is that the absence of NC in NX Gag-Pol causes the protease in the polyprotein to be activated earlier in the assembly and budding process, thereby degrading NX Gag before the virions can be released from the cell. Our cotransfection experiments with various proviruses encoding Gag with NC found that that a gross excess of NX did not inhibit virion production as is predicted by this alternative hypothesis. Instead, the excess of NX proteins increased wild-type and L-domain Gag particle production, apparently assisting the assembly and budding of these Gag molecules by forming mixed virions. In turn, these NC-containing Gags were also able to complement the NX defect, as previously reported (43, 45). Indeed it is possible that protease activation is slower in NX, as suggested by our previous observation that NX processing appears to be more sensitive to protease inhibitors (data not shown) (43).
Our data also ruled out the protease-mediated particle instability hypothesis, as there was no difference in the immediate and longer-term stabilities of NX and wild-type particles. Furthermore, particle production was rescued even by mild decreases in protease activity exhibited by the NX/PRA28S mutant even though this mutant still maintained some NX Gag processing that would have affected virion stability if this were the case. Taken together, these results show that NX particles are not inherently unstable.
Although the NX/PX construct clearly makes particles, the presence of the seven remaining amino acids previously raised the formal possibility that they are important for assembly. Two results demonstrated that this is not the case. The last remaining basic amino acid in NX, lysine 3, was not necessary for NX or NX/PX assembly. Furthermore, the DelNC Gag expression construct, harboring a deletion of the entire NC region, was also able to produce particles. Thus, there are no NC sequences in Gag that are strictly required for particle production.
Another possibility is that efficient NX/PX particle formation requires the higher levels of expression in HEK293T cells or a specific attribute of this cell line. Our experiments in HeLa found that NX/PX Gag still produces particles efficiently even at somewhat lower expression levels. Therefore, cell lines other than HEK293T can support NX/PX particle production.
Even though NC is dispensable in this system, we did detect a 1.3-h delay in the onset of DelNC VLP production yet a near wild-type production rate, reflecting a decrease in assembly initiation and not a slowing in assembly progression once started. This delay has important ramifications for the role of NC in assembly and budding. Based on our previous data, in the absence of NC, Gag relies on a much smaller RNA-binding site in MA for assembly (7, 45). Because this MA-binding site is a smaller and weaker RNA-binding site (comprised of three amino acids spanning five positions) (32, 48), it likely provides this initiating function less efficiently than the stronger RNA-binding properties of NC (15 amino acids spanning 50 positions) (55). Thus, while the deletion of NC does not eliminate particle production, it does delay the initiation of assembly. This is also supported by our previous finding that an MA-RNA-binding site mutant that contains NC (MX/PX) produces virions more efficiently than the NX/PX mutant which possesses an intact MA but no NC (45). Taken together, these data indicate that the delay in DelNC particle production is likely due to the weaker nature of the MA RNA-binding site when used as the sole RNA-binding moiety for Gag (45). Thus, with respect to wild-type Gag, our kinetic data confirm a prediction of the RNA scaffold-bridging model for Gag, namely, that NC primarily provides an efficient initiation of assembly.
Our data clarify and extend our previous findings and those from an in vitro study by Burniston et al. (7): RNA-binding by NC initiates Gag assembly by binding RNA, thereby tethering Gag molecules together on a RNA scaffold to stabilize/facilitate more local-acting Gag-Gag interactions and promote cooperative membrane binding by the growing Gag multimers. In the absence of NC, a weaker RNA-binding site in MA can be used, but it is less efficient. This results in a longer initiation period that delays assembly and disrupts the normal coordination of protease activation and assembly, thereby causing a particle production defect. While NC is not required for membrane binding per se (10, 42), there is likely a coordination between RNA-initiated Gag multimerization and cooperative Gag membrane binding that supports virion assembly. Additional studies should produce a more complete picture of HIV-1 Gag assembly by examining how NC-RNA interactions are coordinated with or induce Gag plasma membrane association, the other major assembly function.
Acknowledgments
We thank Elena Chertova, Robert Gorelick, and Claes Ohlen for helpful comments on the manuscript, Alan Rein for both experimental ideas and manuscript reviews, Eric Freed for sharing the PTAP− clone, and Kunio Nagashima and Adam Harned for expert electron microscopy analysis. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: saquinavir.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contracts HHSN261200800001E and N01-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Adachi, A., S. Koenig, H. E. Gendelman, D. Daugherty, S. Gattoni-Celli, A. S. Fauci, and M. A. Martin. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55347-387. [DOI] [PubMed] [Google Scholar]
- 3.Adamson, C. S., M. Nermut, and I. M. Jones. 2003. Control of human immunodeficiency virus type-1 protease activity in insect cells expressing Gag-Pol rescues assembly of immature but not mature virus-like particles. Virology 308157-165. [DOI] [PubMed] [Google Scholar]
- 4.Arrigo, S. J., and K. Huffman. 1995. Potent inhibition of human immunodeficiency virus type 1 (HIV-1) replication by inducible expression of HIV-1 PR multimers. J. Virol. 695988-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 676487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 706820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 738527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 732270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 696487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, C. Y., Y. F. Chang, S. M. Wang, Y. T. Tseng, K. J. Huang, and C. T. Wang. 2008. HIV-1 matrix protein repositioning in nucleocapsid region fails to confer virus-like particle assembly. Virology 37897-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli, A., and J. Luban. 2000. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J. Virol. 746734-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 743046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin, J., S. Hughes, and H. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 13a.Coffin, J. S., S. Hughes, and H. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.view.showTOC&rid=rv.TOC. [PubMed]
- 14.Crist, R. M., S. A. Datta, A. G. Stephen, F. Soheilian, J. Mirro, R. J. Fisher, K. Nagashima, and A. Rein. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 832216-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251141-157. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 2511-15. [DOI] [PubMed] [Google Scholar]
- 17.Gonda, M. A., S. A. Aaronson, N. Ellmore, V. H. Zeve, and K. Nagashima. 1976. Ultrastructural studies of surface features of human normal and tumor cells in tissue culture by scanning and transmission electron microscopy. J. Natl. Cancer Inst. 56245-263. [DOI] [PubMed] [Google Scholar]
- 18.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 883195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52456-467. [DOI] [PubMed] [Google Scholar]
- 20.Gross, I., H. Hohenberg, and H. G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249592-600. [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 22.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 696810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 7611177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jouvenet, N., P. D. Bieniasz, and S. M. Simon. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 733079-3086. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 686782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan, A. H., and R. Swanstrom. 1991. HIV-1 gag proteins are processed in two cellular compartments. Proc. Natl. Acad. Sci. USA 884528-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193661-671. [DOI] [PubMed] [Google Scholar]
- 29.Kräusslich, H. G. 1991. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc. Natl. Acad. Sci. USA 883213-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kräusslich, H. G. 1992. Specific inhibitor of human immunodeficiency virus proteinase prevents the cytotoxic effects of a single-chain proteinase dimer and restores particle formation. J. Virol. 66567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindhofer, H., K. von der Helm, and H. Nitschko. 1995. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology 214624-627. [DOI] [PubMed] [Google Scholar]
- 32.Lochrie, M. A., S. Waugh, D. G. Pratt, Jr., J. Clever, T. G. Parslow, and B. Polisky. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 252902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luukkonen, B. G., E. M. Fenyo, and S. Schwartz. 1995. Overexpression of human immunodeficiency virus type 1 protease increases intracellular cleavage of Gag and reduces virus infectivity. Virology 206854-865. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 7852-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 765452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Serrano, J., and P. D. Bieniasz. 2003. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 7712373-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 785554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 7611405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikolaitchik, O. A., R. J. Gorelick, M. G. Leavitt, V. K. Pathak, and W. S. Hu. 2008. Functional complementation of nucleocapsid and late domain PTAP mutants of human immunodeficiency virus type 1 during replication. Virology 375539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 745142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, K. Nagashima, R. C. Sowder II, D. T. Poon, and R. J. Gorelick. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 775547-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278111-121. [DOI] [PubMed] [Google Scholar]
- 45.Ott, D. E., L. V. Coren, and T. D. Gagliardi. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 7913839-13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ott, D. E., L. V. Coren, T. D. Gagliardi, and K. Nagashima. 2005. Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1. J. Virol. 799038-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 666304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosé, J. R., L. M. Babé, and C. S. Craik. 1995. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 692751-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudner, L., S. Nydegger, L. V. Coren, K. Nagashima, M. Thali, and D. E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 794055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 747238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, A. J., N. Srinivasakumar, M. L. Hammarskjold, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 672266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 54.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 745845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbaneja, M. A., B. P. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. R. Casas-Finet. 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 28759-75. [DOI] [PubMed] [Google Scholar]
- 56.Wang, S. W., and A. Aldovini. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76:11853-11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, S. W., K. Noonan, and A. Aldovini. 2004. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 78716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xaing, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 686605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral Gag proteins. AIDS 5639-654. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 716765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 721782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]